Abstract
The cytotoxicity of nanoparticles (NPs) and their properties are important issues in nanotechnology research. Particularly, NPs affect the metabolism of microorganisms due to NP interactions with some biomolecules. In order to assess the mechanisms underlying NPs toxicity, we studied the damage caused by copper oxide nanoparticles (CuO-NPs) on Staphylococcus aureus ATCC 24213 and Pseudomonas aeruginosa ATCC 27833. Spherical CuO-NPs characterized by their diameter (13 ± 3 nm) were synthesized with a maximum of 254 nm. These NPs reduced cell viability, with a minimum inhibitory concentration (MIC) of 500 and 700 ppm for Staphylococcus aureus and Pseudomonas aeruginosa, respectively. Surfactant was added to reduce the NP agglomeration, but it did not present any effect. The mechanism of CuO-NPs as antimicrobial agent was assessed by analyzing solubilized Cu2+, quantifying DNA release in the culture media, and measuring intracellular reactive oxygen species (ROS). CuO-NPs induced severe damage on cells as revealed by confocal optical microscopy and scanning electron microscopy (SEM). Our results indicated that CuO-NPs interacted with bacteria, triggering an intracellular signaling network which produced oxidative stress, leading to ROS generation. Finally, we concluded that CuO-NPs exhibited higher antibacterial activity on Gram-negative bacteria than on Gram-positive ones.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Nanomaterials have gained great interest recently because of their novel properties, including a large specific surface area and high reaction activity (Yan et al. 2012; Joh et al. 2011). Due to rapid development of nanotechnology, nanomaterials with various shapes and diameters have been prepared and used in some industrial products and commodities (Laurent et al. 2008). Nevertheless, several studies have suggested that the release of NPs might have negative impacts on many organisms and the environment (Blinova et al. 2010; Handy et al. 2008; Dasari et al. 2013). Metal oxide nanoparticles (NPs) constitute a very attractive family of nanomaterials (Applerot et al. 2012a, b; Baek and An 2011; Karlsson et al. 2013). For example, inorganic metal oxides such as ZnO, MgO, and TiO2 have been extensively used for antimicrobial applications, and they have been replacing the frequently used silver and gold ones, because of their known toxic effect on the environment and human health (Applerot et al. 2012a, b). They have promising application for the future, being manufactured on a large scale for both industrial and household uses (Chang et al. 2012; Rispoli et al. 2010; Moritz and Geszke 2013). CuO-NPs have received great attention owing to their important properties and widespread applications (Askarinezhad and Morsali 2008). They constitute the simplest member of the family of copper compounds which exhibits a wide range of potentially useful physical properties, such as high-temperature superconductivity, electron correlation effects, and spin dynamics (Chen et al. 2008; Zhang et al. 2014). The most important application area of CuO-NPs is electronic technology, specifically in the development of new semiconductors, electronic chips, and heat transfer nanofluids (Li et al. 2007; Guo et al. 2009; Sau et al. 2010; Ebrahimnia-Bajestan et al. 2011). They are an important p-type semiconductor with a narrow band gap (1.4 eV). The key advantage of using materials like copper is that they present high antibacterial activity, chemical stability, long-lasting action period, and thermal resistance compared to organic antibacterial agents (Christy et al. 2013). However, the mechanism underlying the antibacterial activity of these copper oxide NPs has not been unequivocally discerned.
Recently, we reported the effect of CuO-NPs on isolated bacterial strains from agricultural soil suggesting that their antimicrobial mechanism could be attributed to the ionic interactions and oxide-reduction reactions (Concha-Guerrero et al. 2014). NPs are able to affect eukaryotic and prokaryotic biomolecules due to their large specific surface area with high reactive activity and electronic density (Pisanic et al. 2009; Zhang et al. 2014). These interactions produce chemical reactions increasing superoxide radical (O2−) formation and reactive oxygen species (ROS) accumulation, which leads to oxidative stress (De Berardis et al. 2010; Kashmiri and Mankar 2014). ROS are responsible for the major toxicological NPs mechanisms. Large amounts of ROS could be generated even when only small amounts of CuO-NPs are incorporated into the cells (Toduka et al. 2012). In eukaryotic cells, NPs can produce ROS directly, once they are exposed to the acidic environment of lysosomes. Alternatively, they could interact with oxidative organelles such as mitochondria (Bondarenko et al. 2013; Zhang and Gutterman 2007). ROS production affects the biological detoxification system resulting in cell damage (Yang et al. 2009; Xia et al. 2008). The radical ·OH is generally considered as one of the most toxic ROS species because it is able to oxidize almost all the cellular components (Choi and Hu 2009). The extracellular ·OH generated by metal oxide NPs might induce oxidative damage on cell membranes, which can cause toxic effects in the organisms (Kashmiri and Mankar 2014). Excessive oxidative stress may also modify proteins, lipids, and nucleic acids, which further stimulates the anti-oxidant defense system, even leading to cell death. Because of this, it is important to determine the effects of CuO-NPs on prokaryotic microorganisms, specifically on pathogens, for a full characterization of their toxicity mechanisms and antimicrobial capacities. Because toxicity is a direct effect related to size, the agglomeration of NPs plays a crucial role on their toxicity (Wang et al. 2013; Hou et al. 2017). Agglomeration is a consequence of high surface energy that increases when the NP size diminishes (Chávez-Calderón et al. 2016). In order to evaluate the disaggregation, dispersants such as PVP and PEG have been used. Dispersants make a “capping” on the NPs and reduce the agglomerated size increasing the toxic effect (Giannousi et al. 2016; Javed et al. 2017; Kurzhals et al. 2017). The use of Pluronic F-127 has been reported as a drug carrier in iron oxide nanoparticles (Rodrigues et al. 2016; Jain et al. 2005). Pluronic F-127 present high biocompatibility and maintains the magnetic characteristic of magnetic nanoparticles (Kang et al. 2016). Dipex A40, another dispersant that received attention, is a non-toxic polyacrylate dispersant (Garcia-Saucedo et al. 2011; Otero-Gonzalez et al. 2013).
In the present study, CuO-NPs were synthesized and characterized. The influence of dispersants on the agglomeration and hydrodynamic size of CuO-NPs was determined. The toxicity of CuO-NPs agglomeration was examined on two bacterial strains models, Staphylococcus aureus and Pseudomonas aeruginosa, Gram-positive and Gram-negative bacteria, respectively. Growth inhibition, cell wall damage and ROS production were evaluated in order to elucidate the mechanism underlying Cu-ONPs toxicity.
Material and methods
CuO-NPs synthesis and characterization
The synthesis of CuO-NPs was performed according to Lanje et al. (2010) method using copper II acetate [Cu(CH3COO)2] (≥98%, Sigma-Aldrich) as a precursor and sodium hydroxide NaOH (≥98% pellets, Sigma-Aldrich) as a reducing agent. Briefly, 1.08 g of Cu(CH3COO)2 was added into a round-bottomed flask with 300 mL of distilled water, 1 mL of glacial acetic acid (CH3COOH, JT Baker, ACS ≥ 99.7%), and placed on an electric grill and heated up to 95 °C. Then, 0.4 g of NaOH was poured into the flask to maintain the pH 6–7. The color of the solution turned from blue to black immediately because of the reaction between Cu (CH3COO)2 and NaOH. The reaction continued for 15 min at 95 °C. After the reaction was completed, the resulting products were centrifuged at 15,838 RFC for 30 min, washed with water and ethanol (98%, Sigma-Aldrich) for several times. For size measurement, shape, and composition, NPs were sonicated in 1 mL of ethanol for 15 min, placed onto an aluminum support and analyzed by a Scanning Electron Microscope model JEOLJSM 7401F operated at 5.0 kV. All images were taken at the same magnification (×10,000), and 300 individual CuO-NPs in four images with the ImageJ software were analyzed. An average was calculated in order to determine particle size analysis efficiently. A drop of aqueous CuO-NPs suspension was placed onto a carbon-coated copper grid, air dried, and observed by a Transmission Electron Microscopy (TEM) equipment model JEM-2100F, JEOL to characterize CuO-NPs shape. Crystallinity, structure, and crystallite size of CuO-NPs were determined by XRD technique using an X-ray diffractometer model XDR, DP Phillips X’ pert PRO with Cu-Kα radiations (λ = 0.15406 nm) in 2θ range from 20° to 80°. UV-Vis spectrophotometry performed by using a PerkinElmer UV-visible spectrophotometer was used to characterize optical absorption properties of obtained CuO samples. NPs were added into water and sonicated for 20 min to form a homogeneous suspension with the concentration of about 0.1 g/l.
Hydrodynamic size of CuO-NPs and Fourier transform infrared spectroscopy
CuO-NPs were analyzed by using dynamic light scattering (DLS) to determine the hydrodynamic diameter of particles. A Malvern ZetaSizer Nano ZSInstrument operating at a light-source wavelength of 532 nm and a fixed scattering angle of 173° was used. The system ran at a flow rate of 0.5 mL/min, and the dynamic light scattering was analyzed every 3 s, the analyses were done in triplicate. CuO-NPs suspensions were prepared by adding dry particles into M9 medium and into M9 medium with both surfactants: Dispex® and Pluronic F-127, and sonicated (100 W, 40 kHz) for 30 min.
CuO-NPs were characterized by Fourier transform infrared (FT-IR) spectroscopy. Samples of each dispersant were analyzed alone, and the dispersants were prepared with CuO-NPs in the same way as prepared for the toxicity assay. Samples were air-dried and the analyses were carried out on a IRAffinity-1S spectrophotometer (Shimadzu). Each spectrum was obtained in average of 45 interferograms with a resolution of 4/cm.
Bacterial strains and culture conditions
Well-characterized cells of Pseudomonas aeruginosa (ATCC 27833) and Staphylococcus aureus (ATCC 24213) were maintained on solid soy agar. Prior to incubation with NPs, the bacteria were cultured overnight on soy broth (15 g/L peptone from casein, 5 g/L peptone from soy, 3 g/L yeast extract, 5 g/L dextrose, 5 g/L NaCl, 15 g/L bacteriological agar) into a 500-mL round-bottomed flask. In addition, the bacteria were incubated at 37 °C for 12–18 h in a shaker incubator at 200 RPM until the optical density (OD) of the culture reached 3.8 for Staphylococcus aureus and 0.124 for Pseudomonas aeruginosa at 620 nm, which indicates 109 colony-forming units (CFU)/mL. This concentration was determined using a standard curve obtained by plating bacterial suspensions at known different OD onto solid soy agar by broth dilution method. Plates were incubated at 37 °C, and colony-forming units (CFU/mL) were counted after 24 h. All assays were performed in triplicate (Fig. S1).
Determination of minimal inhibitory/lethal doses/and bactericidal concentrations
Minimum inhibitory concentration (MIC) defined as the lowest concentration that inhibits the growth of an organism and bactericidal concentration (BC) of CuO-NPs was determined by the broth dilution method. The overnight cultures were used to inoculate on 96-well plates with a final cell concentration of 1 × 108CFU/mL in minimal medium M9 (86 mM NaCl, 470 mM Na2HPO4, 220 mM KH2PO4, 180 mM NH4Cl, 2 mM MgCl2, 0.4 mM CaCl2, 0.4 mM dextrose, 2% vitamins, 2% trace elements) and CuO-NPs (0–3000 ppm). The CuO-NPs suspension was prepared in a known concentration in water. This suspension was kept in sonication until use. Separately, surfactants were prepared in stock of 10,000 ppm. A mixture of surfactants was made with Pluronic® F-127 (100:0.1, NPs: Pluronic®) and Dispex® A40 (10:1, NPs: Dispex®). The mixtures were sonicated for 30 min after their use in toxicity assays (Wang et al. 2011; Garcia-Saucedo et al. 2011; Simon et al. 2013). Additionally, both surfactants were added to overnight cultures but without NPs in order to assess their effect on the studied pathogen microorganisms. Pluronic® F-127 displays surfactant properties including the ability to interact with hydrophobic surfaces and biological membranes (Batrakova and Kabonov 2008). Dispex® A40 which is an ammonium salt solution of an acrylic polymer in water reduces agglomeration of particles depending on their nature. The plates were incubated at 37 °C for 24 h, and the growth was evaluated by the resazurin method according to Nateche et al. (2009).
For the determination of BC and LD, 10 μL from the culture exposed to the different CuO-NP concentrations was taken from each well and added to 90 μL of sterile water into Eppendorf tubes. The bacteria were plated onto solid soy agar plates by the serial dilution method drip. Plates were incubated at 37 °C and counted after 24 h. The control assays, containing surfactants and bacterial solution without CuO-NPs in round-bottomed flasks, were incubated under the same conditions. From the results obtained for growth inhibition, lethal doses were determined (where 50% of the cells died). For the growth inhibition assay, the concentration inhibiting completely bacterial growth was defined as the bactericidal concentration (BC). All the experiments were done in triplicate, and results are presented as the mean ± standard deviation. Viability test was analyzed by linear regression using Excel.
Chemical analysis of dissolved Cu
In order to determine the dissolved Cu during the test, strains were cultured in soy broth as mentioned above and incubated at 200 RPM for 12 h at 37 °C. Previously, cells exposed and unexposed (control) to CuO-NPs were prepared in 500-mL flasks in triplicate and incubated for 24 h. Each sample was centrifuged at 15,783 RFC for 10 min after 24 h of incubation with CuO-NPs. Clear supernatant was carefully collected and filtered through a 0.22-μm sterilized filter (Millex-GP syringe filter, diameter 33 mm). The ion concentrations were measured by atomic absorption spectrometry (GBC AVANTA Version 2.02 model flame atomic absorption spectroscopy).
Determination of cell wall damage
Fluorescent dyes LIVE/DEAD® BacLight (INVITROGEN) were selected to assess membrane integrity. SYTO® 9 (green fluorescent nucleic acid stain) dye generally dyes all bacteria in a population—those with intact and damaged membranes—presenting an excitation wavelength of 480/500 nm. Propidium iodide (PI) penetrates only cells with damaged membrane. This dye binds to DNA and presents red color; its excitation wavelength is 490/635 nm. The dyes were added according to the manufacturer’s procedures. After incubation with CuO-NP and unexposed cells for 24 h, 1 mL of treated and untreated cell suspension was stained and incubated at 4 °C for 15 min in the dark. A laser scanning confocal microscopy (CLSM, Inverted Zeiss LSM 700 confocal microscopy) with a semiconductor laser was used to examine membrane integrity.
Determination of reactive oxygen species
To identify the production of ROS inside the bacterial cell, 2′,7′-dichlorofluorescin-diacetate (DCFH-DA) was used as a visual indicator of the overall oxidative status of the cell. DCFH-DA can cross the cell membrane and hydrolyzes, by intracellular esterase, to non-fluorescent DCFH. In the presence of ROS, DCFH is oxidized to highly fluorescent dichlorofluorescein (DCF). Therefore, the ROS concentration in the cell is directly proportional to the fluorescent intensity of DCF. Cells exposed to CuO-NPs were incubated for 24 h; once this time elapsed, they were incubated for 30 min in the dark with 0.1 μL DCFH-DA 10 μM. Cells unexposed were used as controls. Finally, the fluorescence was measured at an excitation wavelength of 485 nm and emission wavelength of 535 nm. The stained cells were analyzed in CLSM (Inverted Zeiss LSM 700 confocal microscopy). The ROS generation was obtained by comparing controls and exposed cells. Fluorescence intensity of each cell was measured taking into account the same conditions for each micrograph. Zen black edition software was employed to make the analysis (Release version 7.0© Carl Zeiss MicroImaging GmbH 1997–2011).
Determination of nucleic acids in the culture medium
The concentration of DNA in the biological samples was determined by the diphenylamine colorimetric assay using salmon DNA as standard. A sample of 2 mL diluted with 5% TCA was obtained, 4 mL of diphenylamine reagent was added, and everything was mixed by vortexing. The mix was incubated in boiling water bath for 10 min. The diphenylamine reagent was prepared using 4.5 g of diphenylamine in 300 mL of cold acetic acid and 4.5 mL of concentrated sulfuric acid. In fact, immediately before using the diphenylamine reagent in the biological sample mixture, 1.5 mL of cold acetaldehyde with a concentration of 1.6% was added to the diphenylamine. Then, the sample mixture was rapidly cooled in a cold water bath. Next, in order to perform a quick optical characterization, the absorbance at 600 nm was measured using a UV-Vis spectrometer (PerkinElmer).
Biological sample preparation for scanning electron microscopy
The biological samples were prepared following the method described by Maldonado et al. (2011). A sample of the cultures was fixed in 3% glutaraldehyde for 4 h and washed two times in Milloning buffer phosphate (NaH2PO4 2.26%, NaOH 2.52%, dextrose 5.4%). Then, they were filtered by a polycarbonate membrane (Millex-GP syringe filter, pore diameter 0.22 μm), fixed, and dehydrated in successively increasing gradient concentration of ethanol (30, 50, 70, 90, and 100%). In the next step, they were air dried according to Thomasson and Thomasson (2011). All samples were placed on metal stubs and coated with gold using a Denton Vaccum Desk II. Finally, the samples exposed to CuO-NPs and controls were analyzed by scanning electron microscopy (SEM) using a Jeol JSM 7401F field emission scanning electron microscope.
Results and discussion
Characterization of copper oxide nanoparticles
The CuO-NPs synthesized following Lanje et al. (2010) procedure were spherical (Fig. 1a) forming large agglomerates (Fig. 1b). The size distribution corresponds to an average obtained by analyzing 300 individual CuO-NPs in four SEM images. It has been proved that toxicity has direct relationship with size and shape. For example, ZnO nanoflowers did not show toxicity in the aspheric form (Chávez-Calderón et al. 2016). The primary size of CuO-NPs indicates a nanoscale range. This feature is one of the most important NP characteristics due to its relation to reactivity. As the NPs size decreases, the superficial area increases and more superficial atoms are available to interact and react with biological molecules in a living organism (Wang et al. 2011; Applerot et al. 2012a, b; Chávez-Calderón et al. 2016; Hou et al. 2017). Most NPs were between 10 and 15 nm (average size 13 ± 3 nm. Fig. 1c) in size, approximately twice the size reported by Lanje et al. (2010), who obtained rectangular CuO-NPs with a size range from 5 to 6 nm. In contrast, Zhang et al. (2014) obtained spherical CuO-NPs with a size around 10 nm. The difference in form and size of NPs is probably due to reaction conditions. Indeed, Chen et al. (2008) demonstrated that the morphology of NPs could be controlled by manipulating the synthesis conditions, which allows adapting the properties of NPs during the synthesis procedure. Similarly, Rehman et al. (2011) reported the CuO-NPs synthesis in a size range of 6.67–28.7 nm. It has been reported that agglomeration is a common phenomenon in NP synthesis (Bogoslovskaja et al. 2014). This phenomenon could be explained by the NP charge and size (Chávez-Calderón et al. 2016).
SEM-EDX analysis showed oxygen and copper enrichment (46.62% Cu and 12.85% O. Fig. 1d). These results were consistent with those reported by Kida et al. (2007) and Concha-Guerrero et al. (2014). An absorption band peak between 264 and 308 nm was observed in the UV-VIS analysis (Fig. 1e) indicating that the CuO-NPs were produced at nanometric scale with high purity. This is important as for toxicity because it has been demonstrated that the smaller the nanoparticle is, the more toxic it is. Some NPs have photoactivity as TiO (Joost et al. 2015) or ZnO (Ma et al. 2014) that is activated by UV light, although CuO-NPs do not show this particularity.
These results confirm the primary size of nanoparticles observed by SEM and TEM analysis (Fig. 1a, b), indicating the great reactivity in relation to size (Applerot et al. 2012a, b; Concha-Guerrero et al. 2014). According to Gupta and Ramrakhiani (2009) and Bijanzadeh et al. (2012), the light absorption in the UV-VIS region means that the particle size is in the quantum regime (Vatankhah et al. 2015; Suttiponparnit et al. 2010).
XRD analysis showed two major peaks located at 2θ = 36 and 39 (Fig. 1f), indexed as planes 1, 1, 1, respectively, confirming the monoclinic structure of the CuO (PDF 080–1268). Diffraction peaks become slightly broader due to the small crystallite CuO-NPs size.
Finally, we considered that CuO-NPs reactivity synthesized in the present study could be directly influenced by size, shape, and surface area. It has been shown that the nanomaterial effectiveness is inversely proportional to its size; in this way, a smaller size of the nanomaterial is more effective due to its free electrons located in the surface area (Baek and An 2011; Bondarenko et al. 2012). This information is important in order to fully understand the biological response.
Cell viability
Bacterial growth inhibition was observed in minimal M9 medium when bacteria were exposed to CuO-NPs in the presence or absence of both surfactants (Fig. 2). Pseudomonas aeruginosa growth was inhibited at 30 ppm in medium M9 without surfactants and at 240 ppm when both surfactants were present (Fig. 2a). In contrast, Staphylococcus aureus growth inhibition was observed at 30 ppm in medium M9 in the presence of surfactants and 120 ppm without surfactants (Fig. 2b). It was found that CuO-NPs concentration was directly proportional to inhibition (R 2 = 0.81 to 0.96). The growth of both microorganisms was not inhibited by dispersants (Fig. 2a, b; control without NPs). It was reported that Candida albicans did not present damages in the presence of Dispex® at 100 mg/L (Garcia-Saucedo et al. 2011). Due to Pluronic® biocompatibility, it has drawn attention in drug delivery research (Kang et al. 2016; Rodrigues et al. 2016), nanoparticles synthesis, and solubilization (Simon et al. 2013) among others. The use of dispersants makes a capping around the nanoparticles, and this phenomenon decreases the high surface energy, reducing the attraction between electrostatic forces, increasing steric repulsion (Chávez-Calderón et al. 2016; Rodrigues et al. 2016). Minimum inhibitory concentrations (MIC) and lethal dose 50 (LD50) were determined in order to evaluate CuO-NPs toxicity (Table 1). Pseudomonas aeruginosa had less sensitivity for CuO-NPs than Staphylococcus aureus. The surfactants used did not increase the CuO-NPs toxicity (Table 1), and no differences were observed for the DL 50. These results contrasted with previous reports showing that surfactants promote nanoparticle toxicity (Sager et al. 2009; Javed et al. 2017). The observed inhibition differences between microorganisms could be explained by the cell wall composition as proposed by Chávez-Calderón et al. (2016) who demonstrated that NPs interacted with teichoic acids in the cell wall of Gram-positive bacteria. Our results suggested that Pseudomonas aeruginosa resistance to CuO-NPs could be due to the presence of neutral molecules in the cell wall such as lipopolysaccharides, whose uncharged moiety, sugars, limits the interactions with NPs as previously demonstrated (Ivanov et al. 2011).
Determination of CuO-NPs size distribution in culture medium and Fourier transform infrared (FT-IR) spectroscopy
In order to further explain the effect of surfactants on the CuO-NPs aggregation, the NP size distribution was determined. CuO-NP hydrodynamic size was the smallest (65.71 ± 12.72 nm) with M9 medium (Fig. 3a), whereas the hydrodynamic size was 207.95 ± 40 and 211 ± 47 nm in M9 supplemented by Pluronic® and Dispex®, respectively. These results showed that the primary size was five times smaller than hydrodynamic size and that the addition of surfactants did not diminish the NP agglomeration and size. These observations were in agreement with growth inhibition. Moreover, previous works showed that toxicity increased with the use of dispersants, probably due to the diminution of agglomeration (Sager et al. 2009; Applerot et al. 2012a, b; Otero-Gonzalez et al. 2013; Javed et al. 2017). It has been proposed that the use of dispersant decreases the attraction between electrostatic forces, increasing steric repulsion (Rodrigues et al. 2016).
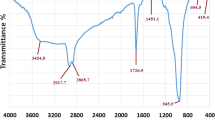
FT-IR showed no interaction of dispersants with CuO-NPs (Fig. 3b). These results are in contrast with those obtained by Garcia-Saucedo et al. (2011) reporting that smaller agglomeration was obtained with Dispex® rather than nanoparticles alone. Other works showed that the use of Plurinic F-127 was efficient for the dispersion of iron oxide nanoparticles with the addition of oleic acid (Jain et al. 2005; Rodrigues et al. 2016) or without addition of a fatty acid (Simon et al. 2013). It has been described that the smaller size of CuO-NPs might favor the interactions with the cell wall (Karlsson et al. 2008, 2013; Cronholm et al. 2013; Ren et al. 2009; Chang et al. 2012). In the same way, many reports have demonstrated that agglomerated NPs showed a less toxic effect (Ji et al. 2010; Song et al. 2010; Ma et al. 2012; Sonia et al. 2016). The size of nanoparticles is the most relevant characteristic in toxicity. The smaller the nanoparticle, the most superficial area. The high superficial area allows the interaction of NPs with cell wall molecules. Due to the size of nanoparticles, the atoms are available in the nanoparticle surfaces (Ivask et al. 2014; Jesuvathy Sornalatha et al. 2015; Anaya et al. 2016). When agglomerations occur, the superficial area decreases and the NP interaction with molecules is lower. An indicative of this agglomeration was the hydrodynamic size, and it could be influenced by the culture medium, dispersants, or organic material (Wang et al. 2011; Applerot et al. 2012a, b; Chávez-Calderón et al. 2016; Hou et al. 2017). In our study, the hydrodynamic size with dispersants showed almost the same for Pluronic® and Dispex® (207.95 ± 40 and 211 ± 47 nm, respectively). For this reason, the MIC and DL50 showed almost the same value. These results were congruent with the results obtained in growth inhibition. Because the smallest hydrodynamic size and growth inhibition were obtained in culture medium M9, all the experiments were performed in this medium.
Chemical analysis of dissolved Cu2+ in bacterial growth media
The available free Cu2+ ions were found to be higher in the Pseudomonas aeruginosa (18.68 ppm) culture than in the Staphylococcus aureus (9.83 ppm) culture (Fig. 4). However, the Cu2+ ion solubilization for both strains was less than 10%, suggesting that the observed toxicity is probably due to CuO-NPs rather than Cu2+ ions. Such observation is in agreement with a previous report indicating that Cu2+ released from CuO-NPs may explain only in part the NPs toxicity (Midander et al. 2009). In contrast, several reports indicated that a possible toxicity mechanism involves Cu2+ solubilization (Bondarenko et al. 2012; Chang et al. 2012) triggering a series of subsequent adverse effects that include ROS generation and DNA damage (Bondarenko et al. 2012). According to these observations, we assume that CuO-NP toxicity on the cell membranes of both microorganisms was mediated by NPs themselves rather than by Cu2+ ions.
Bacterial membrane damage
In order to determine the membrane damage on both bacteria, CuO-NPs were used in M9 medium at sub-lethal dose (333 ± 71 ppm). Both bacteria lost their cellular integrity as indicated by the PI incorporation (Fig. 5), and severe cell wall damage was observed by SEM (Fig. 6). CuO-NPs tended to adhere to the cell surface and to disrupt the cell membrane integrity (Fig.6b, d). Elongated cell, cells that lost their cocci or rod shape, pits and cavities, and even lysed cells were observed. The membrane damage level was further evaluated by DNA quantification in culture media (Fig. 7). Pseudomonas aeruginosa and Staphylococcus aureus showed, respectively, 1.6 and 1.3 times higher DNA release in the presence of CuO-NPs than control cultures in medium without NPs. It is likely that shape deformation contributed with osmolarity loss and DNA leakage in culture medium, which is in agreement with cell viability tests. Many studies have proposed that NP exposure affects the membrane integrity (Baek and An 2011; Azam et al. 2012; Aysaa and Salmanb 2016). In our study, the damage observed on Pseudomonas aeruginosa and Staphylococcus aureus cells could be attributed to physical interaction of CuO-NPs with cellular membranes as proposed by Raffi et al. (2010) who reported that opposite electrical charges between CuO-NPs and cells pro voke oxide-reduction reactions in the bacterial cell wall. This interaction could mediate the cell wall instability and generate holes and the loss of cell integrity. Thus, lost integrity was reflected in PI incorporation (Fig. 5) and DNA release (Fig. 7). This damage was attributed to CuO-NPs, since the ion release was too weak to generate damage. The CuO-NPs damage was attributed to Cu2+ leaching in culture medium (Wang et al. 2011; Bondarenko et al. 2012; Hou et al. 2017). However, in our study, the main damage mechanism was attributed to CuO-NPs interaction with the cell wall. Hence, the toxic differences observed between the two bacterial models could be attributed to the cell wall structure as previously described (Brayner 2008). P. aeruginosa as a Gram-negative bacterium has a thinner peptidoglycan layer than S. aureus as a Gram-positive bacterium, explaining the different CuO-NPs toxicity.
Cell wall damage of Pseudomonas aeruginosa and Staphylococcus aureus determined by confocal microscopy micrographs, ×63. a Control and b Pseudomonas aeruginosa cells exposed to CuO-NPs c control and d Staphylococcus aureus cells exposed to CuO-NPs. The cell samples were dyed with Syto 9 and PI before LSCM observation. The scale of each image is 5 mm
Intracellular ROS concentration
Both microorganisms showed a 1.5 times ROS increase (Fig. 8), compared to the control without exposition to CuO-NPs. Oxidative stress has been established as a biomarker for estimating toxicity of many substances (Junqua et al. 2000; Margaritelis et al. 2016; Karlsson et al. 2013). ROS generation has been proposed as the main mechanism for NP toxicity; however, the results were controversial. Cui et al. (2012) and Concha-Guerrero et al. (2014) reported a decrease in ROS levels when the microorganisms were exposed to CuO-NPs. Several reports related ROS generation to nanoparticles exposition (Dasari et al. 2013; Anaya et al. 2016; Quinteros et al. 2016) for both eukaryotic (Fahmy and Cormier 2009) and prokaryotic cells (Raghupathi et al. 2011). It is suggested that the ROS increase mechanism is mediated by the inhibition of antioxidant defense (Fahmy and Cormier 2009; Mahapatra et al. 2008), lipid peroxidation (Dutta et al. 2012), protein peroxidation (Kashmiri and Mankar 2014), and DNA damage (Chang et al. 2012; Laha et al. 2014). Dimkpa et al. (2011) reported an increase in ROS levels when Pseudomonas chlororaphis O6 was exposed to 500 mg/L of CuO-NPs. In the same study, they found that ROS levels did not increase when the cells were exposed to concentrations below 200 mg/L. In our study, although CuO-NPs were used at sub-lethal doses (333 ppm), the increase of ROS levels was observed for both microorganisms suggesting that ROS production was induced by exposition to CuO-NPs. Several authors propose the leaching of Cu+2 as main toxic CuO-NPs mechanism (Bondarenko et al. 2012; Hou et al. 2017). These ions could generate oxidative stress and DNA damage, lipids, and protein peroxidation. Another proposed mechanism is the ROS generation on the NPs surface (Applerot et al. 2012a, b). The strong interaction observed between the CuO-NPs and the ROS generation in our study suggested that ROS is the main toxic mechanism for both microorganisms. Besides, the production of ROS could exceed the ability of the cell to neutralize the effects of the superoxide radicals. These suppress the ability of enzymes such as SOD, CAT, and GTPx to detoxify ROS. Enzyme suppression could cause an accumulation of pro-oxidants in the cell, which leads to a state of oxidative stress (Kashmiri and Mankar 2014; Elsaesser and Howard 2012).
ROS determination by Confocal microscopy, ×63X. a, d Pseudomonas aeruginosa ATCC 27833 and Staphylococcus aureus ATCC 24213 control stained with DCF respectively. b, e Pseudomonas aeruginosa ATCC 27833 and Staphylococcus aureus ATCC 24213 cells exposed to CuO-NPs stained with DCF. c, f Graphic plot of ROS generation in controls and cells exposed to CuO-NPs. The scale of each image is 5 mm
Conclusion
In this work, we found that the use of a dispersant does not necessarily separate the agglomerated nanoparticles. This phenomenon has been directly related to the toxic effect, as the smaller agglomerated nanoparticles were, the most toxic effect was generated. The less agglomerated CuO-NPs showed the highest growth inhibition. The results showed that the toxicity mechanism was not related to Cu+2 liberation. Incorporation of PI and DNA liberation in the culture medium indicated strong cell wall damage. SEM microscopy showed the CuO-NPs adhesion in the cell wall and morphological change. The influence of the cell wall composition of the microorganisms was evident, the Gram-positive microorganism Staphylococcus aureus being the more sensitive. This result could imply a more potent resistance to stress oxidative cascade in Pseudomonas aeruginosa than in Staphylococcus aureus.
Although this work showed strong evidence that agglomeration was the major factor directly related to toxicity, it is necessary to take into account the difference in the cell wall composition and the metabolic capacities of the microorganisms used to evaluate the toxic effect of NPs. Indeed, the damage probably generates a different response in genes and protein expressions.
References
Anaya MN, Faghihzadeh F, Ganji N, Bothun G, Craver VO (2016) Comparative study between chemostat and batch reactors to quantify membrane permeability changes on bacteria exposed to silver nanoparticles. Sci Total Environ 565:841–848
Applerot G, Lellouche J, Lipovsky A, Nitzan Y, Lubart R, Gedanken A, Banin E (2012a) Understanding the antibacterial mechanism of CuO nanoparticles: revealing the route of induced oxidative stress. Biomed Mater. doi:10.1002/smll.201200772
Applerot G, Lellouche J, Lipovsky A, Nitzan Y, Lubart R, Gedanken A, Banin E (2012b) Understanding the antibacterial mechanism of CuO nanoparticles: revealing the route of induced oxidative stress. Small 8(21):3326–3337. doi:10.1002/smll.201200772
Askarinezhad A, Morsali A (2008) Synthesis and characterization of CdCO3 and CdO nanoparticles by using a sonochemical method. Mater Lett 62:478–482
Aysaa NH, Salmanb HD (2016) Antibacterial activity of modified zinc oxide nanoparticles against Pseudomonas Aeruginosa isolates of burn infections. World Sci News 33:1–14
Azam A, Ahmed AS, Oves M, Khan M, Memic A (2012) Size-dependent antimicrobial properties of CuO nanoparticles against Gram-positive and -negative bacterial strains. Int J Nanomedicine 7:3527–3535
Baek YW, An YJ (2011) Microbial toxicity of metal oxide nanoparticles (CuO, NiO, ZnO, and Sb2O3) to Escherichia coli, Bacillus subtilis, and Staphylococcus aureus. Sci Total Environ 409:1603–1608
Batrakova EV, Kabonov AV (2008) Pluronic block copolymers: evolution of drug delivery concept from inert Nanocarriers to biological response modifiers. Control Release 130(2):98–106
Bijanzadeh AR, Vakili MR, Khordad R (2012) Astudy of the surface plasmon absorption band for nanoparticles. Int J Phys Sci 7(12):1943–1948
Blinova I, Ivask A, Heinlaan M, Mortimer M, Kahru A (2010) Ecotoxicity of nanoparticles of CuO and ZnO in natural water. Environ Pollut 158:41–47
Bogoslovskaja OA, Rakhmetova AA, Ovsyannikova MN, Olkhovskaya IP, Gluschenko NN (2014) Antimicrobial effect of copper nanoparticles with differing dispersion and phase composition. Nanotechnol Russia 9(1):82–86
Bondarenko O, Ivask A, Käkinen A, Kahru A (2012) Sub-toxic effects of CuO nanoparticles on bacteria: kinetics, role of cu ions and possible mechanisms of action. Environ Pollut 169:81–89
Bondarenko O, Juganson K, Ivask A, Kasemets K, Mortimer M, Kahru A (2013) Toxicity of ag, CuO and ZnO nanoparticles to selected environmentally relevant test organisms and mammalian cells in vitro: a critical review. Arch Toxicol 87:1181–1200
Brayner R (2008) The toxicological impact of nanoparticles. Nano Today 3:48–55
Chang Y, Zhang M, Xia L, Zhang J, Xin G (2012) The toxic effects and mechanisms of CuO and ZnO nanoparticles. Materials 5:2850–2871
Chávez-Calderón A, Paraguay-Delgado F, Orrantia-Borunda E, Luna-Velazco A (2016) Size effect of SnO2 nanoparticles on bacteria toxicity and their membrane damage. Chemosphere 165:33–40
Chen PP, Mwakwari SC, Oyelere AK (2008) Gold nanoparticles: from nanomedicine to nanosensing. Review. Nanotechnol Sci Appl 1:45–65
Choi O, Hu Z (2009) Role of reactive oxygen species in determining nitrification inhibition bymetallic/oxide nanoparticles. J Environ Eng 135(12):1365–1370
Christy AJ, Nehru LC, Umadevi M (2013) A novel combustion method to prepare CuOnanorods and its antimicrobial and photocatalytic activities. Powder Technol 235:783–786
Concha-Guerrero SI, Souza-Brito EM, Piñón-Castillo HA, Tarango-Rivero SH, Caretta CA, Luna-Velazco A, Duran R, Orrantia-Borunda E (2014) Effect of CuO nanoparticles over isolated bacterial strains from agricultural soil. J Nanomater 2014:1–13
Cronholm P, Karlsson HL, Hedberg J, Lowe T, Winnberg L, Elihn K, Odnevall WI, Möller L (2013) Intracellular uptake and toxicity of ag- and CuOnanoparticles– a comparison between nanoparticles and their corresponding metal ion. Small 9:970–982
Cui Y, Zhao Y, Tian Y, Zhang W, Lu X, Jiang X (2012) The molecular mechanism of action of bactericidal gold nanoparticles on Escherichia coli. Biomaterials 33(7):2327–2333
Dasari TP, Pathakoti P, Hwang H (2013) Determination of the mechanism of photoinduced toxicity of selected metal oxide nanoparticles (ZnO, CuO, Co3O4 and TiO2) to E. coli bacteria. J Environ Sci 25(5):882–888
De Berardis B, Civitelli G, Condello M, Lista P, Pozzi R, Arancia G, Meschini S (2010) Exposure to ZnO nanoparticles induces oxidative stress and cytotoxicity in human colon carcinoma cells. Toxicol Appl Pharmacol 246:116–127
Dimkpa CO, Calder A, Britt DW, McLean JE, Anderson AJ (2011) Responses of a soil bacterium, Pseudomonas chlororaphisO6 to commercial metal oxide nanoparticles compared with responses tometal ions. Environ Pollut 159(7):1749–1756
Dutta RK, Nenavathu BP, Gangishetty MK, Reddy AVR (2012) Studies on antibacterial activity of ZnO nanoparticles by ROS induced lipid peroxidation. Colloid Surf 94:143–150
Ebrahimnia-Bajestan E, Niazmand H, Duangthongsuk W, Wongwises S (2011) Numerical investigation of effective parameters in convective heat transfer of nanofluids flowing under a laminar flow regime. Int J Heat Mass Transf 54(19–20):4376–4388
Elsaesser A, Howard C (2012) Toxicology of nanoparticles. Adv Drug Deliv Rev 64:129–137
Fahmy B, Cormier SA (2009) Copper oxide nanoparticles induce oxidative stress and cytotoxicity in airway epithelial cells. Toxicol In Vitro 23:1365–1371
Garcia-Saucedo C, Field JA, Otero-Gonzalez L, Sierra-Alvarez R (2011) Low toxicity of HfO2, SiO2, Al2O3 and CeO2 nanoparticles to the yeast, Saccharomyces cerevisiae. J Hazard Mater 192(3):1572–1579. doi:10.1016/j.jhazmat.2011.06.081
Giannousi K, Hatzivassiliou E, Mourdikoudis S, Vourlias G, Pantazaki A, Dendrinou-Samara C (2016) Synthesis and biological evaluation of PEGylated CuO nanoparticles. J Inorg Biochem 164:82–90
Guo Z, Ng HW, Yee GL, Hahn HT (2009) Differential scanning calorimetry investigation on vinyl ester resin curing process for polymer nanocomposite fabrication. J Nanosci Nanotechnol 9(5):3278–3285
Gupta P, Ramrakhiani M (2009) Influence of the particle size on the optical properties of CdSe nanoparticles. Open Nanosci J 3:15–19
Handy RD, Kammer FVD, Lead JR, Hassellov M, Owen R, Crane M (2008) Theecotoxicity and chemistry of manufactured nanoparticles. Ecotoxicology 17:287–314
Hou J, Wang X, Hayat T, Wang X (2017) Ecotoxicological effects and mechanism of CuO nanoparticles to individual organisms. Environ Pollut 221:209–217. doi:10.1016/j.envpol.2016.11.066
Ivanov IE, Kintz EN, Porter LA, Goldberg JB, Burnham NA, Camesano TA (2011) Relating the physical properties of Pseudomonas Aeruginosa lipopolysaccharides to virulence by atomic force microscopy. J Bacteriol 193(5):1259–1266. doi:10.1128/JB.01308-10
Ivask A, Kurvet I, Kasemets K, Blinova I, Aruoja V, Suppi S, Vija H, Kakinen K, Titma T, Heinlaan M, Visnapuu M, Koller D, Kisand V, Kahru A (2014) Size-dependent toxicity of silver nanoparticles to bacteria, yeast, algae, crustaceans and mammalian cells in vitro. PLoS One 9(7):e102108. doi:10.1371/journal.pone.0102108
Jain TK, Morales MA, Sahoo SK, Leslie-Pelecky DL, Labhasetwar V (2005) Iron oxide nanoparticles for sustained delivery of anticancer agents. Mol Pharm 2(3):194–205. doi:10.1021/mp0500014
Javed R, Ahmed M, Haq I u, Nisa S, Zia M (2017) PVP and PEG doped CuO nanoparticles are more biologically active: antibacterial, antioxidant, antidiabetic and cytotoxic perspective. Mater Sci Eng C 79:108–115. doi:10.1016/j.msec.2017.05.006
Jesuvathy Sornalatha D, Bhuvaneswari S, Murugesan S, Murugakoothan P (2015) Solochemical synthesis and characterization of ZnO nanostructures with different morphologies and their antibacterial activity. Optik 126(1):63–67
Ji Z, Jin X, George S, Xia T, Meng H, Wang X, Suarez E, Zhang H, Hoek EM, Godwin H, Nel A, Zink JI (2010) Dispersion and stability optimization of TiO2 nanoparticles in cell culture media. Environ Sci Technol 44(19):7309–7314
Joh DY, Kinder J, Herman LH, Ju SY, Segal MA, Johnson JN, Chan KL, Park J (2011) Single-walled carbon nanotubes as excitonic optical wires. Nat Nanotechol 6:51–56
Joost U, Juganson K, Visnapuu M, Mortimer M, Kahru A, Nommiste E, Joost U, Kisand V, Ivask A (2015) Photocatalytic antibacterial activity of nano-TiO2 (anatase)-based thin films: effects on Escherichia coli cells and fatty acids. J Photochem Photobiol B 142:178–185
Junqua M, Biolley JP, Pie S, Kanoun M, Duran R, Goulas P (2000) In vivo occurrence of carbonyl residues in Phaseolus vulgaris proteins as a direct consequence of a chronic ozone stress. Plant Physiol Biochem 38(11):853–861
Kang EB, Sharker SMd, In I, Park SY (2016) Pluronic mimicking fluorescent carbon nanoparticles conjugated with doxorubicin via acid-cleavable linkage for tumor-targeted drug delivery and bioimaging. J Ind Eng Chem 43:150-157
Karlsson HL, Cronholm P, Gustafsson J, Moller L (2008) Copper oxide nanoparticles are highly toxic: a comparison between metal oxide nanoparticles and carbon nanotubes. Chem Res Toxicol 21:1726–1732
Karlsson HL, Cronholm P, Hedberg Y, Tornberg M, Battice LD, Svedhem S, Wallinder IO (2013) Cell membrane damage and protein interaction induced by copper containing nanoparticles—importance of the metal release process. Toxicology 313:59–69
Kashmiri ZN, Mankar SA (2014) Free radicals and oxidative stress in bacteria. Int J Curr Microbiol App Sci 3(9):34–40
Kida T, Oka T, Nagano M (2007) Synthesis and application of stable copper oxide nanoparticle suspensions for Nanoparticulate film fabrication. J Am Ceram Soc 90(1):107–110
Kurzhals S, Gal N, Zirbs R, Reimhult E (2017) Aggregation of thermoresponsive core-shell nanoparticles: influence of particle concentration, dispersant molecular weight and grafting. J Colloid Interface Sci 500:321–332
Laha D, Pramanik A, Laskar A, Jana M, Pramanik P, Karmakar P (2014) Shape dependent bactericidal activity of copper oxide nanoparticle mediated by DNA and membrane damage. Mater Res Bull 59:185–191
Lanje AS, Sharma SJ, Pode RB, Ningthoujam RS (2010) Synthesis and optical characterization of copper oxide nanoparticles. Adv Appl Sci Res 1(2):36–40
Laurent S, Forge D, Port M, Roch A, Robic C, Van der Elst L, Muller RN (2008) Magnetic iron oxide nanoparticles: synthesis, stabilization, vectorization, physicochemical characterizations, and biological applications. Chem Rev 108:2064–2110
Li Y, Liang J, Tao Z, Chen J (2007) CuO particles and plates: synthesis and gas-sensor application. Mater Res Bull 43:2380–2385
Ma R, Levard C, Marinakos SM, Cheng Y, Liu J et al (2012) Size-controlled dissolution of organic-coated silver nanoparticles. Environ Sci Technol 46:752–759
Ma H, Wallis LK, Diamond S, Li S, Canas-Carrell J, Parra A (2014) Impact of solar UV radiation on toxicity of ZnO nanoparticles through photocatalytic reactive oxygen species (ROS) generation and photo-induced dissolution. Environ Pollut 193:165–172
Mahapatra O, Bhagat M, Gopalakrishnan C, Arunachalam KD (2008) Ultrafine dispersed CuO nanoparticles and their antibacterial activity. J Exp Nanosci 3(3):185–193
Maldonado J, Solé A, Puyen ZM, Esteve I (2011) Selection of bioindicators to detect lead pollution in Ebro delta microbial mats, using high-resolution microscopic techniques. Aquat Toxicol 104(1–2):135–144
Margaritelis NV, Cobley JN, Paschalis V, Veskoukis AS, Theodorou AA, Kyparos A, Nikolaidis MG (2016) Going retro: oxidative stress biomarkers in modern redox biology. Free Radic Biol Med 98:2–12
Midander K, Cronholm P, Karlsson HL, Elihn K, Möller L, Leygraf C, Wallinder IO (2009) Surface characteristics, copper release, and toxicity of nano- and micrometer-sized copper and copper(II) oxide particles: a cross-disciplinary study. Small 5:389–399
Moritz M, Geszke M (2013) Thenewest achievements in synthesis, immobilization and practical applications of antibacterial nanoparticles. Chem Eng J 228:596–613
Nateche F, Martin A, Baraka S, Palomino JC, Khaled S, Portaels F (2009) Application of the resazurinmicrotitre assay for detection of multidrug resistance in mycobacterium tuberculosis in Algiers. J Med Microbiol 55:857–860
Otero-Gonzalez L, Garcia-Saucedo C, Field JA, Sierra-Alvarez R (2013) Toxicity of TiO(2), ZrO(2), Fe(0), Fe(2)O(3), and Mn(2)O(3) nanoparticles to the yeast, Saccharomyces cerevisiae. Chemosphere 93(6):1201–1206. doi:10.1016/j.chemosphere.2013.06.075
Pisanic TR, Jin S, Shubayev VI (2009) Nanotoxicity: from in vivo and in vitro models to health risks. John Wiley & Sons, Ltd., London, pp 397–425
Quinteros MA, Aristizábal VC, Dalmasso PR, Paraje MG, Páez PL (2016) Oxidative stress generation of silver nanoparticles in three bacterial genera and its relationship with the antimicrobial activity. Toxicol in Vitro 36:216–223
Raffi M, Mehrwan S, Bhatti TM et al (2010) Investigations into the antibacterial behavior of copper nanoparticles against Escherichia coli. Ann Microbiol 60(1):75–80
Raghupathi KR, Koodali RT, Manna AC (2011) Size-dependent bacterial growth inhibition and mechanism of antibacterial activity of zinc oxide nanoparticles. Langmuir 27:4020–4028
Rehman S, Mumtaz A, Hasanain SK (2011) Size effects on the magnetic and optical properties of CuO nanoparticles. J Nanopart Res 13:2497–2507
Ren G, Hu D, Cheng EW, Vargas-Reus MA, Reip P, Allaker RP (2009) Characterization of copper oxide nanoparticles for antimicrobial applications. Int J Antimicrob Agents 33:587–590
Rispoli F, Angelov A, Badia D, Kumar A, Seal S, Shah V (2010) Understanding the toxicity of aggregated zero valent copper nanoparticles against Escherichia coli. J Hazard Mater 180:212–216
Rodrigues EC, Morales MA, de Medeiros SN, Suguihiro NM, Baggio-Saitovitch EM (2016) Pluronic® coated sterically stabilized magnetite nanoparticles for hyperthermia applications. J Magn Magn Mater 416:434–440. doi:10.1016/j.jmmm.2016.05.033
Sager TM, Porter DW, Robinson VA, Lindsley WG, Schwegler-Berry DE, Castranova V (2009) Improved method to disperse nanoparticles for in vitro and in vivo investigation of toxicity. Nanotoxicology 1(2):118–129. doi:10.1080/17435390701381596
Sau TK, Rogach AL, Jackel F, Klar TA, Feldmann J (2010) Properties and applications of colloidal nonspherical noble metal nanoparticles. Adv Mater 22(16):1805–1825
Simon T, Boca S, Biro D, Baldeck P, Astilean S, (2013) Gold–Pluronic core–shell nanoparticles: synthesis, characterization and biological evaluation. Journal of Nanoparticle Research 15.
Song W, Zhang J, Guo J, Zhang J, Ding F, Li L, Sun Z (2010) Role of the dissolved zinc ion and reactive oxygen species in cytotoxicity of ZnO nanoparticles. Toxicol Lett 199:389–397
Sonia S, Jayasudha R, Jayram ND, Kumar PS, Mangalaraj D, Prabagaran SR (2016) Synthesis of hierarchical CuO nanostructures: biocompatibleantibacterial agents for Gram-positive and Gram-negative bacteria. Curr Appl Phys 16:914–921
Suttiponparnit K, Jiang J, Sahu M, Suvachittanont S, Charinpanitkul T, Biswas P (2010) Role of surface area, primary particles size, and crystal phase on titanium dioxide nanoparticle dispersion properties. Res Lett 6:27
Thomasson SA, Thomasson JR (2011) A comparison of CPD (critical point drying) and HMDS (Hexamethyldisilazane) in the preparation of Corallorhiza spp. rhizomes and associated mycorrhizae for SEM (scanning electron microscopy). Trans Kans Acad Sci 114(2):129–134
Toduka Y, Toyooka T, Ibuki Y (2012) Flow cytometric evaluation of nanoparticles using side-scattered light and reactive oxygen species—mediated fluorescence—correlation with genotoxicity. Environ Sci Technol 46:7629–7636
Vatankhah C, Saki M, Jafargholineja S (2015) Theoretical and experimental investigation of quantum confinement effect on the blue shift in semiconductor quantum dots. Orient J Chem 31(2):907–912
Wang Z, Li J, Zhao J, Xing B (2011) Toxicity and internalization of CuO nanoparticles to prokaryotic alga Microcystisaeruginosa as affected by dissolved organic matter. Environ Sci Technol 45(14):6032–6040. doi:10.1021/es2010573
Wang, Z, von dem Bussche, A, Kabadi, PK, Kane, AB, Hurt, RH (2013) Biological and environmental transformations of copper-based nanomaterials. ACS Nano 7, 8715-8727
Xia T, Kovochich M, Liong M, Mädler L, Gilbert B, Shi H, Yeh JI, Zink JI, Nel AE (2008) Comparison of the mechanism of toxicity of zinc oxide and cerium oxide nanoparticles based on dissolution and oxidative stress properties. ACS Nano 2:2121–2134
Yan L, Zheng YB, Zhao F, Li S, Gao X, Xu B, Weiss PS, Zhao Y (2012) Chemistry and physics of a single atomic layer: strategies and challenges for functionalization of graphene and graphene-based materials. Chem Soc Rev 41:97–114
Yang H, Liu C, Yang DF, Zhang HS, Xi Z (2009) Comparative study of cytotoxicity, oxidative stress and genotoxicity induced by four typical nanomaterials: the role of particle size, shape and composition. J Appl Toxicol 29:69–78
Zhang DX, Gutterman DD (2007) Mitochondrial reactive oxygen species-mediated signaling in endothelial cells. Am J Physiol Heart Circ Physiol 292:H2023–H2031
Zhang Q, Zhang K, Xu D, Yang G, Huang H, Nie F, Liu C, Yang S (2014) CuO nanostructures: synthesis, characterization, growth mechanisms, fundamental properties, and applications. Prog Mater Sci 60:208–337
Acknowledgments
This work was supported by CONACyT-ConsejoNacional de Ciencia y Tecnología No. 175089 and ECOS NORD 263456. All authors contributed, read, and approved the final manuscript. The authors have declared no conflict of interest. The authors thank W. Antunez-Flores and Ochoa-Gamboa R.A. for technical support in electronic microscopy.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Responsible editor: Philippe Garrigues
Electronic supplementary material
Fig. S1
(DOCX 12 kb)
Rights and permissions
About this article
Cite this article
Ulloa-Ogaz, A.L., Piñón-Castillo, H.A., Muñoz-Castellanos, L.N. et al. Oxidative damage to Pseudomonas aeruginosa ATCC 27833 and Staphylococcus aureus ATCC 24213 induced by CuO-NPs. Environ Sci Pollut Res 24, 22048–22060 (2017). https://doi.org/10.1007/s11356-017-9718-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-017-9718-6