Abstract
Bioremediation of contaminated soils by a combinational approach using specific bacterial species together with ryegrass is a promising strategy, resulting in potentially highly efficient degradation of organic contaminants. The present study tested the combination of strain DXZ9 of Stenotrophomonas sp. with ryegrass to remove DDT and DDE contaminants from soil under natural conditions in a pot experiment. The strain DXZ9 was successfully colonized in the natural soil, resulting in removal rates of approximately 77% for DDT, 52% for DDE, and 65% for the two pollutants combined after 210 days. Treatment with ryegrass alone resulted in slightly lower removal rates (72 and 48%, respectively, 61% for both combined), while the combination of strain DXZ9 and ryegrass significantly (p < 0.05) improved the removal rates to 81% for DDT and 55% for DDE (69% for both). The half-life of the contaminants was significantly shorter in combined treatment with DXZ9 and ryegrass compared to the control. The remediation was mostly due to degradation of the contaminants, as the net uptake of DDT and DDE by the ryegrass accounted for less than 3% of the total amount in the soil. DDT is reductively dechlorinated to DDD and dehydrochlorinated to DDE in the soil; the metabolites of DDE and DDD were multiple undefined substances. The toxicity of the soil was significantly reduced as a result of the treatment. The present study demonstrates that the bioremediation of soil contaminated with DDT and DDE by means of specific bacteria combined with ryegrass is feasible.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
In the past, the insecticide DDT (1,1,1-trichloro-2,2-bis(4-chlorophenyl) ethane) was widely used in a variety of agricultural applications for pest control, as well as to control and reduce the mosquito vector of malaria. The active ingredient of DDT is p,p′-DDT, while its primary metabolites are DDE (1,1-dichloro-2,2-bis (4-chlorophenyl) ethylene) and DDD (1,1-dichloro-2,2-bis (4-chlorophenyl) ethane). Both DDT and its metabolites DDE and DDD pose potential health risks due to their high hydrophobicity, toxicity, persistence, and accumulation in natural food chains. For these reasons they were classified as priority pollutants (Sayles et al. 1997) and persistent organic pollutants (Sudharshan et al. 2012). Though the use of DDT has been prohibited for decades, the residual concentrations of DDT and DDE in the environment remain high (Qiu et al. 2004; Qiu et al. 2005; Wang et al. 2016; Ma et al. 2016). Although abiotic degradation has an effect on in a lot of cases for its removal from contaminated soils, the biodegradation by microorganisms is usually the most dominant process. The biological approaches include bioaugmentation, biostimulation, and attenuation; bioaugmentation is the most promising for the removal of pesticides and its metabolites from soil (Miyazaki et al. 2006; Gao et al. 2015; Cycoń et al. 2017). It should be taken into account that the appropriate strains of bioaugmentation have the ability to withstand higher concentrations of pollutants and to survive in a wide range of environmental conditions. The potential application of microorganisms in the biodegradation of other organochlorine pesticides using bioaugmentation technology has been confirmed (Arshad et al. 2008; Kataoka et al. 2011; Fuentes et al. 2011; Sáez et al. 2012). The promising results of bioaugmentation caused an increasing interest in searching for more effective bioremediation approaches (Raina et al. 2008; Bhalerao 2012; Cao et al. 2013; Cycoń and Piotrowska-Seget 2016; Odukkathil and Vasudevan 2016). For its removal from contaminated soils, bioremediation is considered a more cost-effective technical method compared to physical and chemical removal strategies (Fang et al. 2010; Purnomo et al. 2011; Fan et al. 2013; Chattopadhyay and Chattopadhyay 2015).
Bioremediation by means of specific plant species (phytoremediation) is simple to operate in situ on a large scale, at costs of only a fifth of conventional technology. Phytoremediation is particularly suitable for large areas of contaminated sites with relatively low concentration levels. Plants absorb and partly degrade organic contaminants, while root exudates also degrade the pollutants in the soil (Ahmad et al. 2012; Rostamia et al. 2016; Mitton et al. 2012). In addition, plant roots improve the aeration of the soil, which stimulates the growth of aerobic microorganisms in the rhizosphere (Moubashera et al. 2015). These bacteria further contribute to the degradation of organic pollutants. Thus, the main mechanism involved in phytoremediation is based on the combined effects of absorption and accumulation of pollutants by plants, degradation by root exudates, and degradation by the rhizobia. Ryegrass is a perennial herbaceous plants which can be repeatedly harvested; studies have shown that several types of pollutants, amongst others petroleum products and polyaromatic hydrocarbons, can be efficiently degraded in soil in the presence of ryegrass (Huang et al. 2004; Tang et al. 2010; Yu et al. 2011; Rostamia et al. 2016).
The advantages of bioremediation by combination of microorganism with phytoremediation are obvious, as it combines low operation costs and high safety with the absence of secondary pollution. Nevertheless, when it is dependent on indigenous microorganisms present in the soil, its effectivity may be limited. By inoculation of the soil with specific microorganisms that have a high potential for organic pollutant removal, the efficiency can be enhanced (Yu et al. 2011; Tang et al. 2010; Lu and Zhang 2014; Zhu et al. 2012). This integrated application of microbial remediation and phytoremediation is a promising direction, but the inoculum must be specifically targeted to the nature of the contaminants.
Apart from efficacy of the method, the toxicity of the various metabolites produced during the process need to be carefully considered in a safety evaluation. Unfortunately, the relevant research into this aspect is limited (Kong et al. 2014). Earthworms can be used as key indicator organisms by means of a standardized ecological toxicity test. An assay based on single cell gel electrophoresis (SCGE) using coelomocytes extracted from exposed earthworms can be used to evaluate the toxicity of the remediated soil (Khan et al. 2012; Gandolfi et al. 2010).
A pilot study conducted in our lab had indicated that Stenotrophomonas sp. strain DXZ9 was a suitable candidate for biodegradation of DDT and DDE: in experiments conducted with pure cultures in culture media, removal rates of respectively 55 and 39% were obtained within 5 days. These observations led to the present study, in which strain DXZ9 was combined with ryegrass to degrade DDT from artificially contaminated soil in pot experiments under natural conditions. The colonization of the bacteria in the soil was confirmed by denaturing gradient gel electrophoresis (DGGE). Bioremediation of DDT and DDE was determined and the metabolites that were produced were characterized by gas chromatography/mass spectrometry (GC/MS). Their toxicity was evaluated by the SCGE assay using earthworms.
It has been reported that the strain Stenotrophomonas sp. can degrade HCH and methyl parathion (Zhang et al. 2009), acetamiprid (Tang et al. 2012), and chlorothalonil (Zhang et al. 2014a, 2014). However, there has been no report degrading DDT bacterium, which would be a better bioremediation technology for DDT contaminated sites than the present methods. The objective of the present study was thus to test the suitability of combined microbial- and phytoremediation for the degradation of DDT from contaminated soils.
Materials and methods
Supplies and chemicals
The chemicals DDE (99% pure) and DDT (99% pure) were purchased from Shenyang Research Institute of Chemical Industry, China. Acetone, petroleum ether (60–90 °C), and n-hexane of analytical grade were used; they were distilled prior to use. Acetone was distilled at 56 °C, a petroleum ether distillate was collected at 60~75 °C, and n-hexane was distilled at 69 °C. Anhydrous sodium sulfate (analytical grade) was dried for 6 h at 130 °C and kept in a desiccator before use.
Stenotrophomonas sp. strain DXZ9 was isolated by enrichment culture from DDT-contaminated sludge originating from a pesticide factory; the isolation procedure was according to the reference (Xie et al. 2011). Seeds of ryegrass (Lolium perenne) were commercially purchased. The soil used was sampled from the topsoil of an experimental plot in the South Campus of Shandong Agricultural University, Taian, China (36° 09′ 58.1 N: 117° 09′ 36.6 E). The brunisolic soil contained 17.6-mg kg−1 organic matter, 31.9% sand, 57.7% silt, and 10.4% clay; additional parameters of soil such as pH, organic nitrogenous, readily available phosphorus; CEC was 7.6, 132.3 mg kg−1, 18.4 mg kg−1, and 43.39 cmol kg−1.
Experimental design
The experiment consisted of six treatments with five replicates each: a control of soil treated with distilled acetone solvent only (S), soil spiked with a DDT/DDE mixture dissolved in acetone (S + D), soil with DDT/DDE and DXZ9 bacteria added (S + D + B), soil with ryegrass (S + G), soil with DDT/DDE and ryegrass (S + G + D), and finally soil treated with DDT/DDE, ryegrass, and DXZ9 combined (S + G + D + B). The 30 pots were divided into the six treatment groups, each containing 8-kg soil. The experiment was performed outdoors for 210 days, while the pots were sheltered only during heavy rains. The experiment was conducted from May 29 to December 25, 2012. Because the time period is very long from summer to winter, the temperature was varied from 35 to 8 °C, and the moisture content of the soil commonly remained 11.28~13.50%.
Prior to the experiment, the collected topsoil was passed through a 20-mesh sieve, after which 2% (g/g) fermented cow’s manure was added and thoroughly mixed, to simulate conventional fertilization. Eighty-kilogram soil was mixed with 80-ml acetone for the S and S + G treatments. One hundred sixty-kilogram soil was mixed with 160-ml acetone containing of 2.0-mg ml−1 DDT and 1.0-mg ml−1 DDE to be used for treatments S + D, S + D + B, S + G + D, and S + G + D + B. This resulted in approximate initial soil concentrations of 2-mg kg−1 DDT and 1-mg kg−1 DDE; their exact concentrations were determined in each pot as described below.
The bacterial strain DXZ9 was cultured in MSM (4.0-g NaNO3, 1.5-g KH2PO4, 0.005-g FeCl3, 0.01-g CaCl2, 0.2-g MgSO4, and 0.5-g Na2HPO4 in 1 l of distilled water (pH 7.0)) at 30 °C for 3 days in the experiment, and which were harvested by centrifugation at 8000 r min−1 for 10 min. The bacterial pellet was weighed and 100 g was suspended into 2000-ml sterile water, to be used as inoculum (200 ml per pot); the final concentration (cfu g−1) with an average value of the bacterial strain was of around (2.0 ± 1.0) × 106 cfu per gram of soil. A 5.0 g ryegrass seed was planted per pot. At the end of the experiment, the cultured ryegrass was harvested, dried and weighed, and analyzed for residue content.
Colonization of DXZ9 in the soil
Colonization of DXZ9 bacteria in the soil was determined by denaturant gradient gel electrophoresis (DGGE); the PCR-DGGE analysis was described in the previous study (Zhang et al. 2014, 2014). At days 10, 30, 90, and 210, soil samples were collected (0.5 g) and analyzed as previously described. Briefly, DNA was extracted from the soil using the PowerSoil DNA Isolation Kit (MO BIO LAB) The isolated DNA was used as a template to amplify the 16S rDNA gene by polymerase chain reaction (PCR) using the primers, after which the variable V3 region of Stenotrophomonas was specifically amplified using primers. The amplicon was analyzed by DGGE, and band intensity was compared using strain DXZ9 as a control, to estimate the relative abundance of Stenotrophomonas in the various treatments soils. The ability of inoculated microorganisms to survive in the contaminated soil is vital; it competes with indigenous microbes for nutrients and niches. Studies have shown that the number of microorganisms commonly decreases during the first few days after inoculation (Hong et al. 2007), and the number of inoculants seldom significantly increases; the optimal inoculum size is of great importance to the pesticide degradation (Ramadan et al. 1990; Karpouzas and Walker 2000a), and the inoculum size at a level of 106–1010 cells/g of soil is suitable for the efficient degradation of pesticides by the microorganisms inoculated into the soils (Comeau et al. 1993; Duquenne et al. 1996; Singh et al. 2006). In the present study, the final concentration of inoculum size was of around (2.0 ± 1.0) × 106 cfu per gram of soil; the strain DXZ9 was successfully colonized in the soil.
Extraction and analysis of DDT and DDE residues in the soil and ryegrass biomass
Residual DDT and DDE were extracted from the soil by Soxhlet extraction (EPA 3540C) using sulfuric acid (EPA 3665). The extracted DDT and DDE were quantitatively analyzed by gas chromatography equipped with a 63Ni electron capture detector. An OV-1701 capillary column was used (cross-linked with 14% cyanopropyl phenyl polysiloxane, sized 30 m × 0.53 mm × 0.25 μm) (Australia). The oven temperature was increased from initially 160 to 220 °C at a rate of 40 °C min−1, and then to 250 °C with 4 °C min−1, to finally be kept constant for 2 min. The injector port was maintained at 230 °C and the detector at 280 °C.
The content of DDT and DDE in the dried biomass of harvested ryegrass was extracted using the same method as described for soil.
Mathematical modeling of the dynamics of contaminant removal
Four different degradation models can typically be applied to mathematically describe the dynamics of contaminant removal from an environment: the exponential model (describing a first-order degradation kinetic model, C = C0 · e−kt), a double chamber degradation model (C = A · e−αt + B · e−βt), a first-order absorption model (C = A · (e−αt − e−βt)), and the mathematical model of removal of multiple pesticides (Queyrel et al. 2016). In these equations, C0 is the initial concentration, k is a constant, t is the time, β and α are the degradation constants for the substance, and parameters A and B are its concentrations at the different times. The double chamber degradation model was used here to calculate the half-life of DDT and DDE under the experimental conditions applied.
Detection of the metabolites of DDT and DDE
The extracts obtained as described in “Extraction and analysis of DDT and DDE residues in the soil and ryegrass biomass” were analyzed by GC/MS (PE Clarus 500) equipped with an Elite-5MS capillary column (cross-linked with 5% phenyl-methyl silicone, 30 m × 0.25 mm × 0.25 μm) to determine the nature of metabolites. The oven temperature was increased from initially 70 (1.0-min hold) to 180 °C at a rate of 20 °C min−1 (5-min hold), and then to 260 °C at a rate of 5 °C min−1 (5 min hold). The injector was maintained at 250 °C. Helium was used as the carrier gas at a flow rate of 1 ml min−1. Electron ionization was used at 70 eV. The ion source temperature of the mass spectrometer was 200 °C. Scanning was performed from 40 to 550 m/z. The transfer line temperature was 250 °C. Quantitative analysis was performed by using the selected ion monitoring (SIM) mode.
Toxicity evaluation
At three time points (days 5, 30, and 210), 100-g soil was sampled to test the remaining toxicity. The soil samples were mixed with 30-ml deionized water in conical flasks, and after addition of three earthworms (Eisenia fetida) per sample, the flasks were incubated in the light under atmospheric conditions under constant humidity at 23 ± 2 °C for 48 h. The earthworms were then collected and kept onto filter paper moistened with saline solution (4 °C) for 12 h to clean their digestive tract. The coelomocytes of the animals were collected using a non-invasive extrusion method described elsewhere (Song et al. 2009). These cells were placed on ice and mixed with 1-ml PBS prior to the comet assay, based on SCGE, performed according to the literature (Kong et al. 2014; Song et al. 2009). The extent of DNA damage of the coelomocytes was evaluated by olive tail moment (OTM), which is given by the distance between the tail regions and the centers of the comet head.
Statistical analyses
The mean values of all data were compared using the Statistical Package for Social Sciences (SPSS 20.0 for Windows) package by a multiple comparison test at the 5% probability level. The results were analyzed by variance (ANOVA test). Figures were produced using Sigmaplot 10.0. The removal rate of contaminants was calculated as: \( x=\frac{C_{\mathrm{initial}}-{C}_{\mathrm{residue}}}{C_{\mathrm{initial}}}. \)
With Cinitial being the initial concentration and Cresidue the concentration determined at a particular time point for each contaminant.
Results and discussion
Colonization of DXZ9 bacteria in the soil
First, it was assessed if the inoculated Stenotrophomonas sp. strain DXZ9 was able to colonize the soil. It was evaluated in all treatments by DGGE analysis, targeting the PCR fragment of the variable V3 region of the amplified partial 16S rDNA gene (Fig. 1). Band intensity was compared for the band specific for DXZ9 and the signal representing the dominant population in the soil as determined by DGGE. The target bands of lane B (strain DXZ9) are clearly visible in Fig. 1. This identified that DXZ9 was present during the complete test period, though it predominated in the bacterial population only for the first 30 days. At day 210, the band was less distinct, illustrating that the dominance of strain DXZ9 gradually decreased, but the bacteria were detected throughout the complete experiment. These findings were consistent with results obtained by plate counts (results were not shown), which indicated that the population of bacteria was at a maximum in the treatments with microorganism at day 10; the number of bacteria decreased gradually to a normal level after 30 days. The result suggested that DXZ9 successfully colonized in the soils under the conditions applied. It is vital that the ability of inoculated microorganisms survive in the contaminated soil. Inoculated microorganisms compete with indigenous microbes for nutrients and niches. Studies have shown that the number of microorganisms commonly decreases during the first few days after inoculation (Hong et al. 2007), and the number of inoculants seldom significantly increases; the optimal inoculum size is of great importance to the pesticide degradation (Karpouzas and Walker 2000), and the inoculum size at a level of 106–1010 cells/g of soil is suitable for the efficient degradation of pesticides by the microorganisms inoculated into the soils (Singh et al. 2006). In the present study, the final concentration of inoculum size was of around (2.0 ± 1.0) × 106 cfu per gram of soil; the strain DXZ9 was successfully colonized in the soil.
Fingerprint of DGGE from the DXZ9 and the soil with different treatments. Indicate: B: bacterial Stenotrophomonas strain DXZ9; 10-1: S + D + B-10, 10-2: S + G + D + B-10; 30-1: S + D + B-30, 30-2: S + G + D + B-30; 90-1: S + D + B-90, 90-2: S + G + D + B-90; 210-1: S + D + B-210, 210-2: S + G + D + B-210
Bioremediation of DDT and DDE in the contaminated soil
The remediation fractions of DDT and DDE, as well as their total amounts combined, are shown for the different treatments in Table 1, and the dynamics of the determined degradation are shown in Fig. 2. During the complete course of the experiment, all three remediation treatments (S + D + B for DXZ9 only, S + G + D for ryegrass only, and S + G + D + B for the combined treatment of ryegrass with DXZ9) removed a larger fraction of the pesticide mixture than was observed for the untreated control (S + D). After 210 days of incubation, ryegrass alone had removed approximately 72% DDT and 48% DDE (61% when combined) from the contaminated soil. The inoculum with bacteria resulted in removal of 77% DDT and 52% DDE (65% in combination), while bacteria combined with ryegrass removed 81% DDT and 55% DDE (69% in combination). This is a significant increase compared to single treatments (p < 0.05). Without inoculum, the microbes naturally present in the soil had removed only 30% DDT and 31% DDE. Thus, the combined treatment with ryegrass and bacteria removed the largest fraction of DDT and DDE, reducing the residual concentrations considerably after 210 days.
It could be concluded that microbial remediation was highly efficient initially, but it reduced over time, possibly due to a gradual decrease in viable Stenotrophomonas (Fig. 1). It offers our advice that the efficiency of degradation can be increased if bacterial inocula are repeated at regular time intervals, though this remains to be tested.
The degradation dynamics applying to these experimental conditions were modeled in order to predict the half-life of the contaminants and to be able to predict the concentration of contaminants in the soil at any given time point. Of the four different degradation models that can typically be applied, the dual chamber degradation model (“Materials and methods” section) was found to be most appropriate. The degradation rate constant was calculated using the exponential equation given in the methods. The parameters of the exponential decay are presented in Table 2. The half-life of DDT was calculated as 48 days for treatment with S + G + D + B, 58.5 days for S + D + B, 75.5 days for S + G + D, and 1055 days for the control S + D. Likewise, the half-life of DDE was 162.5, 229, 235, and 745 days for the different treatments, respectively. For the two compounds combined, these were 71.8, 86.5, 114.8, and 1095 days. These calculations demonstrate the considerable reduction in half-life of these pollutants for the combined bacterial/plant treatment compared to the other treatments.
In a previous publication, Zhu has reported that strain DDT-1 (Pseudomonas spp.) could significantly reduce the concentration of DDTs in soil originating from an agricultural field in Chiqi City, Zhejiang Province, China (Zhu et al. 2012). In comparison with those results, the degraded rates obtained in the present study suggest that strain DXZ9 (at least in the soil tested here) resulted in higher removal rates of the pesticide. However, differences between the experiments need to be taken into account. For instance, the soil used by Zhu (Zhu et al. 2012) containing aged DDT and DDE, which may have resulted in derivatives that are less easily biodegraded. Nevertheless, it is possible that the degrading ability of Stenotrophomonas sp. strain DXZ9 is higher than that of Pseudomonas sp. strain DDT-1.
A study using Sedum alfredii resulted in merely 19.9% DDT removal after 180 days (Zhu et al. 2012), which is much less than the removal obtained with ryegrass presented here. Indeed, other works have shown that ryegrass is highly suitable to remediate soils not only containing organic pollutants such as PCBs (Ding et al. 2013), PAH (Meng et al. 2011; Fu et al. 2011), or phenanthrene (Li et al. 2016) but also selenium (Tullo et al. 2015). Performance might even be better if the biomass of growing ryegrass could be further enhanced, or if enzyme activity of the microorganisms living in the soil could be increased. The present results suggest that inoculation with specific bacteria can also increase the efficiency of DDT and DDE phytoremediation.
This interpretation is supported by theoretical considerations. Of the factors affecting remediation, various physicochemical properties of the pollutants need to be taken into account. In general, absorption by plants is affected by the octanol/water partition coefficient (lg Kow) of the pollutant. The lg Kow of DDT is 6.91 and its lg Koc is 6.59. Thus, DDT is strongly lipophilic and undergoes stronger absorption forces compared to the weaker migration forces. As a consequence, this pollutant is less likely removed by root uptake and subsequent metabolic activity in the shoots. In addition, the volatility of a contaminant needs to be considered. This can be evaluated by the Henry constant, which in the case of DDT is 2 × 10−6. As such, DDT can be considered as non-volatile, so that little will be released from the soil this way. Research conducted to investigate this property reported that the volatilization of pesticides mainly occurs in the topsoil, whereby the release of DDT could at most account for 0.43% of the original amount (Gang Ji and Xia 1985). In the present experiments, it may be even less, when the surface is coved with ryegrass. Therefore, we propose that in the experimental setup, DDT was mostly removed by, in order of decreasing magnitude: (1) biodegradation by Stenotrophomonas in the soil, (2) biodegradation by organic secretions and enzymes into the rhizosphere released from the roots of ryegrass, and (3) uptake by the plants (Ahmad et al. 2012; Fu et al. 2011; Anderson et al. 1993). Combining microbial remediation with phytoremediation is the promising strategy; growing hyperaccumulator can confirm a regular supply of nutrients to the soil and thereby enhance the further multiplication of the microorganism (Abhilash et al. 2011); the rhizoremediation has great potential for the treatment of persistent organic chemicals, although the productiveness of the strategy can be influenced by a number of elements that may require adjustment to achieve the best effect (Böltner et al. 2008).
The equilibrium quantity of DDT and DDE in the soil during bioremediation
The success of phytoremediation highly depends on the choice of the plant species, and their accumulation potential of the pesticide could be considered. In the treatment with ryegrass only, the removal rates of the pollutants were relatively high during the first 10 days (the initial concentration of 2.23-mg kg−1 DDT was reduced to 1.53 mg kg−1, and 1.26-mg kg−1 DDE was reduced to 0.84 mg kg−1), and removal rates slowed down after this period, but it remained higher than the treatment without ryegrass. Possibly, the initial high rates were caused by the germination of seeds and the growth of sprout in a short period; during this period, the uptake of amount of pesticides coincided with the uptake of high nutrient. To assess the importance of plant uptake in bioremediation, the fraction of DDT and DDE still residing in the soil was compared to the fraction absorbed by the ryegrass. The balance of DDT and DDE in the soil is shown in Table 3. It is obvious that the net uptake of DDE by the ryegrass exceeded that of DDT. At the end of the experiment, approximately 400-g dry ryegrass was harvested per pot on average, and this amount of plant biomass was so small that the net uptake of DDT and DDE by the ryegrass accounted for only 0.63–0.81 and 1.96–2.78% of the total amount in the soil, respectively.
A number of species have been tested for remediation of insecticides, with variable absorption capacity. Crimson clover, mustard, hairy vetch, and ryegrass can be considered as poor accumulators of DDE, since they accumulated at most two to five times the amount of pollutant present in the soil (White and Kottler 2002). In contrast, willow species represent medium accumulators of DDE and DDT (Mitton et al. 2012), while zucchini and pumpkin plants accumulate high concentrations of DDE in their roots, at levels 10–20 times that of soil (White 2001). The castor bean (Ricinus communis) was also shown to have good potential for removing DDT, as well as cadmium from contaminated soils (Huang et al. 2011). Other tested species include Orychophragmus violaceus, which could remove DDT and HCHs from aged contaminated soil (Sun et al. 2015). Based on our results, we conclude that DDT/DDE-contaminated soil was not significantly remediated via absorption of ryegrass, but rather by degradation under the enhancing action of the microorganisms in the soil.
Metabolites of DDT in the soil
For the samples receiving the combined bacterial/ryegrass treatment, at days 30 and 210, degradation products of DDT in the soil were determined. The results are shown in Fig. 3. DDT is reductively dechlorinated to DDD and dehydrochlorinated to DDE in the soil. DDE can then be further degraded. As a consequence, the overall concentration of DDD remained more or less unchanged, though the concentration of DDE reduced, indicative of its degradation. Nevertheless, its metabolites could not be determined. Although many substances were detected in this analysis, these did not represent metabolites of DDE and DDD as deduced from database searches. Some publications reported that DDE is dechlorinated to DDMU (Quensen Iii et al. 2001; Quensen Iii et al. 1998), and that DDD was converted to DDA, which was again converted to DBP and 4,4-dichlorobenzhydrol (DBH) (Xiao et al. 2011). However, DDMU was not detected in these experiments, so that we have to conclude that the metabolites of DDE were other undefined substances.
TIC of the degradation products of DDT in the soil with “DXZ9-ryegrass” treatment. a p,p′-DDE (23.66 min). b p,p′-DDD (26.09 min). c p,p′-DDT (27.18 min). d 3-Undecano (29.78 and 31.94 min). e 3,4-Dihydro-4,5,6-trimethyl-naphthalenone (8.15 min). f 2,2-Oxybis(1,3-dichloro)-propane (8.95 min). g 1,2-Benzenedicarboxylic acid (18.63 min)
The toxicity of the soil during bioremediation
The safety of soil bioremediation must be carefully determined by measuring the toxicity of the end product, because metabolites resulting from microbial degradation of pollutants may also be carcinogenic or mutagenic. Thus, the toxicity of the soil after bioremediation should be taken into account. Unfortunately, there are few standardized methods available to determine toxicity in an ecological-relevant manner. In the literature, four exposure methods have been described to determine the toxic effects of contaminated soil on earthworms. In the most commonly used method, pollutants are extracted by organic solvents and these are used to expose live animals (Shen et al. 2009). The second method uses such extracts to directly expose isolated coelomocytes of earthworms in vitro. The third method makes use of water extracts (a water/soil mixture of 2.5:1 is typically used) to expose live earthworms, while in the fourth method, the mud (water/soil mixture is made) is also used to expose earthworms; the extent of DNA damage is detected after 48 h of exposure. According to results obtained in our laboratory (Kong 2013), the difference between the control and treatment was most significant for experiments conducted with a soil/water mixture following 48-h exposure, which was considered the most sensitive method. Using this method, all earworms survived when the contaminated soil contained DDT concentrations ranging from 3.11 to 23.80 mg kg−1 (Shi et al. 2016). In the experiment presented here, the test concentration was much lower than this, so that the model was considered appropriate.
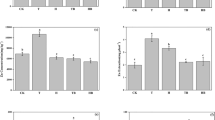
The toxicity of soil samples taken at days 5, 30, and 210 was thus determined. The results are shown in Fig. 4. The extent of DNA damage was evaluated by the value of OTMs, where a longer OTM indicates a higher toxicity. The genetic toxicity of the soil decreased by 58.1% (compared to control) after 210 days of treatment with ryegrass and bacteria (S + G + D + B). Likewise, a 52% reduction was observed with bacterial treatment only (S + D + B) and a 36.9% reduction with ryegrass only (S + G + D). These result showed that the toxicity of soil reduced significantly following microbial remediation and phytoremediation. Thus, the method is considered safe as it reduces toxicity of the soils and does not produce toxic metabolites.
Effect of the metabolites of DDT and DDE in the soil on the coelomocytes comet olive tails moment of earthworm (Eisenia foetida). Error bars represent standard deviations. Bars with the same letter at the same time are not significantly (p < 0.05) different (five replicates) according to the SPSS test
Conclusion
The present work demonstrated that Stenotrophomonas strain DXZ9 successfully colonized in the soil contaminated with DDT and DDE, though inoculation with this strain in the soil enhanced the removal efficiency of DDT and DDE, and planting ryegrass was an effective and prominent phytoremediation in the soil polluted with DDT and DDE; the bioremediation of soil contaminated with DDT and DDE by means of specific bacteria combined with ryegrass is the best and feasible. The degradation of microbes in the soil plays a decisive role in the removal of pollutants; the rhizoremediation has great potential for the treatment of DDT and DDE, while the absorption of ryegrass was contributing less. The toxicity of the soil is rapidly reduced by combined treatment of DXZ9 and ryegrass. However, these observations were based on the results applied in the single soil. Further works are also needed to confirm the degradation of DDT and DDE using soils historically contaminated with DDE and DDT.
References
Abhilash PC, Srivastava S, Singh N (2011) Comparative bioremediation potential of four rhizospheric microbial species against lindane. Chemosphere 82(1):56–63. https://doi.org/10.1016/j.chemosphere.2010.10.009
Ahmad F, Iqbal S, Anwar S, Afzal M, Islam E, Mustafa T, Khan QM (2012) Enhanced remediation of chlorpyrifos from soil using ryegrass (Lollium multiflorum) and chlorpyrifos-degrading bacterium Bacillus pumilus C2A1. J Hazard Mater 237-238:110–115. https://doi.org/10.1016/j.jhazmat.2012.08.006
Anderson TA, Guthrie EA, Walton BT (1993) Bioremediation in the rhizosphere. Environ Sci Technol 27(13):2630–2636. https://doi.org/10.1021/es00049a001
Arshad M, Hussain S, Saleem M (2008) Optimization of environmental parameters for biodegradation of alpha and beta endosulfan in soil slurry by Pseudomonas aeruginosa. J Appl Microbiol 104(2):364–370. https://doi.org/10.1111/j.1365-2672.2007.03561.x
Bhalerao TS (2012) Bioremediation of endosulfan-contaminated soil by using bioaugmentation treatment of fungal inoculant Aspergillus niger. Turk J Biol 36:561–567
Böltner D, Godoy P, Muñoz-Rojas J, Duque E, Moreno-Morillas S, Sánchez L, Ramos JL (2008) Rhizoremediation of lindane by root-colonizing Sphingomonas. Microb Biotechnol 1(1):87–93. https://doi.org/10.1111/j.1751-7915.2007.00004.x
Cao X, Yang C, Liu R, Li Q, Zhang W, Liu J, Song C, Qiao C, Mulchandani A (2013) Simultaneous degradation of organophosphate and organochlorine pesticides by Sphingobium japonicum UT26 with surface-displayed organophosphorus hydrolase. Biodegradation 24(2):295–303. https://doi.org/10.1007/s10532-012-9587-0
Chattopadhyay S, Chattopadhyay D (2015) Remediation of DDT and its metabolites in contaminated sediment. Curr Pollut Rep 1(4):248–264. https://doi.org/10.1007/s40726-015-0023-z
Comeau Y, Greer CW, Samson R (1993) Role of inoculum preparation and density on the bioremediation of 2,4-D contaminated soil by bioagumentation. Appl Microbiol Technol 38:681–6872
Cycoń M, Piotrowska-Seget Z (2016) Pyrethroid-degrading microorganisms and their potential for the bioremediation of contaminated soils: a review. Front Microbiol 7:1463
Cycoń M, Mrozik A, Piotrowska-Seget Z (2017) Bioaugmentation as a strategy for the remediation of pesticide-polluted soil: a review. Chemosphere 172:52–71. https://doi.org/10.1016/j.chemosphere.2016.12.129
Ding N, Xu JM, Schwab P (2013) Accumulation and transformation of PCBs in ryegrass (Lolium multiflorum L.). Functions of Natural Organic Matter in Changing Environment 637–640
Duquenne P, Parekh NR, Gatroux G, Fournier JC (1996) Effect of inoculant density, formulation, dispersion and soil nutrient amendment on the removal of carbofuran residues from contaminated soils. Soil Biol Biochem 28:1805–1811
Fan B, Zhao YC, Mo GH, Ma WJ, Wu JQ (2013) Co-remediation of DDT-contaminated soil using white rot fungi and laccase extract from white rot fungi. J Soil Sediment 13(7):1232–1245. https://doi.org/10.1007/s11368-013-0705-3
Fang H, Dong B, Yan H, Tang FF, Yu YL (2010) Characterization of a bacterial strain capable of degrading DDT congeners and its use in bioremediation of contaminated soil. J Hazard Mater 184(1-3):281–289. https://doi.org/10.1016/j.jhazmat.2010.08.034
Fu DQ, Teng Y, Shen YY, Sun MM, Tu C, Luo YM, Li ZG, Christie P (2011) Dissipation of polycyclic aromatic hydrocarbons and microbial activity in a field soil planted with perennial ryegrass. Front Env Sci Eng 6(3):330–335
Fuentes MS, Sáez JM, Benimeli CS, Amoroso MJ (2011) Lindane biodegradation by defined consortia of indigenous Streptomyces strains. Water Air Soil Pollut 222(1-4):217–231. https://doi.org/10.1007/s11270-011-0818-5
Gandolfi I, Sicolo M, Franzetti A, Fontanarosa E, Santagostino A, Bestetti G (2010) Influence of compost amendment on microbial community and ecotoxicity of hydrocarbon-contaminated soils. Bioresour Technol 101(2):568–575. https://doi.org/10.1016/j.biortech.2009.08.095
Gang Ji WD, Xia ZL (1985) Pesticide of soil and water. Science Press, Beijing, p 156
Gao C, Jin X, Ren J, Fang H, Yu Y (2015) Bioaugmentation of DDT-contaminated soil by dissemination of the catabolic plasmid pDOD. J Environ Sci 27:42–50. https://doi.org/10.1016/j.jes.2014.05.045
Hong Q, Zhang Z, Hong Y, Li S (2007) A microcosm study on bioremediation of fenitrothion-contaminated soil using Burkholderia sp. FDS-1. Int Biodeterior Biodegr 59(1):55–61. https://doi.org/10.1016/j.ibiod.2006.07.013
Huang XD, El-Alawi Y, Penrose DM, Glick BR, Greenberg BM (2004) Responses of three grass species to creosote during phytoremediation. Environ Pollut 130(3):453–463. https://doi.org/10.1016/j.envpol.2003.12.018
Huang H, Yu N, Wang L, Gupta D, He Z, Wang K, Zhu ZQ, Yan XC, Li TQ, Yang XE (2011) The phytoremediation potential of bioenergy crop Ricinus communis for DDTs and cadmium co-contaminated soil. Bioresour Technol 102(23):11034–11038. https://doi.org/10.1016/j.biortech.2011.09.067
Karpouzas DG, Walker A (2000) Factors influencing the ability of Pseudomonas putida epI to degrade ethoprophos in soil. Soil Biol Biochem 32(11-12):1753–1762. https://doi.org/10.1016/S0038-0717(00)00093-6
Kataoka R, Takagi K, Sakakibara F (2011) Biodegradation of endosulfan by Mortieralla sp. strain W8 in soil: influence of different substrates on biodegradation. Chemosphere 85(3):548–552. https://doi.org/10.1016/j.chemosphere.2011.08.021
Khan MI, Cheema SA, Tang XJ, Shen CF, Sahi ST, Jabbar A, Park J, Chen YX (2012) Biotoxicity assessment of pyrene in soil using a battery of biological assays. Arch Environ Contam Toxicol 63(4):503–512. https://doi.org/10.1007/s00244-012-9793-0
Kong LF (2013)Isolation of endosulfan degrading bacteria and its detoxification of endosulfan contaminated soil, Shandong Agricultural University, M.S. Thesis 51–56 (In Chinese)
Kong LF, Zhu SY, Zhu LS, Wei K, Yan TX, Wang J, Wang JH, Wang FH, Sun FX (2014) Colonization of Alcaligenes faecalis strain JBW4 in natural soils and its detoxification of endosulfan. Appl Microbiol Biotechnol 98(3):1407–1416. https://doi.org/10.1007/s00253-013-5033-4
Li WM, Wang DS, Hu F, Li HX, Ma LL, Xu L (2016) Exogenous IAA treatment enhances phytoremediation of soil contaminated with phenanthrene by promoting soil enzyme activity and increasing microbial biomass. Environ Sci Pollut Res 11(23):10656–10664
Lu M, Zhang ZZ (2014) Phytoremediation of soil co-contaminated with heavy metals and deca-BDE by co-planting of Sedum alfredii with tall fescue associated with Bacillus cereus JP12. Plant Soil 382(1-2):89–102. https://doi.org/10.1007/s11104-014-2147-0
Ma J, Pan LB, Yang XY, Liu XL, Tao SY, Zhao L, Qin XP, Sun ZJ, Hou H, Zhou YZ (2016) DDT, DDD, and DDE in soil of Xiangfen County, China: residues, sources, spatial distribution, and health risks. Chemosphere 163:578–583. https://doi.org/10.1016/j.chemosphere.2016.08.050
Meng L, Qiao M, Arp HPH (2011) Phytoremediation efficiency of a PAH-contaminated industrial soil using ryegrass, white clover, and celery as mono- and mixed cultures. J Soils Sediments 11(3):482–490. https://doi.org/10.1007/s11368-010-0319-y
Mitton FM, Gonzalez M, Pena A, Miglioranza Karina SB (2012) Effects of amendments on soil availability and phytoremediation potential of aged p,p′-DDT, p,p′-DDE and p,p′-DDD residues by willow plants (Salix sp.) J Hazard Mater 203-204:62–68. https://doi.org/10.1016/j.jhazmat.2011.11.080
Miyazaki R, Sato Y, Ito M, Ohtsubo Y, Nagata Y, Tsuda M (2006) Complete nucleotide sequence of an exogenously isolated plasmid pLB1, involved in gamma-hexachlorocyclohexane degradation. Appl Environ Microbiol 72(11):6923–6933. https://doi.org/10.1128/AEM.01531-06
Moubashera HA, Hegazya AK, Mohamedc NH, Moustafac YM, Kabiela HF, Hamada AA (2015) Phytoremediation of soils polluted with crude petroleum oil using Bassia scoparia and its associated rhizosphere microorganisms. Int Biodeterior Biodegrad 98:113–120. https://doi.org/10.1016/j.ibiod.2014.11.019
Odukkathil G, Vasudevan N (2016) Residues of endosulfan in surface and subsurface agricultural soil and its bioremediation. J Environ Manag 65:72–80
Purnomo AS, Mori T, Takagi K, Kondo R (2011) Bioremediation of DDT contaminated soil using brown-rot fungi. Int Biodeterior Biodegrad 65(5):691–695. https://doi.org/10.1016/j.ibiod.2011.04.004
Qiu XH, Zhu T, Li PHS, Li QL, Miao GF, Gong JC (2004) Organochlorine pesticides in the air around Taihu Lake, china. Environ Sci Technol 38(5):1368–1374. https://doi.org/10.1021/es035052d
Qiu XH, Zhu T, Yao B (2005) Contribution of Dicofol to the current DDT pollution in China. Environ Sci Technol 39(12):4385–4390. https://doi.org/10.1021/es050342a
Quensen Iii JF, Mueller SA, Jain MK, Tiedje JM (1998) Reductive dechlorination of DDE to DDMU in marine sediment microcosms. Science 280(5364):722–724. https://doi.org/10.1126/science.280.5364.722
Quensen Iii JF, Tiedje JM, Jain MK, Mueller SA (2001) Factors controlling the proportion of DDE dechlorination to DDMU in Palos Verdes margin sediments under anaerobic conditions. Environ Sci Technol 35(2):286–291. https://doi.org/10.1021/es0012873
Queyrel W, Habets F, Blanchoud H, Ripoche D, Launay M (2016) Pesticide fate modeling in soils with the crop model STICS: feasibility for assessment of agricultural practices. Sci Total Environ 542(Pt A):787–802. https://doi.org/10.1016/j.scitotenv.2015.10.066
Ramadan MA, EL-Tayeb OM, Alexander M (1990) Inoculum size as a factor limiting success of inoculation for biodegradation. Appl Environ Microbiol 56:1392–1396
Raina V, Suar M, Singh A, Prakash O, Dadhwal M, Gupta SK, Dogra C, Lawlor K, Lal S, van der Meer JR, Hollinger C, Lal R (2008) Enhanced biodegradation of hexachlorocyclohexane (HCH) in contaminated soils via inoculation with Sphingobium indicum B90A. Biodegradation 19(1):27–40. https://doi.org/10.1007/s10532-007-9112-z
Rostamia S, Azhdarpoorb A, Rostamic M, Samaei MR (2016) The effects of simultaneous application of plant growth regulators and bioaugmentation on improvement of phytoremediation of pyrene contaminated soils. Chemosphere 161:219–223. https://doi.org/10.1016/j.chemosphere.2016.07.026
Sáez JM, Benimelli CS, Amorosso MJ (2012) Lindane removal by pure and mixed cultures of immobilized actinobacteria. Chemosphere 89(8):982–987. https://doi.org/10.1016/j.chemosphere.2012.06.057
Sayles GD, You G, Wang M, Kupferle MJ (1997) DDT, DDD, and DDE dechlorination by zero-valent iron. Environ Sci Technol 31(12):3448–3454. https://doi.org/10.1021/es9701669
Shen CF, Chen YX, Huang SB (2009) Dioxin-like compounds in agricultural soils near e-waste recycling sites from Taizhou area, China: chemical and bioanalytical characterization. Environ Int 35(1):50–55. https://doi.org/10.1016/j.envint.2008.07.005
Shi YJ, Zhang QB, Huang DQ, Zheng XQ, Shi YJ (2016) Survival, growth, detoxifying and antioxidative responses of earthworms (Eisenia fetida) exposed to soils with industrial DDT contamination. Pestic Biochem Physiol 128:22–29. https://doi.org/10.1016/j.pestbp.2015.10.009
Singh BK, Walker A, Wright DJ (2006) Bioremedial potential of fenamiphos and chlorpyrifos degrading isolates: influence of different environmental conditions. Soil Biol Biochem 38(9):2682–2693. https://doi.org/10.1016/j.soilbio.2006.04.019
Song Y, Zhu LS, Wang J, Wang JH, Liu W, Xie H (2009) DNA damage and effects on antioxidative enzymes in earthworm (Eisenia foetida) induced by atrazine. Soil Biol Biochem 41(5):905–909. https://doi.org/10.1016/j.soilbio.2008.09.009
Sudharshan S, Naidu R, Mallavarapu M, Bolan N (2012) DDT remediation in contaminated soils: a review of recent studies. Biodegradation 23(6):851–863. https://doi.org/10.1007/s10532-012-9575-4
Sun GD, Zhang X, Hu Q, Zhang HQ, Zhang DY, Li GH (2015) Biodegradation of dichlorodiphenyltrichloroethanes (DDTs) and hexachlorocyclohexanes (HCHs) with plant and nutrients and their effects on the microbial ecological kinetics. Microb Ecol 69(2):281–292. https://doi.org/10.1007/s00248-014-0489-z
Tang JC, Wang RG, Niu XW, Zhou QX (2010) Enhancement of soil petroleum remediation by using a combination of ryegrass (Lolium perenne) and different microorganisms. Soil Till Res 110(1):87–93. https://doi.org/10.1016/j.still.2010.06.010
Tang HZ, Li J, Hu HY (2012) A newly isolated strain of Stenotrophomonas sp. hydrolyzes acetamiprid, a synthetic insecticide. Process Biochem 47(12):1820–1825. https://doi.org/10.1016/j.procbio.2012.06.008
Tullo PD, Versini A, Bueno M, Hécho IL, Thiry Y, Biron P, Castrec-Rouelle M, Pannier F (2015) Stable isotope tracing: a powerful tool for selenium speciation and metabolic studies in non-hyperaccumulator plants (ryegrass Lolium perenne L.) Anal Bioanal Chem 407(30):9029–9042. https://doi.org/10.1007/s00216-015-9069-4
Wang CF, Wang XP, Gong P, Yao TD (2016) Residues, spatial distribution and risk assessment of DDTs and HCHs in agricultural soil and crops from the Tibetan Plateau. Chemosphere 149:358–365. https://doi.org/10.1016/j.chemosphere.2016.01.120
White JC (2001) Plant facilitated mobilization and translocation of weathered 2,2-bis (p-chlorophenyl)-1,1-dichloroethylene (p,p-DDE) from an agricultural soil. Environ Toxicol Chem 20(9):2047–2052
White JC, Kottler BD (2002) Citrate-mediated increase in the uptake of weathered 2, 2-bis (p-chlorophenyl),1,1-dichloroethylene residues by plants. Environ Toxicol Chem 21(3):550–556. https://doi.org/10.1002/etc.5620210312
Xiao PF, Mori T, Kamei I, Kondo R (2011) A novel metabolic pathway for biodegradation of DDT by the white rot fungi, Phlebia lindtneri and Phlebia brevispora. Biodegradation 22(5):859–867. https://doi.org/10.1007/s10532-010-9443-z
Xie H, Zhu LS, Xu QF, Wang J, Liu W, Jiang JH (2011) Isolation and degradation ability of the DDT-degrading bacterial strain KK. Environ Earth Sci 62(1):93–99. https://doi.org/10.1007/s12665-010-0500-z
Yu XZ, Wu SC, Wu FY, Wong MH (2011) Enhanced dissipation of PAHs from soil using mycorrhizal ryegrass and PAH-degrading bacteria. J Hazard Mater 186(2-3):1206–1217. https://doi.org/10.1016/j.jhazmat.2010.11.116
Zhang H, Yang C, Zhao Q, Qiao CL (2009) Development of an autofluorescent organophosphates-degrading Stenotrophomonas sp. with dehalogenase activity for the biodegradation of hexachlorocyclohexane (HCH). Bioresour Technol 100(13):3199–3204. https://doi.org/10.1016/j.biortech.2009.02.008
Zhang MY, Teng Y, Zhu Y (2014a) Isolation and characterization of chlorothalonil-degrading bacterial strain H4 and its potential for remediation of contaminated soil. Pedosphere 24(6):799–807. https://doi.org/10.1016/S1002-0160(14)60067-9
Zhang QM, Zhu LS, Wang J, Xie H, Wang JH, Wang FH, Sun FX (2014b) Effects of fomesafen on soil enzyme activity, microbial population, and bacterial community composition. Environ Monit Assess 186(5):2801–2812. https://doi.org/10.1007/s10661-013-3581-9
Zhu ZQ, Yang XE, Wang K, Huang HG, Zhang XC, Fang H, Li TQ, Alva AK, He ZL (2012) Bioremediation of Cd-DDT co-contaminated soil using the Cd-hyperaccumulator Sedum alfredii and DDT-degrading microbes. J Hazard Mater 235-236:144–151. https://doi.org/10.1016/j.jhazmat.2012.07.033
Funding
This study was supported by grants from the National Natural Science Foundation of China (No. 41671321) and National Key Research and Development Project of China (2016YFD0800304).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interests
The authors declare that they have no conflict of interest.
Additional information
Responsible editor: Diane Purchase
Rights and permissions
About this article
Cite this article
Xie, H., Zhu, L. & Wang, J. Combined treatment of contaminated soil with a bacterial Stenotrophomonas strain DXZ9 and ryegrass (Lolium perenne) enhances DDT and DDE remediation. Environ Sci Pollut Res 25, 31895–31905 (2018). https://doi.org/10.1007/s11356-018-1236-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-018-1236-7