Abstract
The identification of fecal pollution in aquatic ecosystems is one of the requirements to assess the possible risks to human health. In this report, physicochemical parameters, Escherichia coli enumeration and Methanobrevibacter smithii nifH gene quantification were conducted at 13 marine waters in the coastal beaches of Rio de Janeiro, Brazil. The pH, turbidity, dissolved oxygen, temperature, and conductivity, carried out by mobile equipment, revealed varied levels due to specific conditions of the beaches. The bioindicators’ enumerations were done by defined substrate method, conventional, and real-time PCR. Six marine beach sites (46 %) presenting E. coli levels in compliance with Brazilian water quality guidelines (<2500 MPN/100 mL) showed nifH gene between 5.7 × 109 to 9.5 × 1011 copies. L−1 revealing poor correlation between the two approaches. To our knowledge, this is the first inquiry in qPCR using nifH gene as a biomarker of human-specific sources of sewage pollution in marine waters in Brazil. In addition, our data suggests that alternative indicator nifH gene could be used, in combination with other markers, for source tracking studies to measure the quality of marine ecosystems thereby contributing to improved microbial risk assessment.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Water is essential to life and to the maintenance of ecosystems. The assurance of a plentiful supply of water is essential for all ecological processes with a direct influence on human health, quality of life and development. Thus, maintaining the quality of water resources has been one of the biggest challenges of this century, mainly due to the indiscriminate use and recurrent contamination (Wimalawansa and Wimalawansa 2014). Waterbodies are susceptible to contamination from a range of point and nonpoint sources with potential contributions from wildlife, domesticated animals, and/or humans. Sources of fecal pollution may include discharge of raw sewage, surface runoff, slaughterhouses, and industrial activities (Oliveira and Cunha 2014).
Recreational water users can be exposed to a range of disease-causing microorganisms; however, waters contaminated with human feces are regarded as a greater risk to human health, as they are more likely to contain human-specific enteric pathogens, including Salmonella enterica serovar Typhi, Shigella spp., hepatitis A virus, and Norwalk-group viruses. Despite these, animals can also serve as reservoirs for a variety of enteric pathogens as various serotypes of Salmonella spp., Escherichia coli, and Cryptosporidium spp. (Fewtrell and Kay 2015).
Microbiological water quality has been evaluated frequently by means of fecal indicator bacteria such as total coliforms, thermotolerant coliforms, E. coli and enterococci. However, the ubiquitous nature of these microorganisms and the lack of discriminatory power, and low correlation with pathogens such as Salmonella spp., Campylobacter spp. and human enterovirus, and sub or overestimated counts, especially in tropical environments, brought the need for alternative methods (Field and Samadpour 2007).
Microbial source tracking (MST) is one of the different names given to the process of identifying the particular source of fecal contamination in water (Stoeckel and Harwood 2007). Over the last decade, several MST techniques have been developed to discriminate between human and non-human sources of fecal contamination in recreational waters, including different animal species (Scott et al. 2005; Foley et al. 2009; Eckburg et al. 2005; Johnston et al. 2010).
Several anaerobic microorganisms such as Bifidobacterium spp., Clostridium perfringens, Methanobrevibacter spp., and members of the order Bacteroidales are currently being used as alternative microbial indicators of recent fecal contamination. This is due to their higher abundance compared with traditional indicators, inability to grow in extra-enteric environments, and high specificity (Santo Domingo et al. 2007; Stewart-Pullaro et al. 2006; Ufnar et al. 2006; Ballesté et al. 2010).
The archaeal genus Methanobrevibacter belongs to the order Methanobacteriales and includes 16 known species found in animal intestinal tracts, anaerobic sludge, and sewage treatment plants (Lai et al. 2004). Methanobrevibacter smithii is a methanogen known to colonize human intestine and vaginal tract. Due to its host specificity and high concentrations in human intestine, it may be effective as an indicator for human fecal pollution in environmental waters (Ufnar et al. 2006). The nifH gene of M. smithii is grouped on the pseudo nif cluster found only in methanogens and does not encode a functional nitrogenase. Thus, it can be considered as a specific target for methanogens such as M. smithii (Ufnar et al. 2006).
Several strategies have been developed to detect and quantify fecal pollution in water environments; most of them based on qPCR assays toward for determination of specific microorganisms from animal gut, mainly using 16S rRNA gene (Bernhard et al. 2003; Scott et al. 2005; Fong et al. 2005; McQuaig et al. 2009).
The nifH marker appeared to be highly host-specific and has been used to detect sewage pollution in environmental waters in California, Mississippi, and Florida, in the USA (Ahmed et al. 2012). However, its host specificity in other countries is not well known. Thus, the aim of this study was to detect the presence of human fecal contamination at 13 coastal recreation beaches by standard methods, in combination with the quantification of alternative bioindicator nifH gene in marine waters of Rio de Janeiro, Brazil.
Methods
Environmental water sampling
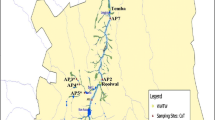
The collection points included the following: Prainha beach (sea and stream water) (23° 04′ 08.97″ S/43° .50′ 57.77″ W), Leblon beach (22° 98′ 75.13″ S, 43° 22′21.1″ W), Leblon channel (22° .98′ 68.32″ S/43° .21′ 54.74″ W), Copacabana beach (22° 58′ 31.2″ S/43° 11′ 12.6″ W), Ramos pool (22° 50′ 22.8″ S/43° 14′ 59.9″ W), São Bento beach (22° 82′ 31.62″ S/43° 18′ 97.7″ W), Bica beach (22° 81′ 93.44″ S/43° 20′ 05.42″ W), Engenhoca beach (22° 82′ 13.22″ S/43° 17′ 05.87″ W), Guanabara beach (22° 79′ 07.42″ S/43° 16′ 50.29″ W), São Francisco beach (22° 92′ 67.77″ S/43° 10′ 42.79″ W), Seca beach (22° 88′ 81.95″ S/42° 34′ 00.43″ W), and Pontinha beach (22° 93′ 94.24″ S/42° 28′ 16.78″ W) (Fig. 1). Five liters from superficial water samples were collected in triplicate at sites that were about 2 m distant from each other, taken from a depth of approximately 30 cm below the water surface and mixed, in order to get a representative sample. All samples were stored in sterile polyethylene bottles on ice and transported to the laboratory within 4 h. Physicochemical parameters, temperature, pH, conductivity, dissolved oxygen (DO), turbidity, and salinity of the samples were analyzed using Water Quality Checker U-10 (HORIBA). The enumeration of E. coli was carried out using the defined substrate method (Colilert, IDEXX), according to the protocol described in Standard Methods for the Examination of Water and Wastewater (APHA 2012). Then, 4 L of each water sample were filtered through a 0.22-μM Stericup® system (Millipore). In the event of filter clogging, additional filters were added. The filters were placed in 2-mL microcentrifuge tubes with 1 mL of PBS and kept at −20 °C overnight.
Geographic coordinates of collection sites: (1) Prainha beach (sea) (2) Prainha beach (stream water), (3) Leblon beach, (4) Leblon channel, (5) Copacabana beach, (6) Ramos pool, (7) São Bento beach, (8) Bica beach, (9) Engenhoca beach, (10) Guanabara beach, (11) São Francisco beach, (12) Seca beach, (13) Pontinha beach
DNA extraction
The procedure used for DNA extraction was a modified version by Bianco et al. 2015 of previously described protocols (Ogram et al. 1987; Smalla et al. 1993). Briefly, tubes containing water sample filters and PBS were submitted to 3 cycles of freezing and thawing (−70 °C/2 min, 65 °C/2 min). Then, an equal volume of glass beads (0.1-mm diameter) was added and the suspension was shaken three times for 80 s at maximum speed in a Bead-Beater. The liquid phase was extracted with phenol–chloroform [1:1 (v/v)] and chloroform–isoamyl alcohol [24:1 (v/v)]. The DNA was precipitated from the aqueous phase with three volumes of ethanol and after drying, the pellet was dissolved in 100 μL of sterile deionized water. For further purification of the DNA, we used the QIAquick® PCR Purification kit (Qiagen GmgH, Hildeitialln, Germany), according to the manufacturer’s instructions to remove possible PCR inhibitors (Clementino et al. 2007). Purified DNA from the water samples was quantified with a Qubit® 2.0 Fluorometer (Invitrogen, Carlsbad, USA), according to the manufacturer’s instructions in order to standardize the DNA concentration for further PCR reactions.
Conventional PCR
PCR analysis was carried out in 50-μL amplification reaction mixtures containing ×1 PCR buffer (Invitrogen Co. Carlsbad, CA, USA), 5 % dimethyl sulfoxide (w/v), 200-μM dNTPs, 2 U of Platinum Taq DNA Polymerase (Invitrogen Co. Carlsbad, CA, USA), 0.5 μM of primer Mnif 342F (5′-AACAGAAAACCCAGTGAAGA-3′) and Mnif 363R (5′-GACGTAAAGGCACTGAAAAACC-3′) (Ufnar et al. 2006), 2.5 mM of MgCl2, and 20 ng of DNA template. The cycling conditions consisted of an initial 95 °C step for 5 min and 35 cycles of amplification at 92 °C for 1 min, 58 °C of annealing temperature for 30 s, 72 °C for 1 min, and a final elongation at 72 °C for 6 min. PCR products were loaded onto a 1.5 % (v/v) agarose gel, and were separated by electrophoresis at 70 V for 2 h in ×1 TAE (40 mmol L-l Tris base, 20 mmol L−1 sodium acetate, 1 mmol L-l EDTA, pH 8.0) buffer with a 100 bp DNA ladder (Invitrogen Co., Carlsbad, CA, USA) as a molecular weight standard. The gels were stained with ethidium bromide, and images were digitalized with the Video Documentation System and analyzed with Image-Master software (Amersham Pharmacia Biotech). In a previous study, we established the host specificity and sensitivity of human-associated marker that were evaluated by screening 49 fecal samples from six host groups. The nifH gene was detected in all (12/12) human fecal DNA samples tested, but not in non-target host groups (0/37) (Bianco et al. 2015). The reproducibility of host-specific archaeal marker amplification was checked by triplicate PCR assays. In addition, genomic DNA (gDNA) from the reference strain M. smithii INCQS A45D/DSM 11975 was included as positive control.
Real-time PCR conditions
The detection and quantification nifH gene of M. smithii was done through a TaqMan-based qPCR assay, using primers Mnif202F (5′-GAAAGCGGAGGTCCTGAA-3′), Mnif353R (5′-CTGAAAAACCTCCGCAAAC-3′), and a hybridization Mnif probe ([FAM]CCGGACGTGGTGTAACAGTAG[BHQ-1]) targeting a 151 bp segment, designed and validated by Johnston et al. (2010). The reactions were processed in optical 96-well reaction plates (Applied Biosystems, Foster City CA, USA), the thermal cycling, and fluorescence detection were carried out in the Applied Biosystems 7500 fast Real-Time PCR System. The optimized reaction was performed in a total volume of 25 μL containing ×1 TaqMan® Universal Master Mix II with UNG (Life Technologies), 800 nmol L−1 of each primer and 240 nmol L−1 of probe (Integrated DNA Technologies, Coralville, IA, USA). The final cycling conditions were an initial denaturation for 10 min at 95 °C, 45 cycles of denaturation for 10 s at 95 °C, and annealing⁄extension for 30 s at 57 °C. gDNA of M. smithii (INCQS A45D/DSM 11975) was used as positive controls; PCR-grade water was evaluated as no-template control.
Standard curve and qPCR inhibition assays
For construction of the standard curve, the stock concentration of M. smithii gDNA was determined with a Qubit® 2.0 Fluorometer (Invitrogen Co. Carlsbad, CA, USA) according to the manufacturer’s instructions; plasmids containing amplified sequences nifH gene of M. smithii strain were constructed with the pGEM-T Easy Vector Systems (Promega), according to the manufacturer’s instructions. Briefly, the fragments of nifH gene, 151 bp were amplified by conventional PCR with the primers already described above. The fragments were cloned into pGEM-T Easy Vector. DH5a E. coli were transformed with these constructions, and after expansion in culture, the plasmids were purified by the Minipreps DNA Purification System (Promega). The stock concentration was determined with a Qubit® 2.0 Fluorometer (Invitrogen Co. Carlsbad, CA, USA), according to the manufacturer’s instructions, and the number of copies was calculated from the molecular weight of the plasmid and insert known, as follows:
DNA (copy=6.02 × 10 23 (copy/mol) × DNA amount (g).
DNA length (dp) × 660 (g/mol/dp).
The CT values were plotted against the logarithm of their initial template copy numbers. Each standard curve was generated by a linear regression of the plotted points. From the slope of each standard curve, PCR amplification efficiency (E) was calculated according to the equation (Rasmussen 2001): E = 10−1/slope−1. These plasmids were used as external standards and 11-point tenfold serial dilution was run in triplicate to a final concentration, ranging from 4.5 × 105 to 4.5 × 1015 copies L−1. Besides, the standard curve also served as positive controls for all the samples analyzed; for negative control, a strain of Klebsiella pneumoniae was used INCQS 629 (ATCC BAA-1706). The commercially available TaqMan® Exogenous Internal Positive Control Reagent was added to verify the presence of inhibitors in reactions according to the manufacturer’s instructions. The internal positive control DNA is measured with VIC-labeled probe, allowing it to be discriminated from the gDNA M. smithii detected with the FAM-labeled probe. Data acquisition and analysis were done using sequence detection system software (version Software v2.0, Applied Biosystems).
Results
Physicochemical parameters
The physicochemical parameters showed variations among the different samples. The pH of the 13 seawater samples analyzed ranged between 5.1–8.8; turbidity showed high values in Ramos pool and São Francisco beach that may be related to the very low depth (Ramos pool) and dispersion of particulate matter as a result of sea currents, or churning of the sea during the tidal cycle (São Francisco beach). Dissolved oxygen levels were highest in Leblon channel and Ramos pool, probably due to aeration, which is essential for organic matter biodegradation. The temperature of the sampled sites remained between 20 and 27 °C favoring the presence of mesophilic forms. Salinity was maintained between 2.0 and 4.0 %. Low salinity values may be related to the discharge of sewage. Conductivity was highest in Seca beach and lowest in Engenhoca beach. Conductivity values are directly proportional to the amount of ionized substances dissolved in the water column (Table 1).
Enumeration of E. coli
According to the auditing standards (CONAMA, 357/2005), Prainha seawater and Prainha stream water, Ramos pool, San Francisco Beach, Seca beach, and Pontinha beach were considered suitable according to water quality guidelines (<2500 MPN/100 mL), presenting 1119.9, 1299.7, 1119.9, 791.5, 99.2, and 211.4 MPN/100 mL, respectively. The other seven beach water samples were classified as inappropriate for recreational activities, with E. coli levels above the acceptable limits (Table 2).
M. smithii Detection by conventional PCR
The nifH gene was detected in 11 out of the 13 samples analyzed showing a fragment of approximately 222 bp compatible with M. smithii positive control. The samples from Ramos pool and São Bento beach did not produce amplification for the expected fragment (Table 2).
Detection and quantification of nifH gene M. smithii by qPCR
The amplification of nifH gene showed a fragment of 151 bp as expected. The slope of the standard curves varied between −3.32 and −3.37 for all real-time PCR assays performed, and the correlation coefficient was always higher than 0.99 while the efficiency ranged from 98 to 107 %. The efficiency of the IPC curve reactions was similar to that obtained with standard curve of M. smithii. As expected, the IPC indicated a wide range of inhibition in the majority seawater samples; thus, the DNA samples were diluted and reanalyzed by a correction factor to calculate the real target number. The nifH gene was detected and quantified in 12 seawater samples, ranging from 5.7 × 109 to 9.5 × 1011 copies L−1, on Leblon beach and Prainha (stream water) beach, respectively. São Bento beach sample showed no amplification in qPCR reactions (Table 2).
Discussion
Fecal pollution of recreational waters remain a significant public health issue, especially in developing countries, but with particular attention to human fecal sources due the host specificity of the pathogens.
In this study, 46 % of the marine water samples presenting E. coli counts in compliance with the Brazilian federal water quality guidelines showed the presence of nifH gene copies between 109 and 1011 L−1. qPCR yielded higher human fecal pollution marker values than either culture method, likely reflecting that qPCR measures the presence of genetic material while the culture methods measure viable cells. However, Haugland et al. (2005) found significant correlations between the enumeration methods when fecal contamination is recent and in high concentrations, probably because there is reduced time for decoupling of cellular metabolism and DNA presence. In addition, DNA is not as sensitive to sunlight as culturable cells, and does not degrade as quickly after UV exposure (Walters et al. 2009).
A study in northwest France showed a significant correlation between E. coli counts and the presence of human markers, whereas no correlation was observed for other animal markers (Gourmelon et al. 2007). This result is not surprising, since many alternative indicators and human pathogens have shown low correlations with traditional indicator bacteria, usually attributed to fecal differential inputs, persistence, and survival of E. coli, among others (Savichtcheva and Okabe 2006; Cole et al. 2003; Shanks et al. 2006; Boehm et al. 2009). The traditional microorganisms used for many years as water quality indicators are not limited only to humans, but are also present in the intestines of other warm-blooded animals (Harwood et al. 2014). This way, although the correlation between fecal indicators is not clear, fecal indicators are significant predictor of potential risk, despite that their concentrations rarely correlate perfectly with other pathogens.
Our results showed that nifH marker was detected in the marine water samples analyzed by both endpoint PCR and qPCR, respectively. However, it could not be detected in São Bento beach by qPCR but was positive for conventional PCR, while the opposite was observed in Ramos pool. On the other hand, nifH gene was revealed in all environmental water samples (16/16) with known or spiked sewage inputs (Johnston et al. 2010), but the same was not observed in Southeast Queensland, Australia, where it was not detected in 181 (96 %) of 188 fecal and wastewater samples (Ahmed et al. 2012). These discrepancies may be attributed to the variability in nucleic acid extraction and sample inhibition and the sensitivity among PCR endpoint and qPCR methods.
Regarding physical/chemical parameters, our data revealed higher temperatures in some samples relating to the shallow depth and the degree of turbulence of the water where these samples were collected. A reduction in salinity and conductivity levels was also observed probably due to an increase of rainfall, evaporation and, particularly, to the release of organic matter due to the mixing of fresh water and salt water (Schmiegelow 2004). High acidification levels in both collection points of Prainha beach could be related to the discharge of organic matter. It is a recognized fact that in the presence of sufficient nutrient levels, E. coli grows in seawater almost as well as it does in rich culture media. In fact, it has been shown that in nutrient-enriched seawater, E. coli competed successfully with other marine bacterial strains (Jannasch 1968). In addition, an acidic pH (5.1–5.4) was found to be most favorable for E. coli survival (in the 5.0–9.0 range) in seawater and pH around 8, contributes to the deleterious effects on E. coli survival (Rozen and Belkin 2001). However, this has not been observed in this study, where the samples with higher E. coli counts presented higher pH levels (7.0–8.8). The evaluation of physical and chemical conditions of marine waters allows us to profile current environmental conditions, revealing potential environmental problems (Lebaron et al. 2015).
Our results demonstrated that 46 % of the marine beach presenting E. coli levels in compliance with water quality guidelines (<2500 MPN/100 mL) showed nifH gene between 5.7 × 109 and 9.5 × 1011 copies L-1 revealing poor correlation between the approaches adopted. A recent work pooling data from 36 sites across the USA has suggested that an empirical relationship can be developed between qPCR and culture-based measurements (Whitman et al. 2010). These data suggest this correlation may be influenced by environmental factors like pollution, abiotic factors, climatic alterations, and bioindicator behavior, under these different conditions.
The application of multiple markers to detect any potential source is essential, as each way has its particular strengths and weaknesses, with regard to other issues that can limit the usefulness of MST. For example, the most thoroughly vetted marker, Bacteroides HF183, is not completely specific for human waste, although it was found to be the most effective marker human fecal contamination in the California-based study (Layton et al. 2013).
However, it has the advantages of being broadly distributed among human populations, and to be present at a relatively high concentration in sewage. Other human-associated microorganisms, such as HPyVs and pathogenic viruses, are less concentrated in sewage and are therefore difficult to detect in dilute samples even at levels where pathogens may be present. However, their specificity engenders great confidence in the finding of human sewage pollution, when detected (Harwood et al. 2014).
Tropical environment, such as Brazil, research in this area is still scarce and to our knowledge, this is the first qPCR assay using M. smithii as a biomarker in marine waters. Although M. smithii is found only in 30 % of the population, its concentration in human feces can vary between 107 and 1010 cell per gram (dry weight) and it is highly prevalent in mixed sewage (Ufnar et al. 2006). Thus, it is possible to conclude that for each 4 L of seawater analyzed in this study, we observed 1 g of M. smithii.
Furthermore, our data reinforces that application of the nifH marker could be an alternative indicator of environmental degradation of marine beaches that have nonpoint sources of human fecal pollution once traditional fecal indicators were not associated with health risks. Although microbial source tracking tools are widely accepted as alternative methods to evaluate sources of pollution, threshold values have not yet been entirely determined to assess the microbial quality of waterbodies (Scott et al. 2002). Therefore, the occurrence of fecal pathogens and markers can increase your likelihood of predicting their occurrence in aquatic systems and serves as the basis for source water protection and the control of pathogenic organisms in the environment. Thus, the knowledge of the source of fecal pollution is essential to assess possible health risks, as well as determine preventive or remedial measures to resolve the issue.
In the light of the importance of the subject area for health, we conclude that the adoption of nifH gene detection as a marker needs the development of new standards and should focus on extensive field tests of real-time approaches at different beaches. This should increase confidence in management guidelines based on risk of fecal pollution and contribute to the implementation of better monitoring and remediation programs, in order to improve the quality of human health and ecosystems.
References
Ahmed W, Sidhu JPS, Toze S (2012) Evaluation of the nifH gene marker of Methanobrevibacter smithii for the detection of sewage pollution in environmental waters in Southeast Queensland, Australia. Environmental science & technology 46(1):543–550
American Public Health Association–APHA; American Water Works Association–AWWA; Water Environment Federation–WEF (2012). Standard methods for the examination of water and wastewater. 22th ed. Washington DC.
Ballesté E, Bonjoch X, Belanche LA, Blanch AR (2010) Molecular indicators used in the development of predictive models for microbial source tracking. Appl Environ Microbiol 76(6):1789–1795
Bernhard AE, Goyard T, Simonich MT, Field KG (2003) Application of a rapid method for identifying fecal pollution sources in a multi-use estuary. Water Res 37:909–913
Bianco K, Barreto C, Oliveira SS, Pinto LH, Albano RM, Miranda CC, Clementino MM (2015) Fecal pollution source tracking in waters intended for human supply based on archaeal and bacterial genectic markers. J Water Health 13(4):985–995
Boehm AB, Ashbolt NJ, Colford JM, Dunbar LE, Fleming LE, Gold MA, Hansel JA, Hunter PR et al (2009) A sea change ahead for recreational water quality criteria. J Water Health 7:9–20
Clementino MM, Fernandes CC, Vieira RP, Cardoso AM, Polycarpo CR, Martins OB (2007) Archaeal diversity in naturally occurring and impacted environments from a tropical region. J Appl Microbiol 103(1):141–151
Cole D, Long SC, Sobsey MD (2003) Evaluation of Fþ RNA and DNA coliphages as source-specific indicators of fecal contamination in surface waters. Appl Environ Microbiol 69(11):6507–6514
Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlesfsen L, Sargent M, Gill SR, Nelson KE et al (2005) Diversity of the human intestinal microbial flora. Science 308:1635–1638
Fewtrell L, Kay D (2015) Recreational water and infection: a review of recent findings. Current Environmental Health Reports 2(1):85–94
Field KG, Samadpour M (2007) Fecal source tracking, the indicator paradigm, and managing water quality. Water Res 41(16):3517–3538
Foley SL, Lynne AM, Nayak R (2009) Molecular typing methodologies for microbial source tracking and epidemiological investigations of Gram-negative bacterial foodborne pathogens. Infect Genet Evol 9(4):430–440
Fong TT, Griffin DW, Lipp EK (2005) Molecular assays for targeting human and bovine enteric viruses in coastal waters and their application for library-independent source tracking. Appl Environ Microbiol 71(4):2070–2078
Gourmelon M, Caprais MP, Ségura R, Le Mennec C, Lozach S, Piriou JY, Rincé A (2007) Evaluation of two library-independent microbial source tracking methods to identify sources of fecal contamination in French estuaries. Appl Environ Microbiol 73(15):4857–4866
Harwood VJ, Staley C, Badgley BD, Borges K, Korajkic A (2014) Microbial source tracking markers for detection of fecal contamination in environmental waters: relationships between pathogens and human health outcomes. FEMS Microbiol Rev 38(1):1–40
Haugland RA, Siefring SC, Wymer LJ, Brenner KP, Dufour AP (2005) Comparison of Enterococcus measurements in freshwater at two recreational beaches by quantitative polymerase change reaction and membrane filter culture analysis. Water Res 39:559–568
Jannasch HW (1968) Competitive elimination of Enterobacteriaceae from seawater. Appl Microbiol 16:1616–1618
Johnston C, Ufnar JA, Griffith JF, Gooch JA, Stewart JR (2010) A real-time qPCR assay for the detection of the nifH gene of Methanobrevibacter smithii, a potential indicator of sewage pollution. J Appl Microbiol 109(6):1946–1956
Lai MC, Lin CC, Yu PH, Huang YF, Chen SC (2004) Methanocalculus chunghsingenisis sp. Nov., isolated from an estuary and a marine fishpond in Taiwan. Int J Syst Evol Microbiol 54:183–118
Layton BA, Cao Y, Ebentier DL, Hanley K, Ballesté E, Brandão J, Byappanahalli M, Converse R, Farnleitner AH, Gentry-Shields J, Gidley ML, Gourmelon M, Lee CS, Lee J, Lozach S, Madi T, Meijer WG, Noble R, Peed L, Reischer GH, Rodrigues R, Rose JB, Schriewer A, Sinigalliano C, Srinivasan S, Stewart J, Werfhorst LC, Wang D, Whitman R, Wuertz S, Jay J, Holden PA, Boehm AB, Shanks O, Griffith JF (2013) Performance of human fecal anaerobe-associated PCR-based assays in a multi-laboratory method evaluation study. Water Res 47(Issue 18):6897–6908
Lebaron P, Cournoyer B, Lemarchand K, Nazaret S, Servais P (2015) Environmental and human pathogenic microorganisms. In: Environmental microbiology: fundamentals and applications. Springer, Netherlands, pp. 619–658
McQuaig SM, Scott TM, Lukasik JO, Paul JH, Harwood VJ (2009) Quantification of human polyomaviruses JC virus and BK virus by TaqMan quantitative PCR and comparison to other water quality indicators in water and fecal samples. Appl Environ Microbiol 75:3379–3388
Ogram A, Sayler GS, Barkay T (1987) The extraction and purification of microbial DNA from sediments. J Microbiol Methods 7:57–66
Oliveira BSSD, Cunha ACD (2014) Correlation between water quality and precipitation variability in the southern state of Amapá. Rev Ambient Água 9(2):261–275
Rasmussen R (2001) In: Meuer S, Wittwer C, Nakagawara K (eds) Quantification on the LightCycler. Springer-Verlag, Berlin, pp. 21–34
Rozen Y, Belkin S (2001) Survival of enteric bacteria in seawater. FEMS Microbiol Rev 25(5):513–529
Santo Domingo JW, Bambic DG, Edge TA, Wuertz S (2007) Quo vadis source tracking? Towards a strategic framework for environmental monitoring of fecal pollution. Wat Res 41:3539–3552
Savichtcheva O, Okabe S (2006) Alternative indicators of fecal pollution: relations with pathogens and conventional indicators, current methodologies for direct pathogen monitoring and future application perspectives. Water Res 40:2463–2476
Schmiegelow, J. M. M. (2004). The Blue Planet: An Introduction Marine Science. Brazil.
Scott TM, Rose JB, Jenkins TM, Farrah SR, Luhasik J (2002) Microbial source tracking: current methodology and future directions. Appl Environ Microbiol 68:5796–5803
Scott TM, Jenkins TM, Lukasik J, Rose JB (2005) Potential use of a host associated molecular marker in Enterococcus faecium as an index of human fecal pollution. Environ Sci Technol 1(39):283–287
Shanks OC, Nietch C, Simonich M, Younger M, Reynolds D, Field KG (2006) Basin-wide analysis of the dynamics of fecal contamination and fecal source identification in fecal pollution source tracking in waters intended for human supply journal of water and health in Tillamook Bay, Oregon. Appl Environ Microbiol 72(8):5537–5546
Smalla K, Cresswell N, Mendonca-Hagler LC, Wolters A, van Elsas JD (1993) Rapid DNA extraction protocol from soil for polymerase chain reaction-mediated amplification. J Appl Bacteriol 74:78–85
Stewart-Pullaro J, Daugomah JW, Chestnut DE, Graves DA, Sobsey MD, Scott GI (2006) F+ RNA coliphage typing for microbial source tracking in surface waters. J Appl Microbiol 101(5):1015–1026
Stoeckel DM, Harwood VJ (2007) Performance, design, and analysis in microbial source tracking studies. Appl Environ Microbiol 73(8):2405–2415
Ufnar JA, Wang SY, Christiansen JM, Yampara-Iquise H, Carson CA, Ellender RD (2006) Detection of the nifH gene of Methanobrevibacter smithii: a potential tool to identify sewage pollution in recreational waters. J Appl Microbiol 101(1):44–52
Walters SP, Yamahara KM, Boehm AB (2009) Persistence of nucleic acid markers of health-relevant organisms in seawater microcosms. Water Res 43:4929–4939
Whitman RL, Ge Z, Nevers MB, Boehm AB, Chern EC, Haugland RA, White EM (2010) Relationship and variation of qPCR and culturable enterococci estimates in ambient surface waters are predictable. Environ Sci Technol 44:5049–5054
Wimalawansa SA, Wimalawansa SJ (2014) Impact of changing agricultural practices on human health: chronic kidney disease of multi-factorial origin in Sri Lanka. Wudpecker J Agric Res 3(5):110–124
Acknowledgments
We gratefully acknowledge the Genome Sequencing Core-PDTIS/FIOCRUZ. This work was supported by Coordenação de Aperfeiçoamento de Pessoal de Ensino Superior (CAPES), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ), and INCQS/FIOCRUZ.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Philippe Garrigues
Rights and permissions
About this article
Cite this article
Oliveira, S.S., Sorgine, M.H.F., Bianco, K. et al. Detection of human fecal contamination by nifH gene quantification of marine waters in the coastal beaches of Rio de Janeiro, Brazil. Environ Sci Pollut Res 23, 25210–25217 (2016). https://doi.org/10.1007/s11356-016-7737-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-016-7737-3