Abstract
The bacterial community structure and diversity were assessed at the scale of rotating biodisk procedure (RB) in a semi-industrial pilot plant. As well, the Salmonella community was particularly monitored, and the effects of ultraviolet (UV-C254) on the bacterial community were studied. The identification of dominant bacteria revealed the presence of beneficial and useful species that could play an important role in the process of wastewater purification. Several species as Enterobacter agglomerans, Cronobacter sakazakii, and Pantoea agglomerans known for their bioremediation activities were revealed in the majority of biofilm samples. Common detection of Salmonella community provides evidence that the RB system did not seriously affect Salmonella. Furthermore, the investigation on the (UV)-C254 inactivation of the whole bacterial community, in secondary treated wastewater, showed variable UV resistance results. No Salmonella detection was registered at a dose of around 1440 mW s cm−2 since a total disappearance of Salmonella was recorded.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The rotating biological contactor (RBC) or rotating biodisk system (RB) is a biological treatment process that involves the biological purification of the wastewater. While the engineering aspects of the RB were well understood, little was known about the composition of the accountable microbial community acting in the process. The identification and the diversity of the bacterial community could be assessed through the usage of either culture-dependent or culture-independent methods. However, culture-dependent methods are frequently time consuming and pricey. Moreover, species occurring in low numbers are often out-competed in vitro by numerically more abundant microbial species (Hengenholtz et al. 1998), and some may be unable to grow in vitro (Head et al. 1998). Hence, culture-dependent methods are biased by the selection of species that obviously do not represent and indicate the actual status of the microbial dominance in the field (Ding et al. 2011). Community-level subject areas are relying more and more on cultivation-independent methods based on the direct analysis of DNA or RNA without any culturing stage (Jany and Barbier 2008). Matched with a global analysis, these methods make it possible and likely to investigate complex microbial communities.
Denaturing gradient gel electrophoresis (DGGE) technique based on the 16S rDNA gene was commonly used for a direct comparison of structural diversity among different microbial communities (Muyzer et al. 1993). Knowledge of the dynamics and of the structural diversity of microbial communities in wastewater treatment plant and an understanding of the contributions of major members of these communities to overall degradation and purification of various pollutants are likely to provide unprecedented control over the biological treatment of sewage (Ding et al. 2011).
Moreover, with increased demands on water resources in the recent years, problems of surface water contamination by enteric microorganisms have become an increasing concern. Sewage contains pathogenic microorganisms like Salmonella that could be transmitted by the fecal-oral route. Members of the Salmonella enterica species are public health difficulties and challenges that are interrelated with significant morbidity and mortality in those infected with the pathogens (Foley et al. 2008; Turki et al. 2014). Consequently, the sanitary quality of the effluent must be sufficiently guaranteed before discharge of treated wastewater in the various natural environments. In this setting, the detection of Salmonella taken as a model of pathogenic organism in the effluent is of great importance and could give an accurate idea about the bacteriological wastewater quality. Halatsi et al. (2006) proposed a set of primers that could target the Salmonella quorum sensing sdiA gene.
When released into natural receiving environments, untreated or insufficiently treated wastewaters could cause several various problems, such as microbial contamination, eutrophication, oxygen uptake, and toxicity (Ding et al. 2011). To improve the microbiological water quality, UV-C254 irradiation is typically recognized as inducing a good germicidal effect on microorganisms. Indeed, the UV-C254 absorbed by the DNA of the microbial cells induce damage in this DNA through altering the nucleotide base pairing, thereby creating new linkages between adjacent nucleotides on the same DNA strand (Ben said et al. 2011).
In the present study, we aimed to assess at first, the bacterial community structure and diversity in wastewater and biofilm at the scale of rotating biodisk process by using DGGE and 16S rRNA methods; secondly to determine the contribution of predominant bacteria on the wastewater purification; thirdly, to assess the dynamic of Salmonella community taken as referring pathogens in the RB system, and finally to study the influence of UV-C254 taken as a tertiary treatment of disinfection on the main bacterial pathogenic communities.
Material and methods
Experimental RB treatment system and sample collection
The biodisk reactor consists of plastic filling disks on a horizontal axis. The drum, half-immersed in water, rotates at a constant speed of two rev/min around this axis. A biofilm develops on the plastic support, and the rotation of the disks ensures both oxygenation and contact with the wastewater. The biomass excess is detached gradually, and it will be separated in the secondary settling tank. The performance study of the process was conducted by Ben Rajeb et al. (2011) and confirmed that the reactor is performed as an advanced treatment system for DBO, COD, SS, NH4+-N, and NO3−-N. Disinfection performances reduce microbial load; however, chlore, ozone, or UV disinfection should be considered.
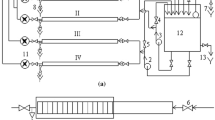
Wastewater and biofilm samples were collected from a semi-industrial wastewater treatment, pilot plant situated at Mutuelle-ville, zone of El Menzah 1, an important residential and business area of the Tunis city, Tunisia, which receives essentially municipal and hospital wastewater of three neighboring hospitals (Fig. 1). At seven points of the different treatment lines, biofilm and wastewater samples were collected during summer (June–July: temperature range between 30 and 37 °C) and winter (October, November: temperature range between 20 and 25 °C). The physical and geometric characteristics of the rotating biodisk system studied here were described in Table 1.
Experimental location site and schematic plan of the semi-industrial pilot plant. EB entrance of the primary settlement tank, REP distribution tank input, D1 disk tank 1, D2 disk tank 2, SD outlet part of the secondary settling tank, BIO1 and BIO2 biofilm samples taken from the first and the second disk, respectively. Raw wastewater (EB), REP (distribution tank input), and in disk tank 1 (D1), disk tank 2 (D2)
UV-C photoreactor system
The laboratory UV-tool was created in collaboration with Guy Daric S. A. (Aubervilliers, France) (Saidi et al. 2011). Germicidal doses were calculated as the product of the intensity of irradiation and the time of exposure. In order to evaluate the bacterial community resistance to UV-C254 irradiation in wastewater, a wastewater treated water sample collected at the exit of the secondary settling tank was irradiated at the following several different exposure times: 60, 120, 180, 240 s, using the photoreactor described earlier by Hassen et al. (2000).
DNA extraction and PCR amplification of 16S rDNA
Total DNA was extracted from biofilm and wastewater samples as described previously by Turki et al. (2013). Extracted DNA was visualized by electrophoresis in 0.5× TBE (Tris-Borate-EDTA buffer) on a 0.8 % agarose gel. PCR amplification was achieved using universal primers: 907R and 357F attached to 40-bp GC-rich sequence. PCR was prepared as previously described (Sass et al. 2001).
DGGE and DNA sequence analysis
DGGE electrophoresis was achieved using the system INGENY phorU-2 (Leiden, Netherlands) as described previously by Turki et al. (2013). DGGE profiles were exploited to create matrices indicating the presence or absence of bands, and a program was built using the unweighted pair group method with arithmetic mean (UPGMA) using MVSP 3.13 (Multi Variate Statistical Package).
DGGE gel images were examined with Image J1.45 software, and the position and the intensity of bands were considered. Principal component analysis (PCA) was produced to perceive the relationships among patterns by using PAST 2.17c software Package (Paleontological Statistics Software Package for education and data analysis).
The DNA of DGGE bands selected from the gel was analyzed and amplified as described previously by Turki et al. (2013) and then sequenced. Obtain sequences were compared to the 16S rDNA sequences in the GenBank database by applying the basic local alignment search tool (BLAST, National Centre for Biotechnology Information, US National Library of Medicine).
Detection of Salmonella sdiA gene
The total DNA of each sample was studied by PCR to detect the presence of Salmonella sdiA gene in treated and untreated wastewater and biofilm samples, using sdiA1 and sdiA2 primers (Halatsi et al. 2006). The amplicon size was around 274 bp. PCR analyses were done as previously reported (Turki et al. 2012).
Results
DGGE fingerprinting of wastewater and biofilm samples
The DGGE pattern is viewed as an “image” of the total bacterial community. Analysis of wastewater and biofilm sample DGGE patterns is based on the number and intensity of the bands reflecting the bacterial diversity and density, respectively. During summer (Fig. 2A) and winter (Fig. 3A), DGGE patterns enclosing from 2 to 13 visible bands were noticed for biofilm and wastewater samples. The results showed that biofilm and wastewater obtained in summer were more complex than those acquired in the winter. This latter result means that bacterial communities revealed in the hot season (summer) were more abundant, more complex, and more variable than those obtained in winter, and consequently, the purification RD procedure was more efficient and successful during the hot season of summer. On the other side, important differences in microbial community structure were observed among the different treatment stages in the plant. This effect was indicated by the lessening of the band number and the complexity of the banding patterns according to the progress in the steps of treatment.
A DGGE patterns of V3-V5 region of the 16S rDNA generated from the wastewater and biofilm samples collected in summer from different steps in the RBC system. a–d Four replicates of samples collected in summer. EB raw input, Rep distribution tank input, D1 disk tank 1, BIO1 biofilm of disk 1, D2 disk tank 2, BIO2 biofilm of disk tank 2, and SD output secondary clarifier. Bands marked were excised and sequenced. M Salmonella amplification product. The gradient of urea and formamide ranged from 40 to 60 %. B Dendrogram [UPGMA] of PCR-DGGE analysis of the communities in wastewater and biofilm samples during the summer. C Score plot of principal component analysis of PCR-DGGE profile
A DGGE patterns of V3-V5 region of the 16S rDNA generated from the wastewater and biofilm samples collected in winter from different steps in the RBC system. a–d Four replicates of samples collected in winter. EB raw input, Rep distribution tank input, D1 disk tank 1, BIO1 biofilm of disk 1, D2 disk tank 2, BIO2 biofilm of disk tank 2, and SD output secondary clarifier. Bands marked were excised and sequenced. M Salmonella amplification product. The gradient of urea and formamide ranged from 40 to 60 %. B Dendrogram [UPGMA] of PCR-DGGE analysis of the communities in wastewater and biofilm samples during winter. C Score plot of principal component analysis of PCR-DGGE profile
DGGE patterns were exploited to construct UPGMA dendrograms according to Pearson correlation (Figs. 2B and 3B) that allowed the bacterial community structure analysis during the different stages of wastewater treatment. Granting to the diversity perceived among treated and non-treated wastewater and biofilm samples, 87 and 85 % of similarity were obtained during the summer from samples of raw wastewater (EB) and the repartition tank (REP), and of disk tank 1 (D1) and disk tank 2 (D2), respectively. For biofilm samples (Bio1 and Bio2) and treated wastewater (SD), the cluster analysis presented 90 and 65 % of similarity, respectively. However, treated wastewater (EB) revealed 65 % of similarity. In winter, treated wastewater (SD) showed only 45 % of similarity. Still, 65 and 85 % of similarity were obtained in samples of raw wastewater (EB) and repartition tank (REP), and of disk tanks (D1 and D2) and biofilm disk tanks (BIO1) and (BIO2), respectively. The principal factor analysis based on the relative intensities of bands in DGGE profiles provided more information about the microbial community structure (Figs.2C and 3C). During summertime, the plots of samples from the treated wastewater (SDc, SDb, SDd) were circulated on the right side and could be distinguished from the other samples. The plots of untreated wastewater samples (EDc, REPc, EBd, REPd) were distributed on the right and left side of the scatter, respectively, and plots of (EBa, REPa and EBb, REPb) took a middle position in the diagram, which mean that the bacterial communities of untreated wastewater were variable over time. Plots of biofilm samples (BIO1, BIO2) and wastewater samples (D1, D2) were relatively adjacent and took a middle position in the diagram. During winter, all plots appeared to be distributed randomly; however, we could see that biofilm sample plots (BIO1a, BIO2a and BIO1c, BIO2c) were distributed on the left side, which indicates that the bacterial communities in the biofilms were relatively stable. Moreover, the plots of treated wastewater, SDa and SDb, could obviously be differentiated from each other and from other samples. These results indicated that the community structure was variable throughout the four seasons and according to the different stages of treatment. Besides, the RB process was effective in bacterial reduction and abatement.
Sequence analysis of the dominant bacteria
To identify the dominant bacterial groups in the wastewater and biofilms, 18 DNA bands were removed from DGGE gel, reamplified, sequenced, and subjected to BLAST GeneBank analysis. The homology results and the closest relative origin of the sequences obtained were shown in Table 2.
Sequence analysis of DGGE bands has shown that the dominant groups existing in this biological system were phylogenetically associated to 4 major clusters of the bacteria domain: beta and gamma-Proteobacteria (the majority of sequences), the Bacteroidetes phylum (two sequences, B6 and B11), and the Cyanobacteria phylum (one sequence, B5).
The DNA sequence of DGGE bands B1, B2, B13, B15, and B17 showed 99 % of homology with Chromobacterium sp. and Chromobacteriumviolaceum. Bands B3, B7, and B9 presented 99 % of similarity on the 16S rRNA gene level with S. enterica. The recovered fragment sequences of bands B5 and B14 were closely related (99 %) to an uncultured Cyanobacterium and Aquitalea sp., respectively. The 16S rDNA sequence of band B8 showed 100 % of similarity with Cronobacter sakazakii. However, the two bands, B16 and B18, were exclusively detected in samples collected in winter, and they showed 99 % of similarity with Burkholderia sp. and 100 % of homology with Pantoea agglomerans, respectively. It is almost certainly that the corresponding organism of band B16 is Burkholderia xenovorans that could grow at low temperature of around 25 °C. All these bands were frequently found equally in the untreated and treated wastewater and the biofilm. These results highlighted the significance of the season on the bacterial community stability and on the operating performance of the RB process.
The 16S rDNA sequence of the band B4 was closely related to Enterobacter sp., and it was exclusively identified in the biofilms (BIO1, BIO2) and in the treated wastewater (SD). Sequence analysis of the DGGE B6 and B11 bands showed 100 % of homology with Flavobacterium sp. (HQ538672) and Fluviicola taffensis, respectively. These two last species were affiliated to Bacteroidetes phylum, and they were equally detected in treated and raw effluent. The band B12 matched 100 % of similarity to Methylobacillus sp. (EU194898) that was held at the same time in treated and non-treated wastewaters (EB, REP, D1, SD).
DGGE pattern analysis of the UV-irradiated wastewater
The analysis of DGGE patterns revealed a clear lessening in the number and in the band intensity according to the increase of exposure time to UV-C254, and consequently to the UV-C254 dose (Fig. 4). In summer, wastewater profile enclosed 8 bands including B5, B11, B13, B14, and B17. After 60 s of UV-C254 irradiation exposure (720 mW s cm−2 UV-C254 dose), the bands B5, B11, and B17 matching to Cyanobacterium, to Fluviicola taffensis, and to Chromobacterium sp., respectively, have a tendency to completely disappear from the profile. For the lasting band B13, high UV-C254 dose (≥2160 mW s cm−2) reduced noticeably the band intensity, and it may suggest the probable diminution and/or the total disappearance (total inactivation) of these populations of bacteria from wastewater. On the other side, high intensity of the band B14 was observed at UV-C254 dose around 1440 mW s cm−2 (Fig. 4A).
In winter, DGGE pattern found for the secondary treated wastewater enclosed 3 visible clear bands (B9, B16, and B18). The band B9 matching to Salmonella sp. disappeared after a UV-C254 exposure dose of around 1440 mW s cm−2. However, the band B16 (corresponding to Burkholderia sp.) and B18 (corresponding to P. agglomerans) persisted and continued existing in the banding profile even after the high UV-C254 exposure dose of around 2160 mW s cm−2 (Fig. 4B).
Monitoring of Salmonella community in wastewater and biofilms according to the season
The dynamic of the Salmonella community was performed using the amplified sequence of the quorum sensing sdiA gene. In winter, the sdiA-PCR analysis showed that Salmonella community detection was not steady. With all, in summer, Salmonella community was regularly found in all samples (REP, BIO2, and SD). The detection of Salmonella in the biofilm samples (BIO2) and at the exit of the secondary settling tank (SD) suggested that biofilm excess is stripped off the media as sloughs and transported by the wastewater flow to a secondary settling tank (Fig. 5). Consequently, the RB system did not severely affect the wastewater Salmonella community.
Graphs show the PCR products of the Salmonella quorum sensing gene sdiA. A (a–d) Four replicates of samples collected in summer; B (a–d) Four replicates of samples collected in winter; lane 1 Rep, distribution tank input, lane 2 BIO2, biofilm of disk tank 2, lane 3 SD, output secondary clarifier. M 100 bp marker
Discussion
In order to avoid the deterioration of the surface water quality, aerobic biological procedures are frequently used to treat hospital, municipal, and industrial wastewaters containing various soluble and particulate materials. Particularly, the hospital wastewater constituted a major discharge of pathogenic microorganisms and several chemicals and organic compounds, such as pharmaceutical partially metabolized, radioactive elements, chemicals for cancer treatment, hormones, and other medicine residues. Aerobic biological procedures support mixed consortia of microorganisms that can concurrently convert a broad range of compounds into new cells. In spite of the important role of these processes, there is a restricted and a limited understanding of the bacterial community structure and function that is largely due to the usage of culture-dependent methods (Lapara et al. 2000). In this context, cultivation-independent techniques such as DGGE allow considerably more exhaustive and precise analysis, and at the same a faster assessment of the main whole microbial communities (Lapara et al. 2000; Giannino et al. 2009). In this study, examination of DGGE profile based on the comparison of dominant bacteria in biofilm and wastewater showed an important diversity both between different treatment steps and along seasons. Changes in the microbial community structure were confirmed by a modification, both in number and in intensity of bands that will be well established by PCA analysis. Based on the results of PCA, we can confirm that the bacterial community structure in wastewater and in biofilm was remarkably different and diversified. Some PCA plots obtained in the case of biofilm analysis (BIO1, BIO2) looked close, either in summer and in winter. This result points out that these bacterial communities were relatively stable and unchangeable, suggesting the existence of an active and dynamic microbial consortium in the biofilm. This finding argues in favor of the treatment system stability despite that the scrubber microorganisms are always immersed in a random and changeable environment.
All these found results demonstrated and affirmed that different and diversified microbial groups were implicated in the biofilm formation and in the wastewater purification. So, the majority of these bacteria could have the potential to degrade and to remove several pollutants from wastewater of different sources. Similar results were also reported by Ding et al. (2011) and demonstrated equally that DGGE bands of dominant bacterial in a wastewater treatment plant include sequences of potential nitrogen remover. The study of Kulikowska et al. (2010) on the same topic found comparable results when they investigated municipal landfill leachate nitrification in RB biofilm.
Thus, the identification of dominant bacteria in some sewage samples revealed at the same time the presence of beneficial species that played certainly an important function in the wastewater treatment purification and of harmful species like Salmonella community that can severely affect the treated wastewater quality.
In summation, some bacteria could play an indispensable part and an important role in the course of degradation of varied chemical and organic pollutants likely to be found in the sewage. In this context, Chromobacterium violaceum is commonly found in soil and water and was associated with animals and plants (Bazylinski et al. 1986). This species was found in the majority of wastewater treatment steps and biofilm. Different studies demonstrated that Chromobacterium species are known as a very active denitrifying bacteria (Bazylinski et al. 1986) and are involved in cyanate detoxification for example (Duran and Menck 2001; Carepo et al. 2004).
Also, Tezuka (1969) confirmed that members of Flavobacterium genus are a good flocculent growth in activated sludge and it is related to denitrification in wastewater systems (Horn et al. 2005). Moreover, Aslam et al. (2005) isolated Flavobacterium granuli from granules used in the wastewater treatment plant. Lo et al. (1998) identified the role of Flavobacterium species in the Pentachlorophenol biodegradation. Other studies reported that the application of Cyanobacterium species showed immense potentialities in wastewater and industrial effluent treatment, bioremediation of aquatic and terrestrial eco-systems, chemical industries, biofertilizers, and fuel production (Gardea-Torresdey et al. 1998; El-Bestawy 2008; Dubey et al. 2011).
In this study, Cronobacter sakazakii was detected in all wastewater and biofilm samples. This species represented a relatively stable and accommodated population throughout the different stages of treatment since it forms and adheres to the biofilm matrix (Hurrell et al. 2009; Hartmann et al. 2010; Chandra et al. 2011). In addition, Fluviicola taffensisas represented by the weak band (B11) was likewise found in all wastewater and biofilm samples. It was shown that this species consumed and degraded low and high-molecular-weight dissolved organic matter (Cottrell and Kirchman 2000; O’Sulivan et al. 2005). Aquitalea species corresponding to the band (B14) denoted by strong bands were present in wastewaters at all the stages of treatment and in all biofilm samples studied. Besides, it was reported that Aquitalea species using acetate cultivate at high-organic loading rate and to be found in aerobic sludge granules (Aday et al. 2010). On the other hand, Methylobacillus (B12) was reported to degrade polycyclic aromatic hydrocarbons in integrated microbial consortium and to degrade microcystin toxins of cyanobacteria (Vinas et al. 2005; Hurrell et al. 2009). In this work, this bacterium was only found in wastewater and not detected in biofilm samples. Several studies proved the role of Burkholderia in PCB, 2,4-dichlorophenoxyacetic acid (2,4-D), and 2,4,5-trichlorophenoxyacetic acid (2,4,5-T) biodegradation, and in the use of 2,3,4,5,6-pentafluorobiphenyl and 4,4′-difluorobiphenyl as sole sources of carbon and energy (Huong et al. 2007; Rehmann and Dauguli 2008; Hughes et al. 2011).
Several species as Enterobacter agglomerans (B4), Cronobacter sakazakii (B8), and Pantoea agglomerans (B18) well known for their bioremediation activities were revealed in the majority of biofilm samples. Numerous studies have shown the role of these bacteria in the degradation process of some organic pollutants such as kerosene, olive oil, and sodium dodecyl sulfate (SDS) (Francis et al. 2000; Abboud et al. 2007; Jacobucci et al. 2009) and also in the production of enzymes and antibiotics, and as an antagonist organism against many phytopathogens (Chernin et al. 1995). Similarly, biofilm pattern analysis divulged the existence of some various bands, suggesting the existence of an active microbial consortium that could develop diverse interactions acting in synergy or in antagonism (Abboud et al. 2007; Jacobucci et al. 2009; Darvishi et al. 2011). Indeed, in the full-scale wastewater treatment process, it appeared that there was a well-balanced selection for either predominance of a single bacterial population or several dominant bacterial ones. It was suggested that different sewage plants might support varied populations and changed levels of species productivity (Rowan et al. 2003; Hallin et al. 2005).
In addition, enduring process leads to the selection phenomenon of different microbial populations and of different bacterial community compositions (Ding et al. 2011). Previous studies reported that the disturbance caused by the climatic alterations, especially the temperature variation and the volume of solar radiation, may be the primary causes of community changes (Aloice and Tatsuya 1996; Dytczak et al. 2008) and a limiting factor to exploit the whole function of the efficient bacterial groups (Ding et al. 2011). In fact, variations in temperature affect all biological processes. Temperature variation has effect on physiochemical and microbiological wastewater characteristics (Metcalf and Eddy 2003; Christa et al. 2007; Hammadi et al. 2008; Mogens et al. 2011; Mehri et al. 2013). Thus, obtained results in summer and winter discern the bacterial community activities. Burkholderia sp. (B16) and Pantoea agglomerans (B18) tolerant to low temperature were exclusively detected in samples collected in winter. However, high temperature contributes to the proliferation of other species as (Salmonella, Enterobacter, Aquitalea, Flavobacterium, Chromobacterium, etc.) that grew optimally at 30–37 °C. Salmonella community was identified in summer and winter patterns. This result was corroborated by sdiA-PCR analysis. Frequent detection of Salmonella in raw and treated wastewater and biofilm samples can be explained by the proximity of the wastewater treatment plant to three hospitals that discharged wastewater containing pathogenic microorganisms, especially in summer time during outbreak of enteric disease (Khattabi et al. 2007) and provide evidence that the RB system did not seriously affect the wastewater Salmonella community. Several studies demonstrated that besides chemicals, hospitals are sources of pathogenic microorganisms as antibiotic resistant bacteria, viruses, and prions. Clinical wastewater represents at least a factor of 2–10 higher than those conventionally detected in domestic wastewater (Moura et al. 2009).
Actually, the wastewater treatment is intended to eliminate or to inactivate pathogenic microorganisms and to avoid waterborne transmission. Therefore, ultraviolet disinfection system was implemented on the final stage of the rotating biodisk procedure. The results obtained showed inter-specific variability in the bacterial tolerance to UV-C254 irradiation. Hassen et al. (2000) demonstrated that UV-C254 is more efficient on the Gram-negative strains as compared to the Gram-positive ones.
Conclusion
In conclusion, the RBC process used to treat municipal wastewater supported 4 distinct microbial communities of the domain bacteria: β and δ-Proteobacteria, the Bacteroidetes phylum and the Cyanobacteria phylum. Identification of dominant bacteria revealed the presence of beneficial species in the majority of biofilm samples. These species known for their bioremediation activities could play an important role in the process of degradation of chemical and organic pollutants. Environmental conditions such as temperature contribute to proliferation of some species than others and result in selection of bacterial communities. Furthermore, common detection of Salmonella in biofilm, untreated and treated wastewater samples, provides evidence that the RBC system did not seriously affect the wastewater Salmonella community. The UV-C wastewater treatment process showed inter-specific variability in bacterial tolerance to UV253.7, and total disappearance or inactivation of Salmonella sp. was observed at a dose of around 1440 mW s cm−2.
In this study, identified bacteria are important members of aquatic and terrestrial bacterial communities, showing variations in lifestyle, geographical distribution, and genome size. They are ubiquitous in soil and water and were sometimes associated with animals and plants. To determine the functional capacity of dominant bacteria in wastewater, biofilms, and environmental niches and the characteristics associated with their relative abundance, further studies were conducted.
References
Abboud MM, Khleifat KM, Batarseh M, Tarawneh KA, Mustafa AA, Madadhah MA (2007) Different optimization conditions required for enhancing the biodegradation of linear alkylbenzosulfonate and sodium dodecyl sulfate surfactants by novel consortium of Acinetobacter calcoaceticus and Pantoea agglomerans. Enzyme and Microb Technol 41:432–439
Aday SS, Lee DJ, Lai JY (2010) Potential cause of aerobic granular sludge breakdown at high organic loading rates. Appl Microbiol Biotechnol 85:1601–1610
Aloice WM, Tatsuya N (1996) Effects of temperature and pH on the growth of heterotrophic bacteria in waste stabilization ponds. Water Res 30:447–455
Aslam Z, Im WT, Kim MK, Lee ST (2005) Flavobacterium granuli sp. nov., isolated from granules used in a wastewater treatment plant. Int J Syst Evol Microbiol 55:747–751
Bazylinski AD, Palome E, Blakemore NA, Blakemore RP (1986) Denitrification by Chromobacterium Violaceum. Appl Environ Microbiol 52:696–699
Ben Rajeb A, Kallali H, Saidi N, Abidi S, Jedidi N, Hassen A (2011) Physicochemical and microbial Caracteristics Performency in wastewater treated under aerobic reactor. Am J Environ Sci 7:254–262
Ben Said M, Khefacha S, Maalej L, Hassen A (2011) Effect of ultraviolet, electromagnetic radiation subtype C (UV-C) dose on biofilm formation by Pseudomonas aeruginosa. Afr J Microbiol Res 5:4353–4358
Carepo MSP, De Azevzdo JSN, Porto JIR, Bentes-Sousa AR, Da Silva BJ, Da Silva ALC, Schneider PC (2004) Identification of Chromobacterium violaceum genes with potential biotechnological application in environmental detoxification. Genet Mol Res 3:181–194
Chandra R, Bharagaya RN, Kapley A, Purohito HJ (2011) Bacterial diversity, organic pollutants and their metabolites in two aeration lagoons of common effluent treatment plant (CETP) during the degradation and detoxification of tannery wastewater. Bioressource Technol 102:2333–2341
Chernin L, Ismailov Z, Haran S, Chet I (1995) Chitinolytic Enterobacter agglomerans antagonistic to fungal plant pathogens. Appl Environ Microbiol 61:1720–1726
Christa L, Cruikshank PE, David G, Gilles PE (2007) Temperature modeling and control for biological wastewater treatment design. Water Environ Fed (Industrial Water Quality 2007): 120–132
Cottrell MT, Kirchman DL (2000) Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low-and high-molecular-weight dissolved organic matter. Appl Environ Microbiol 66:1692–1697
Darvishi P, Ayatollahi S, Mowla D, Niazi A (2011) Biosurfactant production under extreme environmental conditions by an efficient microbial consortium, ERCPPI-2. Coll and surf. B: Biointerfaces 84(2):292–300
Ding L, Zhou Q, Wang L, Zhang Q (2011) Dynamics of bacterial community structure in a fullscale wastewater treatment plant with anoxic-oxic configuration using 16S rDNA PCR-DGGE fingerprints. Afr J Biotechnol 10:589–600
Dubey SK, Dubey J, Mehra S, Tiwari P, Bishwas AJ (2011) Potentiel use of cyanobacterial species in bioremediation of industrial effluents. Afr J Biotechnol 10:1125–1132
Duran N, Menck CF (2001) Chromobacterium violaceum: a review of pharmacological and industrial perspectives. Crit Rev Microbiol 27:201–222
Dytczak MA, Londry KL, Oleszkiewicz JA (2008) Activated sludge operational regime has significant impact on the type of nitrifying community and its nitrification rates. Water Res 42:2320–2328
El-Bestawy E (2008) Treatment of mixed domestic-industrial wastewater using cyanobacteria. J Ind Microbiol Biotechnol 35:1503–1516
Foley SL, Lynne AM, Nayak R (2008) Salmonella challenges: prevalence in swine and poultry and potential pathogencity of such isolates. J Anim Sci 86E:E149–E162
Francis CA, Obraztsova AY, Tebo BM (2000) Dissimilatory metal reduction by the facultative anaerobe Pantoea agglomerans SP1. Appl Environ Microbiol 66:543–548
Gardea-Torresdey JL, Arenas JL, Francisco NMC, Tiemann KJ, Webb R (1998) Abilityof immobilized cyanobacteria to remove metal ions from solution and demonstration of the presence of metallothionein genes in various strains. J Hazard Subt Res 1:1–18
Giannino ML, Marzotto M, Dellaglio F, Feligini M (2009) Study of microbial diversity in raw milk and fresh curd used for Fontina cheese production by culture-independant methods. Intern. J. Food Microbiol 130:188–195
Halatsi K, Oikonomou I, Lambiri M, Mandilara A, Vatopoulos A, Kyriacou A (2006) PCR detection Salmonella spp. using primers targeting the quorum sensing gene sdiA. FEMS Microbiol Lett 259:201–207
Hallin S, Lydmark P, Kokolj S, Hermansson M, Sorensson F, Jarvis A, Lindgren PE (2005) Community survey of ammonia-oxdizingbacteria in full-scale activated sludge process with different solids retention time. J Appl Microbiol 99:629–640
Hammadi L, Ponton A, Belhadri M (2008) Effet de traitement thermique sur le comportement physico-chimique et rhéologique des boues activées de station d’épuration. Rev Energ Renouv 11:465–472
Hartmann I, Carranza P, Lehner A, Stephan R, Eberl L, Riedel K (2010) Genes involved in Cronobacter sakazakii formation. Appl Environ Microbiol 76:2251–2261
Hassen A, Mahrouk M, Ouzari H, Cherif A, Boudabous A (2000) UV disinfection of treated wastewater in a large scale pilot plant and inactivation of selected bacteria in a laboratory UV device. Bioresour Technol 7:141–150
Head IM, Saunders JR, Pickup RW (1998) Microbial evolution, diversity and ecology: a decade of ribosomal RNA analysis of uncultivated microorganisms. Microbiol Ecol 35:1–21
Hengenholtz P, Goebel BM, Pace NR (1998) Impact of cultred-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol 180:4765–4774
Horn MA, Ihssen J, Matthies C, Schramm A, Acker G, Drake H (2005) Dechloromonas denitrificans sp. nov., Flavobacterium denitrificans sp. nov., Paenibacillus anaericanus sp. nov. and Paqenibacillus terrae strain MH72, N2O-producing bacteria isolated from the gut of the earthworm Aporrectodea caliginosa. Int J Syst Evol Microbiol 55:1255–1265
Hughes D, Clarck B, Murphy CD (2011) Bioremediation of polyfluorinated biphenyl in bacteria. Biodegradation 22:41–749
Huong N, Itoh K, Suyama K (2007) Diversity of 2, 4-dichlorophenoxyacetic acid (2, 4-D) and 2, 4,5-trichlorophenoxyacetic acid (2 4, 5-T)-degrading bacteria in Vietnams soils. Microbes Environ 22:243–256
Hurrell E, Kucerova E, Loughlin M, Caubilla-Barron J, Forsythe SJ (2009) Biofilm formation on enteral feeding tubes by Cronobacter sakazakii, Salmonella serovars and other Enterobacteriaceae. Int J Food Microbiol 136:227–231
Jacobucci DF, Oriani MR, Regina L (2009) Reducing COD level on oil effluant by utilizing biosurfactant-producing bacteria. Braz Arch Biol Technol 52:1037–1042
Jany JL, Barbier G (2008) Culture_independent methods for identifying microbial communities in cheese. Food Microbiol 25:839–848
Khattabi H, Belle B, Servais P, Aleya L (2007) Changes in spatial and temporal patterns of bacterial abundance in four basins of Étueffont landfill leachate treatment (Belfort, France). (Variations spaciale et temporelle des abondances bactériennes dans quatre bassins de traitement du lixiviat de la décharge d’Etueffont) (Belfort, France). Comptes Rendus Biologies 330:429–438
Kulikowska D, Jozwiak T, Kowal P, Ciesielski S (2010) Municipal landfill leachate nitrification in RBC biofilm-process efficiency and molecular analysis of microbial structure. Biores Technol 101:3400–3405
LaPara TM, Nakatsu CH, Pantea L, Alleman JE (2000) Phylogenetic analysis of bacterial communitues in mesophilic and thermophilic bioreactors treating pharmaceutical wastewater. Appl Environ Microbiol 66:3951–3959
Lo KV, Zhu CM, Cheuk W (1998) Biodegradation of pentachlorophenol by Flavobacterium species in batch and immobilized continuous reactors. Environ Technol 19:91–96
Mehri I, Turki Y, Cherif H, Khessairi A, Hassen A, Gtari M (2013) Influence on biological treatment and ultraviolet disinfection system on pseudomonas spp. diversity in wastewater as assessed by DGGE. CLEAN 4:1–7
Metcalf and Eddy (2003) Wastewater engineering: treatment and reuse, 4th edn. McGraw-Hill, Boston, p. 55
Mogens H, van Loosdrecht MCM, Ekama GA, Brdjanovic D (2011) Biological wastewater treatment principles, modeling and design. IWA Publishing, London, pp. 18–19
Moura A, Tacao M, Heneriques I, Dias J, Ferreira P, Correia A (2009) Characterization of bacterial diversity in two aerated lagoons of a wastewater treatment plant using PCR-DGGE analysis. Microbiol Res 164:560–569
Muyzer G, Dewaal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700
O’Sulivan LA, Rinna J, Humphreys G, Weightman AJ, Fry JC (2005) Fluviicola taffensis gen. Nov., sp. nov., a novel freshwater bacterium of the family Cryomorphaceae in the phylum ’Bacteroidetes’. Int J Syst Evol Microbiol 55:2189–2194
Rehmann L, Dauguli AJ (2008) Biodegradation of PCBs in two-phase partitioning bioreactors following solid extraction from soil. Biotechnol Bioeng 99:1273–1280
Rowan AK, Snape JR, Fearnside D, Barer MR, Curtis TP, Head IM (2003) Composition and diversity of ammonia-oxidising bacerial communities in wastewater treatment reactors of different design treating identical wastewater. FEMS Microbiol Ecol 43:195–206
Saidi N, Kouki S, Mehri I, Ben Rejeb A, Belila A (2011) Biofilm and siderophore effects on secondary waste water disinfection. Curr Microbiol 63:337–340
Sass AM, Sass H, Coolen MJ, Cypionka H, Overmann J (2001) Microbial communities in the chemocline of a hypersaline deep-sea basin (Urania basin, Mediterranean Sea). Appl Environ Microbiol 67:5392–5402
Tezuka Y (1969) Cation-dependent flocculation in a Flavobacterium species predominant in activated sludge. Appl Microbiol 17:222–226
Turki Y, Ouzari H, Mehri I, BenAissa R, Hassen A (2012) Biofilm formation, virulence gene and multidrug resistance in Salmonella Kentucky isolated in Tunisia. Food Res Int 45:940–946
Turki Y, Mehri I, Cherif H, Hassen A, Ouzari H (2013) Effect of biological treatment and ultraviolet (UV)-C radiation disinfection process on wastewater bacterial community as assessed by denaturing gradient gel electrophoresis (DGGE) fingerprints. Afr J Microbiol Res 42:4927–4933
Turki Y, Mehri I, Ouzari H, Khessairi A, Hassen A (2014) Molecular typing, antibiotic resistance, virulence gene and biofilm formation of most prevalent Salmonella enterica serotypes isolated in Tunisia. J Gen Appl Microbiol 60:123–130
Vinas M, Sabaté J, Guasp C, Lalucat J, Solana AM (2005) Culture-dependent and -independent approaches establish the complexity of a PAH-degrading microbial consortium. Can J Microbiol 51:897–909
Acknowledgments
This work is one of the outputs Researches Program Contract (2010-2012) under the title “Monitoring and Treatment of Water Pollution”, financed by the Tunisian Ministry of Higher Education and Scientific Research. Thanks to the experimental WWT staff for their help during all the experimentation.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Robert Duran
Rights and permissions
About this article
Cite this article
Yousra Turki, Mehri, I., Lajnef, R. et al. Biofilms in bioremediation and wastewater treatment: characterization of bacterial community structure and diversity during seasons in municipal wastewater treatment process. Environ Sci Pollut Res 24, 3519–3530 (2017). https://doi.org/10.1007/s11356-016-8090-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-016-8090-2