Abstract
Purpose
Dissimilatory iron-reducing bacteria have been described by both culture-dependent and -independent methods in various environments, including freshwater, marine sediments, natural wetlands, and contaminated aquifers. However, little is known about iron-reducing microbial communities in paddy soils. The goal of this study was to characterize iron-reducing microbial communities in paddy soil. Moreover, the effect of dissolved and solid-phase iron (III) species on the iron-reducing microbial communities was also investigated by enrichment cultures.
Methods
Ferric citrate and ferrihydrite were used respectively to set up enrichment cultures of dissimilatory iron-reducing microorganisms using 1% inoculum of soil samples, and the iron reduction was measured. Moreover, bacterial DNA was extracted and 16S rRNA genes were PCR-amplified, and subsequently analyzed by the clone library and terminal restriction fragment length polymorphism (T-RFLP).
Results
Phylogenetic analysis of 16S rRNA gene sequences extracted from the enrichment cultures revealed that Bradyrhizobium, Bacteroides, Clostridium and Ralstonia species were the dominant bacteria in the ferric citrate enrichment. However, members of the genera Clostridium, Bacteroides, and Geobacter were the dominant microorganisms in the ferrihydrite enrichment. Analysis of enrichment cultures by T-RFLP strongly supported the cloning and sequencing results.
Conclusions
The present study demonstrated that dissimilatory iron-reducing consortia in As-contaminated paddy soil are phylogenetically diverse. Moreover, iron (III) sources as a key factor have a strong effect on the iron (III)-reducing microbial community structure and relative abundance in the enrichments. In addition, Geobacter species are selectively enriched by ferrihydrite enrichment cultures.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Microbial iron reduction is considered to be a ubiquitous and important redox process, in which dissimilatory iron-reducing bacteria can gain energy for growth by coupling the oxidation of organic compounds or hydrogen to the reduction of Fe (III) oxides (Lovley et al. 2004). This biochemical reaction has a profound influence on the global biogeochemical cycling of elements, including carbon (C), sulfur (S), nitrogen (N), and phosphorus (P), in addition to heavy metals and arsenic (As) (Lovley and Phillips 1986; Stipp et al. 2002; Kappler and Straub 2005). Furthermore, numerous dissimilatory iron-reducing bacteria belonging to different phylogenetic groups have been described by both culture-dependent and -independent methods in various environments, including freshwater (Lovley and Phillips 1988), marine sediments (Roden and Lovley 1993), natural wetlands (Weiss et al. 2003), and contaminated aquifers (Scala et al. 2006; Lin et al. 2007).
The diversity of soil bacteria and fungi (Ge et al. 2008; He et al. 2008) and the abundance and community composition of some soil functional microorganisms such as ammonia oxidizers (Chen et al. 2008a), methanotrophs (Zheng et al. 2008), and sulfate reducers (Liu et al. 2009) have been investigated in paddy soils. However, little is known about the microorganisms responsible for anaerobic iron respiration. Paddy soil is intermediate between upland systems and true aquatic systems, and the alternation between anoxic and oxic conditions causes periodically occurring redox reactions. Because of the unique characteristics of paddy soil, it can provide iron (III)-reducing bacteria with abundant electron acceptors and decomposition of organic matter for growth, and it has been estimated that iron (III)-reducing bacteria accounted for 12% of total bacteria cells in the rhizosphere of wetland plants (Weiss et al. 2003). Thus, we assume that dissimilatory iron-reducing bacteria are abundant in paddy soils.
Arsenic is a toxic metalloid widely distributed in the Earth’s crust, most notably in the groundwater of Southeast Asia, which subsequently polluted rice fields and rice by irrigation (Williams et al. 2006); additionally, mining activities also resulted in As contamination in paddy soil and As accumulation in rice (Zhu et al. 2008). In order to remediate As-contaminated soils, and reduce potential risks for humans, it is extremely important to understand the factors affecting As mobility in subsurface environments. In addition to geochemical parameters, such as pH, it has been demonstrated that dissimilatory iron reduction plays a key role in As release into the waters from shallow reducing aquifers and paddy soils by reducing and dissolving iron (III) minerals (Horneman et al. 2004; Islam et al. 2004). Although previous studies have indicated the important role of the iron-reducing bacteria, the structure and abundance of these organisms are not completely elucidated and understood in As-contaminated paddy soils.
In this study, typical paddy soil samples were taken from Chenzhou, which is one of the major cities for the production of different metals, and As is present at high levels in soils impacted by mining and smelting activities (Liao et al. 2005). The goal of the present study was to characterize iron-reducing microbial communities in the paddy soil. Moreover, in order to determine the effect of dissolved and solid-phase iron (III) species on iron-reducing microbial communities, enrichment cultures were obtained by two forms of iron sources. We used 16S rRNA gene and terminal restriction fragment length polymorphism (T-RFLP) methods to investigate the diversity of iron-reducing bacteria in enrichment cultures.
2 Materials and methods
2.1 Soil sampling
Soil samples were taken from a mining area, Bailutang in Shizhuyuan (25º48′N, 113º02′E), located in Chenzhou, Hunan. Soil samples were collected from the top 20 cm of paddy soil, kept in polyvinylchloride bottles and submerged in water mimicking the field conditions, and then transported back to the laboratory. Analytical methods for soil were referred to the previous publication (Lu 1999). The physico-chemical properties of the soil are shown in the Table 1.
2.2 Enrichment conditions
An anoxic carbonate-buffered freshwater medium was used for enrichment of iron-reducing bacteria in the As-contaminated paddy soil. The basal medium contained 1 g L−1 NaCl, 0.4 g L−1 MgCl2•6H2O, 0.1 g L−1 CaCl2•2H2O, 0.25 g L−1 NH4Cl, 0.2 g L−1 KH2PO4, and 0.5 g L−1 KCl, The basal medium was autoclaved and cooled to room temperature under an atmosphere of N2/CO2 (90/10, v/v) and 30 ml L−1 bicarbonate solution (1 mol L−1, autoclaved under CO2), 1 ml L−1 vitamin, 1 ml L−1 trace elements mixture, and 1 ml L−1 selenite–tungstate solution were added (Widdel and Bak 1992). In order to limit the growth of sulfate-reducing bacteria, sulfate and sulfide were substituted by 100 μmol L−1 cysteine as a sulfur source. A widdel flask was used for batch preparation of anoxic medium and dispensation into culture serum bottles (100 ml), which were capped with thick butyl rubber stoppers and aluminum caps.
A fresh soil sample (10 g) was put into 90 ml anoxic water, mixed homogeneously; then, 5 ml of soil suspension was added to 45 ml mineral medium containing 30 mmol L−1 ferric citrate/ferrihydrite and 10 mmol L−1 acetate under an atmosphere of N2–CO2 (90/10, v/v). Ferrihydrite was prepared using FeCl3 and NaOH as described previously (Straub et al. 2005), and identified by X-ray diffraction (data not shown). One enrichment was designated as HN, referring to the iron source of ferric citrate, and the other was HN-HFO, referring to ferrihydrite used as an electron acceptor. There were control treatments with sterilized soil suspension in the defined medium. Each enrichment had three replicates. The cultures were incubated at 30°C in the dark. Standard anaerobic culturing techniques were used throughout the experiments (Widdel and Bak 1992).
2.3 DNA extraction
DNA of the microbial community was collected when iron reduction tended to approach a plateau. The presence of iron oxides in enrichment cultures inhibited DNA isolation. Therefore, the iron oxides were dissolved before genomic DNA extraction: 4 ml of the enrichment cultures was collected anoxically using a 5-ml sterile syringe and 21-gage needle, dispensed into centrifuge tubes and spun at 16,000×g for 3 min. The supernatant was decanted and the iron oxide–bacteria floc was resuspended in 1 ml of a filter-sterilized solution of ammonium oxalate (28 g L−1) and oxalic acid (15 g L−1) and incubated until complete solubilization of iron was achieved. Total genomic DNA was extracted from the remaining cells using standard procedures (Marmur 1961).
2.4 Construction of 16S rRNA gene clone library and amplified ribosomal DNA restriction analysis
The DNA was used as a template for amplification of nearly the entire 16S rRNA gene (~1,500 bp) with the bacteria-specific primers 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and R1492 (5′-TACGGYTACCTTGTTACGACTT-3′) as previously described (Lane 1991). The PCR program was as follows: 95°C for 5 min and then 32 cycles at 95°C for 30 s, 55°C for 30 s and 72°C for 30 s, followed by a 10-min extension time at 72°C. After amplification, gel slices containing the PCR products were excised and purified using Wizard® SV Gel and PCR Clean-Up System (Promega, Madison, WI, USA). PCR products were ligated into pGEM-T easy vector (Promega) and then cloned into Escherichia coli JM109 (TaKaRa, Shiga, Japan) in accordance with the manufacturer’s instructions. Transformants were grown on LB agar containing ampicillin (100 µg ml−1) and 40 µl of 40 mg ml−1 of X-Gal, and positive white clones were randomly picked to confirm the right insert using PCR with primers (T7 and SP6), which were complementary to the flanking regions of the PCR insertion site of the pGEM-T easy vector. The PCR program was described above. All clones containing inserts of the correct size were stored in LB medium at 4°C.
The PCR products were digested with restriction endonucleases, HhaI, at 37°C for 4 h. The restriction enzyme digests were separated on a 2% agarose gel running in 1×TAE buffer at 100 V for approximately 1 h. Fragments shorter than 80 bp were not taken into consideration because they were very close to the detection threshold. According to amplified ribosomal DNA restriction analysis (ARDRA) patterns, clones were grouped into OTUs. One or more clones representing each OTU were sequenced. The coverage for each clone library was calculated using the formula C = (1 − n 1/N) × 100%, where n 1 is the number of OTUs containing only one clone, and N is the total number of clones in the clone library. Rarefaction curves were calculated using the freeware program aRarefact Win (Holland 2003).
2.5 T-RFLP fingerprinting
The primers were used as described above; however, 27F primer was labeled with 5-hexachloroflurescein (HEX). PCR conditions and PCR products were purified as described above. Fifteen microliters of the PCR products were digested separately with restriction enzymes (HhaI, MspI, TaqI, HaeIII) at 37°C overnight. The digested DNA was precipitated with 0.1 vol 3 mol L−1 sodium acetate and 2.0 vol 95% ethanol followed by spinning at 16,000×g for 15 min. The DNA pellet was washed with 70% ethanol, dried, and resuspended in a mixture of 14.5 μl deionized formamide and 0.5 μl of DNA fragment length internal standard. The mixture was denatured for 2.5 min at 94°C and then immediately placed on ice. Electrophoresis was performed on an ABI 373 DNA sequencer (Applied Biosystems, Foster City, CA, USA).
2.6 Sequencing and phylogenetic analyses
Clones representing each distinct ARDRA pattern were sent to Sangon (Shanghai, China, http://www.sangon.com/) for sequencing (ABI PRISM 3730 sequencer). The presence of possible chimeric sequences was investigated by using the CHIMERA_CHECK program of the Ribosomal Database Project (RDP)II. The obtained sequences were manually proofread and corrected if necessary, edited, and aligned using BioEdit version 4.8.5. Nucleotide sequences generated in this study have been deposited in GenBank database under accession numbers FJ269043 to FJ269081 and FJ269081 to FJ269108 (HN and HN-HFO clone libraries, respectively). All sequences were analyzed by NCBI BLASTN search program, and the closest relatives to the clone sequences were extracted and aligned by Molecular Evolutionary Genetics Analysis (MEGA 3.1) software for phylogenetic analyses. Phylogenetic trees were constructed using neighbor-joining method. Robustness of derived groupings was tested by bootstrap using 1,000 replications.
3 Results
3.1 Physiological characterization in iron (III)-reducing enrichment
Two types of iron (III) were used to set up enrichment cultures: one was ferric citrate, and the other was ferrihydrite. The accumulative iron (II) content in the enrichment was shown in the Fig. 1. The iron (II) production by iron-reducing microorganisms (HN) increased sharply, and iron (II) content reached 18.41 mmol L−1 (see Fig. 1a). While iron-reducing microorganisms (HN-HFO) reduced 3.90 mmol L−1 iron (III) in 7 days, the iron reduction curve tended to reach a plateau (see Fig. 1b). No iron reduction was observed in the control inoculated with sterilized soil suspension.
3.2 Screening of 16S rRNA gene clones by ARDRA
ARDRA was performed on 134 and 138 clones, respectively, randomly chosen from HN and HN-HFO clone libraries. The ARDRA patterns were obtained with HhaI digestion, and OTU was defined as a group of clones that had identical banding patterns. According to ARDRA patterns, 134 clones were grouped into 33 OTUs in the HN clone library, and 138 clones were grouped into 31 OTUs in the HN-HFO clone library. Most of the OTUs were represented by two to three clones, with some OTUs containing a single clone. Then, approximately 120 clones were screened from 33 OTUs and 31 OTUs for sequencing. Clone libraries were statistically evaluated by rarefaction analysis in, which the expected number of different OTUs vs. the number of clones in the library was calculated. Rarefaction curves tended to approach the saturation plateau (Fig. 2), indicating that the clones used in each library’s screening could well cover the diversity of 16S rRNA genes.
3.3 Community of profiling of iron (III)-reducing enrichment
The distribution of the taxonomic groups differed between HN and HN-HFO enrichments based on clone library (Fig. 3) and T-RFLP results (Table 2). Bacteroidetes constituted 33.8% of 16S rRNA gene sequences from HN enrichment containing dissolved iron (III). Twenty nine percent of 16S rRNA gene sequences were related to α-Proteobacteria and 24.8% of the sequences belonged to β-Proteobacteria. Firmicutes constituted 9.8% of 16S rRNA gene sequences and 1.5% and 0.8% belonged to γ-Proteobacteria and Planctomycetes, respectively (see Fig. 3a). In contrast to the HN enrichment, no α-Proteobacteria and Planctomycetes-related sequences were detected in the HN-HFO enrichment containing an iron (III) mineral as the electron acceptor. However, δ-Proteobacteria-related sequences were obtained and accounted for 17.4% of the whole clones. In addition, 44.2% of 16S rRNA gene sequences were related to Bacteroidetes, 34.1% of the clones were related to Firmicutes and 2.9% and 1.4% of the clones belonged to γ-Proteobacteria and β-Proteobacteria, respectively (see Fig. 3b).
Four restriction enzymes were used to create different fingerprints for each of the initial enrichment culture samples in T-RFLP analysis. HhaI and MspI consistently yielded the highest number of resolvable peaks and were used in subsequent analyses. TaqI and HaeIII produced significantly lower number of resolvable peaks compared with HhaI and MspI, which was most likely due to conservation of enzyme recognition site sequences. The T-RFs size for each clone sequence was also generated by the restriction enzymes HhaI and MspI via silico restriction digestion. Table 2 shows the impact of dissolved and solid-phase iron (III) on the composition and relative abundance of bacterial groups, and T-RF size generated by the restriction enzymes HhaI and MspI is also listed. Analysis of the HN fingerprint generated by digestion with HhaI/MspI revealed the presence of 9/8 peaks above background fluorescence. The HN-HFO fingerprint included 7/7 resolvable peaks as generated by digestion with HhaI/MspI. Two T-RFs (86, 150) dominated the HN enrichment and three T-RFs (86, 92, 162) were dominant in the HN-HFO enrichment by the restriction enzymes MspI. Five T-RFs (64, 92, 94, 96, 198) and three T-RFs (91, 97, 99) dominated the HN enrichment and HN-HFO enrichment, respectively, by the restriction enzymes HhaI (see Table 2).
3.4 Phylogenetic analysis of iron (III)-reducing enrichment
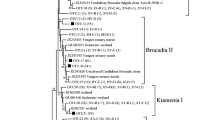
The phylogenetic trees of the clone sequences and their most similar GenBank sequences are shown in Figs. 4 and 5. There were marked differences in the phylotypes observed in iron-reducing bacteria enrichments between HN and HN-HFO. Bradyrhizobium, Bacteroides, Clostridium, and Ralstonia species were the dominant bacteria in the HN enrichments (see Fig. 4). Thirty five clones from HN enrichment represented by HN1 and HN2 sequences were closely related to Bradyrhizobium elkanii and Bradyrhizobium sp, respectively. HN11 and HN56 sequences representing 24 clones were closely related to Bacteroides sp., and HN3 representing 15 clones were closely related to uncultured Clostridiaceae. HN4 representing nine clones showed high sequence similarity to Ralstonia sp., and a single sequence (HN122) showed high sequence similarity to uncultured Planctomycete. However, members of the genera Clostridium, Bacteroides, and Geobacter were the dominant microorganisms in the HN-HFO enrichments (see Fig. 5). Thirty nine clones from HN-HFO enrichment belonged to the genera Clostridium, and 38 clones belonged to the genera Bacteroides. HN-HFO16 and HN-HFO140 representing 19 clones were closely related to Geobacter uraniumreducens, while HN-HFO54 representing five clones was closely related to Geobacter sulfurreducens.
Phylogenetic tree of 16S rRNA gene sequences (~1,500 bp) of iron-reducing bacteria, retrieved from HN enrichment. A phylogenetic tree was constructed from neighbor-joining method using MEGA 3.1 software. Robustness of derived groupings was tested by bootstrap using 1,000 replications. The scale bar represents 0.02 substitutions per nucleotide site
Phylogenetic tree of 16S rRNA gene sequences (~1,500 bp) of iron-reducing bacteria, retrieved from HN-HFO enrichment. A phylogenetic tree was constructed from neighbor-joining method using MEGA 3.1 software. Robustness of derived groupings was tested by bootstrap using 1,000 replications. The scale bar represents 0.02 substitutions per nucleotide site
4 Discussion
In the present study, clone library analysis, combined with T-RFLP method, was used to assess the diversity of iron (III) reducers in the paddy soil. Both approaches are important molecular genetic tools with which to monitor the diversity, structure, and dynamics of microbial populations. The abundance of iron-reducing bacteria was calculated based on clone library data and T-RFLP data, which are shown in Fig. 3 and Table 2. In the HN enrichment, for example, 29.3% of 16S rRNA gene sequences were related to α-Proteobacteria, while T-RFLP data showed that 6.7% and 45.6% T-RFs were related to α-Proteobacteria by using digestion enzymes HhaI/MspI. These differences could be due to insufficient clonal library screening or to biases introduced during the clone process. In addition, the T-RFLP technique is inherently biased for underestimating the diversity in a given sample owing to the conservation of enzyme recognition site sequences in a given gene across different organism (Marsh et al. 2000). Although there were no identical data from two methods, that can obtain similar conclusions. Our results suggest that community composition of bacteria can be better investigated using these two approaches together.
As can be seen from Fig. 1, much more iron was reduced with ferric citrate than with ferrihydrite. It is easier to reduce ferric citrate than ferrihydrite by iron-reducing bacteria, and ferrihydrite reduction by iron-reducing bacteria can be stimulated by secondary bacteria (Straub and Schink 2004). Weiss et al. (2003) found that there was high content of poorly crystalline iron (III) in the soil, and the percentage of poorly crystalline iron (III) was significantly correlated with the percentage of iron-reducing bacteria. Moreover, plant root exudates and plant debris can release organic acids (e.g., citrate), which are known to chelate Fe (III); thus, high levels of dissolved iron (III) were observed in paddy soils (Ratering and Schnell 2001). Therefore, ferric citrate and ferrihydrite were used in our experiments to investigate iron-reducing microbial communities following the natural inhibits; no such report has been published as we are aware of. We observed a significant effect of the different types of iron (III) on the microbial community structures and relative abundance in the enrichments. This might be due to the fact that these compounds have different redox potentials; the reduction of ferric citrate to the ferrous form occurs at a much higher redox potential (+372 mV) than ferrihydrite (−100 to +100 mV). Owing to the complexity of natural environments and the wealth of microbial capacities, it is not surprising that different microorganisms developed different strategies in reducing diverse iron (III) species under various conditions. For example, Geobacter metallireducens can cope with the difficulty of transferring electrons from the cell to the surface of barely soluble electron acceptors by physical contact (Nevin and Lovley 2000). In addition to direct electron transfer to iron minerals, some evidence indicates that Shewanella algae and Geothrix fermentans produce and release both iron (III)-chelators and electron shuttles (Lovley et al. 2004). Furthermore, diverse membrane-bound and soluble carriers, such as c-type cytochromes, quinines, and multicopper oxidases, can play a role in the reduction of insoluble and soluble substrates (Gralnick and Newman 2007). For example, evidence in Geobacter spp. indicates that different cellular compounds are involved in the reduction of dissolved ferric-citrate and ferrihydrite (Straub and Schink 2004). Therefore, in one case, we preferentially stimulate the microbes that prefer dissolved iron (III) species; in the other case, we get the ones that prefer the iron (III) mineral. However, the difference in microbial community structures may also be attributed to citrate, which is a fermentable substrate for bacteria, in addition to the fate of iron to a certain extent.
Our findings demonstrate that the actual assemblage of iron (III) reducers in the paddy soil is more complex. Besides the well-known Geobacter species, several dominant species were observed in the enrichments. The presence of Ralstonia species in the HN enrichment has been described in environments contaminated with toxic metals (Konstantinidis et al. 2003; Mergeay et al. 2003), and they have also been shown to be able to reduce selenite to elemental selenium (Roux et al. 2001). Thus, the relatively high abundance of Ralstonia may be related to the relatively high level of trace metals (e.g., Cd and Pb) in the Chenzhou paddy soil.
Clostridia species were dominant in both HN and HN-HFO enrichments. No significant effect of iron (III) substrates on consortia in the enrichments was observed, suggesting that the Clostridia are flexible in the use of electron acceptors. These microbes anaerobically reduce iron primarily through fermentative processes, although this reduction represents only a minor pathway for electron flow (Lovley et al. 2004). In the present study, acetate was present as the sole electron donor in the enrichments; however, the residual organic matter from soil inocula may have provided sufficient organic substrates to support fermentation. While these microbes themselves may not be important iron reducers, they can channel electrons from anaerobic oxidation via humic acids towards iron reduction (Benz et al. 1998). In addition, there have been recent reports on the implication of Clostirdium species in As (V) reducing activity (Rhine et al. 2005). Therefore, the high As level in the Chenzhou paddy soil might be a key factor that impacts the abundance of these organisms.
Bacteroides species were also dominant in the HN and HN-HFO enrichment culture. Microorganisms belonging to the Bacteroides were mostly derived from human fecal and oral sources, as well as from other mammalian organs such as the rumen (Holdeman et al. 1984; Paster et al. 1994). However, several strains have been isolated from rice residue in irrigated rice-field soil recently (Ueki et al. 2006a, b, 2007, 2008). In addition, 16S rRNA gene sequences affiliations to Bacteroides were also detected in iron-reducing enrichment (Lin et al. 2007). Although iron reduction of these organisms was not identified in these studies, these strains could utilize a range of substrates to produce acetate, propionate, and succinate for dissimilatory iron-reducing bacteria, which demonstrated that these bacteria affiliated with Bacteroides play an important role in paddy soil.
Interestingly, one clone belonging to Planctomycetes was obtained in the present study (see Fig. 4). Planctomycetes were considered to be solely aquatic microorganisms until they were discovered unexpectedly in soil by culture-independent 16S rRNA methods (Liesack and Stackebrandt 1992). Meanwhile, they have been found in many habitats, such as brackish water lagoon, activated sludge, and wastewater (Neef et al. 1998). Moreover, three Candidatus genera were recently suggested for the not-yet cultured anaerobic planctomycetes performing anaerobic ammonium oxidation (Schmid et al. 2000; Jetten et al. 2003). Furthermore, it was assumed that dissimilatory reducing bacteria can use NH +4 as an energy source under anaerobic and reduced conditions (Clément et al. 2005). Recent study also showed that ammonium can enhance Fe (II) production in As-contaminated paddy soil (Chen et al. 2008b). Thus, we hypothesize that the members of Planctomycetes may use iron (III) as an electron acceptor while oxidizing NH +4 for energy production. However, nitrite, nitrate, or nitrogen should be measured to lend further support to the hypothesis.
To our knowledge, some sequences identified in our study were closely related to species such as Bradyrhizobium that have not been reported to perform dissimilatory iron reduction. Although results from our enrichment cultures and molecular approaches provide some information regarding microbial community members catalyzing iron reduction in the As-contaminated paddy soil, it must be stressed that isolation of such organisms and further characterization of their capacity for dissimilatory iron (III) reduction, particularly in relation to metal bioavailability, are essential. In addition to bacterial community, specific archaea (e.g., methanmogenic archaea) may have also contributed to iron reduction in the soil; therefore, the archeal community should be investigated in future research. In addition, the effect of As on iron-reducing microbial community should be answered in further studies.
To sum up, results from the present study indicated that dissimilatory Fe (III)-reducing consortia in As-contaminated paddy soil are phylogenetically diverse. Knowledge of microbial community may further our understanding of C/N/Fe biogeochemical processes and microbial functions in paddy soil.
5 Conclusions
The present study demonstrated that the dissimilatory iron-reducing consortia in As-contaminated paddy soil are phylogenetically diverse, and phylogenetic diversity of dissimilatory ferric iron reducers can be better investigated by clone library analysis combined with T-RFLP method. Moreover, iron (III) sources as a key factor have a strong effect on the iron (III)-reducing microbial community structure and relative abundance in the enrichments. In addition, Geobacter species are selectively enriched by ferrihydrite enrichment cultures.
References
Benz M, Schink B, Brune A (1998) Humic acid reduction by Propionibacterium freudenreichii and other fermenting bacteria. Appl Environ Microbiol 64:4507–4512
Chen XP, Zhu YG, Xia Y, Shen JP, He JZ (2008a) Ammonia-oxidizing archaea: important players in paddy rhizosphere soil? Environ Microbiol 10:1978–1987
Chen XP, Zhu YG, Hong MN, Kappler AN, Xu YX (2008b) Effect of different forms of nitrongen fertilizers on arsenic uptake by rice plants. Environ Toxicol Chem 27:881–887
Clément JC, Shrestha J, Ehrenfeld JG, Jaffé PR (2005) Ammonium oxidation coupled to dissimilatory reduction of iron under anaerobic conditions in wetland soils. Soil Biol Biochem 37:2323–2328
Ge Y, He JZ, Zhu YG, Zhang JB, Xu ZH, Zhang LM, Zheng YM (2008) Differences in soil bacterial diversity: driven by contemporary disturbances or historical contingencies? ISME J 2:254–264
Gralnick JA, Newman DK (2007) Extracellular respiration. Mol Microbiol 65:1–11
He JZ, Zheng Y, Chen CR, He YQ, Zhang LM (2008) Microbial composition and diversity of an upland red soil under long-term fertilization treatments as revealed by culture-dependent and culture-independent approaches. J Soils Sediments 8:349–358
Holdeman LVR, Kelly RW, Moore WEC (1984) Genus I. Bacteroides Castellani and Chalmers. In: Krieg NR, Holt JG (eds) Bergey’s manual of systematic bacteriology, vol 1. Williams and Wilkins, Baltimore, pp 604–631
Holland SM (2003) Analytic Rarefaction 1.3. http://www.uga.edu/~strata/software/anRareReadme.html
Horneman A, van Geen A, Kent DV, Mathe PE, Zheng Y, Dhar RK, O’Connell S, Hoque MA, Aziz Z, Shamsudduha M, Seddique AA, Ahmed KM (2004) Decoupling of As and Fe release to Bangladesh groundwater under reducing conditions. Part I: evidence from sediment profiles. Geochim Cosmochim Ac 68:3459–3473
Islam FS, Gault AG, Boothman C, Polya DA, Charnock JM, Chatterjee D, Lloyd JR (2004) Role of metal-reducing bacteria in arsenic release from Bengal delta sediments. Nature 430:68–71
Jetten MS, Sliekers O, Kuypers M, Dalsgaard T, van Niftrik L, Cirpus I, van de Pas-Schoonen K, Lavik G, Thamdrup B, Paslier DL, Op den Camp HJM, Hulth S, Nielsen LP, Abma W, Third K, Engström P, Kuenen JG, Jørgensen BB, Canfield DE, Sinninghe Damsté JS, Revsbech NP, Fuerst J, Weissenbach J, Wagner M, Schmidt I, Schmid M, Strous M (2003) Anaerobic ammonium oxidation by marine and freshwater planctomycete-like bacteria. Appl Microbiol Biotechnol 63:107–114
Kappler A, Straub KL (2005) Geomicrobiological cycling of iron. Rev Mineral Geochem 59:85–108
Konstantinidis KT, Isaacs N, Fett J, Simpson S, Long DT, Marsh TL (2003) Microbial diversity and resistance to copper in metal-contaminated lake sediment. Microbial Ecol 45:191–202
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, Chichester, pp 115–175
Liao XY, Chen TB, Xie H, Liu YR (2005) Soil As contamination and its risk assessment in areas near the industrial districts of Chenzhou City, Southern China. Environ Int 31:791–798
Liesack W, Stackebrandt E (1992) Occurrence of novel groups of the domain bacteria as revealed by analysis of genetic material isolated from an Australian terrestrial environment. J Bacteriol 174:5072–5078
Lin B, Hyacinthe C, Bonneville S, Braster M, Cappellen PV, Roling WFM (2007) Phylogenetic and physiological diversity of dissimilatory ferric iron reducers in sediments of the polluted Scheldt estuary, Northwest Europe. Environ Microbiol 9:1956–1968
Liu XZ, Zhang LM, Prosser JI, He JZ (2009) Abundance and community structure of sulfate reducing prokaryotes in a paddy soil of southern China under different fertilization regimes. Soil Biol Biochem 41:687–694
Lovley DR, Phillips EJP (1986) Organic matter mineralization with reduction of ferric iron in anaerobic sediments. Appl Environ Microbiol 51:683–689
Lovley DR, Phillips EJP (1988) Novel mode of microbial energy metabolism: organic carbon oxidation coupled to dissimilatory reduction of iron or manganese. Appl Environ Microbiol 54:1472–1480
Lovley DR, Holmes DE, Nevin KP (2004) Dissimilatory Fe (III) and Mn (IV) reduction. Adv Microb Physiol 49:219–286
Lu RK (1999) Analytical methods for soil and agricultural chemistry. China Agricultural Science and Technology Press, Beijing
Marmur J (1961) A procedure for the isolation of DNA from microorganisms. J Mol Bio 3:208–218
Marsh T, Saxman P, Cole J, Tiedje J (2000) Terminal restriction fragment length polymorphism analysis program, a web-based research tool for microbial community analysis. Appl Environ Microbiol 66:3616–3620
Mergeay M, Monchy S, Vallaeys T, Auquier V, Benotmane A, Bertin P, Taghavi S, Dunn J, van Der Lelie D, Wattiez R (2003) Ralstonia metallidurans, a bacterium specifically adapted to toxic metals: towards a catalogue of metal-responsive genes. FEMS Microbiol Rev 27:385–410
Neef A, Amann R, Schlesner H, Schleifer KH (1998) Monitoring a widespread bacterial group: in situ detection of planctomycetes with 16S rRNA-targeted probes. Microbiol 144:3257–3266
Nevin KP, Lovley DR (2000) Lack of production of electron-shuttling compounds or solubilization of Fe (III) during reduction of insoluble Fe (III) oxides by Geobacter metallireducens. Appl Environ Microbiol 66:2248–2251
Pan YM, Yang ZG (1988) Hunan background values for heavy metals and research methods. Publishing House, China Environmental Science, Beijing
Paster BJ, Dewhirst FE, Olsen I, Fraser GJ (1994) Phylogeny of Bacteroides, Prevotella, and Porphyromonas spp and related bacteria. J Bacteriol 176:725–732
Ratering S, Schnell S (2001) Nitrate-dependent iron (II) oxidation in paddy soil. Environ Microbiol 3(2):100–109
Rhine ED, Dominguze EG, Phelps CD, Young LY (2005) Environmental microbes can speciate and cycle arsenic. Environ Sci Technol 39:9569–9573
Roden EE, Lovley DR (1993) Dissimilatory Fe (III) reduction by the marine microorganism Desulfuromonas-Acetoxidans. Appl Environ Microbiol 59:734–742
Roux M, Sarret G, Pignot-Paintrand I, Fontecave M, Coves J (2001) Mobilization of selenite by Ralstonia metallidurans CH34. Appl Environ Microbiol 67:769–773
Scala DJ, Hacherl EL, Cowan R, Young LY, Kosson DS (2006) Characterization of Fe(III)-reducing enrichment cultures and isolation of Fe(III)-reducing bacteria from the Savannah River site, South Carolina. Res Microbiol 157:772–783
Schmid M, Twachtmann U, Klein M, Strous M, Juretschko S, Jetten M, Metzger JW, Schleifer KH, Wagner M (2000) Molecular evidence for genus level diversity of bacteria capable of catalyzing anaerobic ammonium oxidation. Syst Appl Microbiol 23:93–100
Stipp SLS, Hansen M, Kristensen R, Hochella MF, Bennedsen L, Dideriksen K (2002) Behaviour of Fe-oxides relevant to contaminant uptake in the environment. Chem Geol 190:321–337
Straub KL, Schink B (2004) Ferrihydrite reduction by Geobacter species is stimulated by secondary bacteria. Arch Microbiol 182:175–181
Straub KL, Kappler A, Schink B (2005) Enrichment and isolation of ferric-iron- and humic-acid-reducing bacteria. Method Enzymol 397:58–77
Ueki A, Akasaka H, Suzuki D, Ueki K (2006a) Paludibacter propionicigenes gen. nov., sp nov., a novel strictly anaerobic, Gram-negative, propionate-producing bacterium isolated from plant residue in irrigated rice-field soil in Japan. Int J Syst Evol Microbiol 56:39–44
Ueki A, Akasaka H, Suzuki D, Hattori S, Ueki K (2006b) Xylanibacter oryzae gen. nov., sp nov., a novel strictly anaerobic, Gram-negative, xylanolytic bacterium isolated from rice-plant residue in flooded rice-field soil in Japan. Int J Syst Evol Microbiol 56:2215–2221
Ueki A, Akasaka H, Satoh A, Suzuki D, Ueki K (2007) Prpvotella paludivivens sp nov., a novel strictly anaerobic, Gram-negative, hemicellulose-decomposing bacterium isolated from plant residue and rice roots in irrigated rice-field soil. Int J Syst Evol Microbiol 57:1803–1809
Ueki A, Abe K, Kaku N, Watanabe K, Ueki K (2008) Bacteroides propionicifaciens sp nov., isolated from rice-straw residue in a methanogenic reactor treating waste from cattle farms. Int J Syst Evol Microbiol 58:346–352
Weiss JV, Emerson D, Backer SM, Megonigal JP (2003) Enumeration of Fe (II)-oxidizing and Fe (III)-reducing bacteria in the root zone of wetland plants: Implications for a rhizosphere iron cycle. Biogeochemistry 64:77–96
Widdel F, Bak F (1992) Gram-negative mesophilic sulfate reducing bacteria. In: Balows A, Trüper HG, Dworkin M, Harder W, Schleifer KH (eds) The prokaryotes. Springer, New York, pp 3352–3378
Williams PN, Islam MR, Adomako EE, Raab A, Hossain SA, Zhu YG, Feldmann J, Meharg AA (2006) Increase in rice grain arsenic for regions of Bangladesh irrigating paddies with elevated arsenic in groundwaters. Environ Sci Technol 40:4903–4908
Zheng Y, Zhang LM, Zheng YM, Di HJ, He JZ (2008) Abundance and community composition of methanotrophs in a Chinese paddy soil under long-term fertilization practices. J Soils Sediments 8:406–414
Zhu YG, Sun GX, Lei M, Teng M, Liu YX, Chen NC, Wang LH, Carey AM, Deacon C, Raab A, Meharg AA, Williams PN (2008) High percentage inorganic arsenic content of mining impacted and nonimpacted Chinese rice. Environ Sci Technol 42:5008–5013
Acknowledgements
This study was financially supported by the National Natural Science Foundation (40671102 and 20720102042). We thank Dr. Karrie Weber for her critical comments on previous versions of this paper.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Ji-Zheng He
Rights and permissions
About this article
Cite this article
Wang, XJ., Yang, J., Chen, XP. et al. Phylogenetic diversity of dissimilatory ferric iron reducers in paddy soil of Hunan, South China. J Soils Sediments 9, 568–577 (2009). https://doi.org/10.1007/s11368-009-0113-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11368-009-0113-x