Abstract
Various antibiotics have been extensively used to treating infectious diseases in hospitals. In this study, the abundance and diversity of antibiotics and antibiotic resistance genes (ARGs) were observed in the wastewater samples from five hospitals in Xinjiang, China. The total concentrations of tetracyclines, sulphonamides, and quinolones in hospital influents ranged from 363.4 to 753.3 ng/L, 285.5 to 634.9 ng/L, and 1355.8 to 1922.4 ng/L, respectively. However, the removal efficiency of tetracyclines, sulphonamides, and quinolones in wastewater treatment processes ranged from 72.4 to 79.3 %, 36.0 to 52.2 %, and 45.1 to 55.4 %, respectively. The contamination levels of the selected ARGs varied in all wastewater samples. The highest relative concentrations of sul1, sul2, tetQ, and qnrS were significantly higher than those of other ARGs in this study. Significant positive correlations between the relative abundance of partial ARGs and concentrations of certain antibiotics were observed in hospital wastewaters. Results show that integrons played an important role in disseminating and distributing ARGs in microorganism systems. Furthermore, strong correlations were observed between tetQ, sulphonamide resistance genes (except sulA) and intI1. This study aimed to determine the contamination levels of antibiotics and ARGs and analyze the relationships among ARGs, and antibiotics and integron genes in hospital wastewaters.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Antibiotics play an important role in treating and preventing diseases in the medical industry since penicillin was introduced by Fleming. The overuse of antibiotics is inevitable in preventing and treating infectious diseases in humans and animals. Antibiotics are also used as promoters in livestock and aquaculture. For example, approximately 210 million kilograms of antibiotics are produced annually in China (Su et al. 2012) and 16 million kilograms of antibiotics are used annually in the USA for human or agriculture use (Sarmah et al. 2006). Antibiotics are only partially absorbed after administration, and nearly 75 % of antibiotics are excreted as metabolites by humans and animals (Luo et al. 2011). Eventually, antibiotics and antibiotic metabolites are released into different environment systems by media. Antibiotic residues generate selective pressure to the bacteria in the environment, thus contributing to the proliferation of antibiotic resistance genes (ARGs) and antibiotic-resistant bacteria in microorganisms. The occurrence of antibiotic resistance changes the composition and structure of the microbial community and increases the potential risks to human health and the environment (Kim et al. 2004; Akinbowale et al. 2007). The environment contaminants of antibiotics contribute to the increase in the proportion of antibiotic-resistant bacteria. Once bacteria acquired antibiotic resistance (even multiple antibiotic resistance) to protect themselves, antibiotic resistance can compromise the effectiveness of antibiotic therapy.
Tetracyclines, sulfonamides, and quinolones were the most widely used in treating infectious diseases. The tetracyclines and sufanomides were used to treat human disease for a longer history and resistant gram-negative bacteria and gram-positive bacteria. The same class drugs of tetracyclines and sufanomides have cross-resistance characteristics. So far, more than 40 different tetracycline resistance genes have been described and at least 22 tetracycline resistance genes have been observed in various environment systems (Zhang and Zhang 2011). Tetracycline resistance genes were divided as the efflux pump genes (tetA, tetB, tetC, tetD, tetE, and tetL), ribosomal protection protein genes (tetO, tetW, tetM, and tetQ) and modification enzyme gene (tetX) by different resistance mechanisms of ARGs. The sulfonamide resistance genes mainly included sul1, sul2, sul3, and sulA. Quinolones were one of synthetic broad-spectrum antibiotics, which mainly were resistant against gram-negative bacteria. Quinolone resistance genes belong pentapeptide repeat family and plasmid-mediated genes. Among, qnrA, qnrB, qnrC, qnrD, qnrS, qepA, parC, and gyrA were identified and detected in the microbial cells. The occurrence and spread of ARGs was closely associated with diverse resistance mechanisms and dissemination mechanisms. The plasmids, transposons, and integrons contributed to the increase of type and concentration of ARGs in various environment media. The relationships between integrons and different ARGs were specially concerned by previous studies (Belinda et al. 2013; Zhang et al. 2009).
Hospital wastewater is one major source of antibiotic residues, as well as normal human microbiota and pathogenic bacteria, are introduced into ecological systems. These environmental factors inevitably create an advantageous condition for the enrichment and dissemination of antibiotic-resistant bacteria and ARGs. Thus, hospital wastewater has a high potential to be a threat to human and animal health because of its infectious and toxic characteristics. Investigating the distribution, effect, and removal mechanism of antibiotic resistance in hospital wastewater will be significant in protecting human health and the environment.
Since economic and medical were in underdevelopment condition in some areas of Xinjiang, tetracyclines and sulfonamides still have not been completely eliminated in the treatment of disease. Meanwhile, quinolones are still commonly used in medical. Hospital wastewater samples in northern Xinjiang were selected as the research objects in this study. Three classes of antibiotics containing tetracyclines, sulphonamides, and quinolones, and their corresponding resistance genes and two integron genes were considered and monitored. This study mainly aims to occurrence and contamination levels of targeted antibiotics and corresponding ARGs and integron genes in hospital wastewater and to evaluate the relationship between them in the community.
Materials and methods
Sample collection
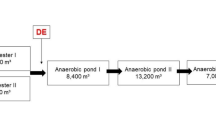
Water samples were collected from the five hospitals in northern Xinjiang during July 2014. The five hospitals were provided with comprehensive wastewater treatment facilities, which were mainly used to treat medical and domestic wastewater. Sampling sites included the raw influent and final effluent of the WWTPs of hospitals. Detailed information on the five hospitals is shown in Table 1.
All surface samples were collected in triplicate from the top 50 cm of the surface in sterile containers and then mixed together. The samples were stored in an icebox, immediately delivered to the laboratory and processed within 12 h.
Antibiotic detection from wastewater samples
Before 11 antibiotics were determined, the selected samples were analyzed in triplicate following the described protocol. Each wastewater sample was filtered through 0.45-μm glass fiber filters. The pH of each water sample was then adjusted to pH 3.0 with 5 M formic acid. Thereafter, 2 mL 5 % (w/v) Na2EDTA was added as a chelating agent to 200 mL water samples to mitigate the effects of antibiotic binding to residual heavy metal ions. Before extraction, the oasis hydrophilic lipophilic balance extraction cartridges were preprocessed with 4 mL methanol, 4 mL ultrapure water, and 4 mL 2 g/L Na2EDTA with a pH value of 3.0 in sequence. The samples were loaded and passed through the cartridges at an appropriate speed of approximately 3 mL/min. After the samples were loaded, the cartridges were rinsed with 4 mL ultrapure water and 4 mL 10 % (v/v) methanol in sequence to eliminate the interference matrix components. The cartridges were then vacuum-dried for 30 min. Finally, the analytes were eluted with 4 mL methanol, followed by 4 mL methanol containing 2 % ammonium hydroxide (v/v, 2 %). The eluates were concentrated under a gentle stream of nitrogen and were dissolved in 1 mL methanol containing 0.1 % formic acid for high-performance liquid chromatography electrospray ionization tandem mass spectrometry analysis. The HPLC separation was performed on an Agilent 1200 series. The antibiotics were separated on a Waters Symmetry-C18 column that operated at 40 °C, with injection volumes of 10 μL. The mobile phase was composed of acetonitrile (phase A) and ultrapure water with 0.3 % formic acid (v/v) (phase B). The separation was maintained at a flow rate of 0.2 mL/min with the following flow gradient: phase A was linearly increased from 15 to 20 % in 1 min and held constant for 1 min; phase A was then increased to 30 % over 8 min. The cycle was completed by returning phase A to the initial percentage (15 %) over 7 min and maintaining for 8 min until the next injection.
Limit of detection (LOD) and limit of quantification (LOQ) were defined as a minimum detectable amount of an analyte corresponding to a signal-to-noise (S/N) ratio of 3 and 10, respectively. Mixed working solutions were used to calculate calibration curves (r 2 > 0.99). LODs of antibiotics in wastewater samples ranged from 0.06 to 0.80 ng/L, respectively. SA, QN, and TC recoveries in water samples were 89.3 to 107 %, 72.5 to 97.4 %, and 69.3 to 84.2 %, respectively. The relative standard deviations (RSDs) of all of the analytes were <14 %.
DNA extraction from wastewater samples
Wastewater samples (500 mL) were filtered through a 0.45-μm filter by using a vacuum filtration apparatus. The filters were placed in the extraction tubes of the Power Water DNA Isolation Kit (MoBio Laboratories, Inc.).
DNA extraction was performed following the manufacturer’s protocol. The quality and concentration of the purified DNA were evaluated by spectrophotometry analysis (NanoDrop ND-2000c, MA, USA) and 2 % agarose gel electrophoresis. The purified DNA was stored at −20 °C prior to real-time qPCR analysis.
Real-time PCR of ARGs and integron genes
Real-time qPCR was applied in this study to quantify the presence of tetracycline resistance genes (tetM, tetO, tetQ, and tetW), sulphonamide resistance genes (sul1, sul2, sul3, and sulA), quinolone resistance genes (qnrS, qnrC, qnrD, and qepA) and integron genes (intI1 and intI2). The relative quantification of PCR was applied in a process using 16S rRNA genes as reference genes. The relative concentrations of ARGs and integron genes were defined as the number of copies of genes normalized to the copy number of 16S rRNA genes.
All reactions were performed in a 25 μL volume containing 12.5 μL of SYBR Premix Ex Taq™ (TaKaRa), 0.3 μL of ROX reference dye, 0.5 μL of 0.2 μM concentration of each primer, 2 μL of template DNA, and 9.2 μL of ddH2O. qPCR amplification and quantification were conducted by using a StepOnePlus™ real-time PCR system (ABI7900, USA). The qPCR protocol was then followed: 30 s at 95 °C, 40 cycles of 5 s at 95 °C, 30 s at different annealing temperatures (Table 2), extension for 30 s at 72 °C, fluorescence acquisition step at 72 °C, and a final melt curve stage with temperature ramping from 60 to 95 °C. All reactions were performed in triplicate for DNA samples from hospitals. Additional information on qPCR primers is shown in Table 3.
Statistical analysis
Statistical analysis was performed by using Origin 8.5 and SPSS 17.0. Correlation analysis was used to calculate Pearson’s bivariate correlation and p values. One-way ANOVA was used to assess the homogeneity of variance with a significance level of 5 % (p < 0.05).
Results and discussion
Occurrence and abundance of antibiotics
The concentrations of four fluoroquinolone, three sulphonamide, and four tetracycline antibiotics detected in the influent and effluent of the WWTPs of hospitals are summarized in Table 4. The 11 antibiotics were detected in the influent and effluent of all the WWTPs of hospitals, except that chlortetracyclin (CTC) was not observed in the effluent of hospital R. The high detection frequency showed that hospital wastewater could be a major source of antibiotic contamination. The concentrations of all target antibiotics in the collected samples were typically in the ng/L level. By contrast, the concentrations of sulfonamides and tetracyclines were notably lower than those of quinolones in wastewater from hospitals. The concentrations of sulphonamides in all collected samples were 285.5 to 634.9 ng/L in the raw influent and 139.4 to 406.9 ng/L in the final effluent. The highest concentrations were observed for sulfamethazine (31.2 ng/L) in hospital S, sulfamonomethoxine pyridazine (77.3 ng/L) in hospital S, and sulfadiazine (SDZ) (574 ng/L) in hospital M. The contamination concentration of SDZ in the samples of the five hospitals was comparable to that in the WWTP effluents of the Haihe watershed (Luo et al. 2011). Similarly, the concentrations of tetracyclines were 363.4 to 753.3 ng/L in the raw influent and 104.7 to 178.3 ng/L in the final effluent. These findings were significantly higher than the concentrations of four tetracyclines in the influent and effluent of a WWTP in the USA (Karthikeyan and Meyer 2006). The concentrations of quinolones in the present study were the highest in all detected antibiotic families. The concentrations ranged from 1355.8 to 1922.4 ng/L in the hospital influent and 619.9 to 925.1 ng/L in the hospital effluent. The results were higher than the contamination levels of quinolones in a WWTP in the Jiulongjiang River Basin (Jiang et al. 2013a, b). Relatively, 14,378 ng/L of ofloxacin (OFL) and 13,780 ng/L of ciprofloxacin (CIP) were observed in a hospital effluent in Spain (Rodriguez-Mozaz et al. 2015).The quinolones had a noteworthy therapeutic effect to treat infectious diseases, which contributed to their medical consumption was a significant value in hospitals probably (MacDougall et al. 2005).
The removal efficiency and concentrations of different types of antibiotics in wastewater systems were affected by the physicochemical property of the antibiotics and the wastewater treatment process. The average removal efficiencies were 36.0 to 52.2 % for sulfonamides, 72.4 to 79.3 % for tetracyclines, and 45.1 to 55.4 % for quinolones in the different wastewater treatment processes. The removal efficiencies of sulphonamides and quinolones were consistent with those in a conventional treatment process (Li et al. 2013). The high removal efficiency of tetracyclines at 87.9 % was reported in a WWTP (Xu et al. 2015). The finding suggested that the trend of antibiotic removal efficiencies was tetracyclines > quinolones > sulphonamides in the present study. The obvious difference in removal efficiency was strongly connected to the elimination mechanism of antibiotics. The antibiotic partition between water and sediments was a dynamic process. However, the importance of adsorption as an attenuation mechanism could be determined by normalizing the antibiotic concentrations in the water to the corresponding concentrations in the underlying sediments. The adsorption and biodegradation of the wastewater treatment process were mainly contributed to the attenuation and elimination of antibiotics. Tetracyclines and quinolones with the polar/ion groups exhibited high adsorption ability, which is important to remove them during the sewage treatment process in the WWTPs of hospitals (Wu et al. 2009; Dorival-García et al. 2013). Although sulphonamides possessed high solubility and chemical stability in water systems, biodegradation play a significant role in the removal of sulphonamides (Yang et al. 2012). Overall, the residual antibiotics persisted and migrated in various environments through the WWTPs, which would provide a selective pressure for the establishment and amplification of antibiotic-resistant reservoirs.
Quantification of 12 ARGs and two integron genes
The occurrence and distribution of the selected ARGs and integron genes in the samples from different hospitals were investigated, and the qualitative occurrence data of ARG families and integron genes by conventional PCR analysis are listed in Table 5. The number of copies of genes was normalized to the 16S rRNA genes copy number, as shown in Figs. 1, 2, 3, and 4.
Four sulfonamide resistance genes (sul1, sul2, sul3, and sulA) were present in all wastewater samples. However, sulA was not detected in the influent sample of hospital R. The relative occurrences of sul1, sul2, and sul3 ranged from 1.79 × 10−1 to 6.67 × 10−1, 7.33 × 10−2 to 3.38 × 10−1, and 9.22 × 10−2 to 5.90 × 10−1, respectively, which were approximately four orders of magnitudes higher than the relative abundances of sulA. For tetracycline resistance genes, the highest relative abundance was 2.80 × 10−1 to 7.47 × 10−1 for tetQ in all samples, followed by tetO, tetW, and tetM. However, the four quinolone resistance genes were observed in all influents and effluents. The relative concentration of qnrS was the maximum value in targeted quinolone resistance genes. The following trend of the relative concentrations was observed: qnrS > qepA > qnrD > qnrB. Two integron genes (intl and intI2) were detected in all collected samples from the five hospitals. The ratio of concentration of intI1 normalized with 16S rRNA genes ranged from 8.07 × 10−3 to 3.02 × 10−2. The contamination level of intI2 was slightly lower than the relative concentration of intI1 in this study.
Although the use of sulphonamides was restricted in recent decades for resistance and dissemination among treated patients, sulfonamide resistance genes sul1, sul2, and sul3 were detected in the various aquatic ecosystems, including hospital wastewater (Berglund et al. 2015), livestock lagoons (Sayah et al. 2005), and even freshwater rivers (Jiang et al. 2013a, b). Sulphonamide antibiotic resistance was acquired by mutations in the enzyme dihydropteroate synthase (DHPS) in the folic acid pathway. Sul1, sul2, sul3, and sulA were four alternative DHPS genes. Perhaps the degree of difficulty in gene mutations was different, and sulA was rarely detected in previous reports. The contamination levels of sul1 and sul2 were comparable to those in the wastewater treatment systems of swine farms (Tao et al. 2014). The relative concentrations of sul1 and sul2 genes were 2.81 × 10−5 to 3.33 × 10−3 and 1.04 × 10−5 to 3.80 × 10−3 in a lake ecology. These concentrations were significantly higher than those in the present observations (Zhou et al. 2014). However, the relative concentrations of sul1 genes were slightly higher than those of sul2, and this result was inconsistent with the result in aquaculture environments (Gao et al. 2012). The contamination levels of sul3 genes were similar to those reported by Xu et al. (2015). The relative concentrations of sul1 and sul2 were the highest in selected ARGs in different hospitals, which suggested the dissemination of sulphonamide resistance genes was likely associated with mobile genetic elements (Zhou et al. 2014).
The detection frequency of selected tetracycline resistance genes was 100 % by conventional PCR analysis, and this result demonstrated the prevalence of tetracyclines in all the WWTPs of hospitals. Four ribosomal protection protein (tetO, tetM, tetQ, and tetW) were selected as the objects in this study. The ribosomal protection protein genes were identified in various bacterial species, including gram-positive and gram-negative, thus contributing to their high concentrations in different environmental systems in previous reports (Aminov et al. 2001; Adelowo and Fagade 2009). The conjugative transposons and wide host range contributed to the prevalence of RPP genes. The contamination level of tetQ was slightly lower than that in the aquaculture environment of the WWTPs of pig farms (Cheng et al. 2013). However, the relative concentrations of tetO and tetW in the wastewater of hospitals were comparable to the contamination levels in the livestock lagoons of various operation types (McKinney et al. 2010). The relative abundance of tetM was significantly lower than that reported in a vertical flow constructed wetlands (Liu et al. 2014).
The main mechanism of quinolone resistance was related to the mutations that affect the quinolone-resistant determination region of genes that encode topoisomerase II or IV (Robicsek et al. 2006). Quinolone resistance was disseminated by chromosomal mutations or plasmid-mediated quinolone resistance genes. Plasmid-mediated quinolone resistance genes were originally reported in a Klebsiella pneumoniae clinical isolate from the USA in 1998. These genes were detected in genes from bacteria or bacteriophage in numerous studies (Dahmen et al. 2010). However, the contamination distribution of quinolone resistance genes was rarely reported. qnrS showed a significant contamination level in a horizontal subsurface flow wetland (Nõlvak et al. 2013). The relative concentration of qnrD in this study was comparable to that in a sewage treatment plant and its receiving river in a recent field study (Xu et al. 2015). The finding suggested that the prevalence of quinolone resistance genes was a serious threat to human health.
Integrons can transfer and disseminate antibiotic resistance by capturing and integrating one or more gene cassettes and converting them into functionally expressed genes to permit the rapid acquisition of diverse resistance genes (Mazel 2006). The high relative concentrations of intI1 and intI2 contributed to the distribution and dissemination of ARGs in various environment systems. The ratio of concentration of intI1 that normalized with the 16S rRNA genes was significantly higher than that reported in the Haihe River (Luo et al. 2010).
Relationship between ARGs and antibiotics and integron genes
The increase of ARGs in hospital effluents was primarily caused by the extensive use of antibiotics. The relative concentration of ARGs should be closely linked to the levels of residual antibiotics that generated a selection pressure on the microorganisms. Table 6 summarizes the correlations between the concentration of antibiotics and the relative concentration of ARGs.
The majority of the correlations between antibiotics and ARGs showed no statistical significance (p > 0.05). However, these correlations were probably convincing evidence that explained the relationship between antibiotics and ARGs. Strong correlations existed between antibiotics and their corresponding resistance genes, such as NOR and qepA (r = 0.707, p < 0.05), ENR and qnrB (r = 0.751, p < 0.05), and SMR and sulA (r = 0.740, p < 0.05). However, the positive and statistically significant correlations between tetracycline and their noncorresponding antibiotic genes (qnrB, qnrD, and sul3, p < 0.05) were 0.681, 0.724, and 0.638, respectively. All the selected ARGs and total antibiotics exhibited weak positive correlations in this study.
Consistent with previous studies, the observations suggested that exposure to residual antibiotics could lead to selective pressure for ARG. Li et al. (2012) suggested that the relative abundance of total quinolone resistance genes and the measured concentrations of total quinolones were significantly correlated in wastewater and soil (r = 0.71, p < 0.05). Strong correlations between tetracycline resistance genes and antibiotics were not observed, and this result was inconsistent with the results of ribosomal protection protein genes, which showed significant correlation with tetracyclines in an improved AAO WWTP (Huang et al. 2015). The positive and significant correlations between tetracycline and their noncorresponding antibiotic genes suggested that the selective pressure of increasing ARGs was not only from the corresponding classes of antibiotics but also from the noncorresponding classes of antibiotics. The various environmental behavior and propagation mechanisms of ARGs and antibiotics in the environment probably contributed to the weak correlations between them. Along with the effect of microorganisms and other factors, antibiotics would gradually be degraded. However, ARGs can persistently be disseminated and migrated within the microbial community. Different classes of antibiotics and heavy metal ions in the wastewater may affect the correlations between ARGs and antibiotics (Aktan et al. 2013). Integrons and other mobile genetic elements could play an important role in developing resistance among microorganisms by transferring various resistance genes simultaneously.
The high contamination levels of antibiotics, ARGs were observed in selected hospital wastewater systems. Obviously, antibiotic resistance was complexed associated with antibiotics and ARGs in microbial matrices of hospital wastewater. The reason why antibiotic resistance was universal concerned was that it has been increasingly difficult to treat and cure pathogen infectious diseases by antibiotic prescriptions. If antibiotic-resistant pathogens were permeated into human organs, which can weaken the effect of the treatment of disease and prolong the healing time of infection, even threat life safety. Hospital wastewater was one of the main microbial matrices including various medical antibiotics and pathogens. As accelerators, smaller molecular units played an invaluable role during the spread of antibiotic resistance. Intuitively, antibiotics provided selective pressure and induction for bacteria and pathogens in hospital wastewater. Most of survived pathogens were resistant against low concentration of antibiotics. The antibiotic-sensitive strains were likely to be eliminated by inhibiting their growth and reproduction. It was potential possible that antibiotic resistance pathogens were preserved and multiply propagated as dominant strains. Beneficial strains to human and ecosystem were eliminated for sensitive to antibiotics.
This study showed the positive correlations between intI1 and all ARGs. The positively strong correlations between intI1 and sulphonamide resistance genes, except for sulA, were observed in this study. The correlations between intI1 and sul1, sul2, and sul3 were 0.970, 0.881, and 0.786, respectively, which were the highest in all correlation coefficients between selected antibiotics and ARGs. Class 1 integron can facilitate the proliferation of different ARGs. Luo et al. (2010) reported that the correlation between intI1 and sul1 was 0.889 (p < 0.01) in the Haihe River. The correlations between the relative abundance of intI1, sul1, and sul2 in the water collected in the Pearl River and Pearl River estuary were 0.94 and 0.90 in summer (p < 0.01) (Chen et al. 2015). Furthermore, the results in present suggested that intI1 gene could be associated with the dissemination of sulphonamide resistance genes in the wastewater from hospitals. A positively strong correlation between the relative abundance of tetracycline-resistant gene tetO and intI1 (r 2 = 0.868, p < 0.01) was observed. Although no tetracycline resistance genes were detected in any gene cassette of intI1, some reports demonstrated that intI1 could accrete noncassette resistance genes outside the cassette region by the insertion sequence common region, a newly highlighted complex integron element usually linked to intI1 (Liu et al. 2012). This finding suggested that class 1 integron contributed to the proliferation of tetracycline resistance genes. Overall, classes 1 and 2 integrons contributed to the propagation of ARGs.
It is imperative to understand the behaviors of antibiotic resistance genes to enable the development of strategies to reduce their abundance and dissemination. Current knowledge does not provide a sufficiently detailed portrait of the evolution and movement of antibiotic resistance genes to enable reliable predictions about their behavior or the design of precise strategies to manage them (Udikovic-KoLic et al. 2014). Horizontal gene transfer perhaps breaks the boundaries that genes were diffused in the genetic strains, which resulted in the spread routes of ARGs becoming more complex. The modes of horizontal gene transfer included transduction, transformation, and conjugation. Furthermore, integrons, transposons, plasmids, and integron-gene cassette systems played irreplaceable roles during horizontal gene transfer. Transposons can freely inserted in or leaped from other DNA molecule and transfer information of antibiotic resistance among chromosomes, plasmids, and phages. Integron-gene cassette systems may integrate genes to contribute themselves into antibiotic resistance gene clusters (Su et al. 2012). Antibiotic resistance persistently disseminated in and was difficult to be eliminated in pathogens for their complex resistance mechanisms and spread mechanisms. The selective pressure and induction of antibiotics contributed to the gene mutations of genetic material in bacteria, which resulted in antibiotics no-effected strain cells. For example, some hydrolytic enzymes and inactivated enzymes leaded to inactivation of antibiotics, change of targeted sits, and even evolution of bacteria’s characteristics. Antibiotics, ARGs, antibiotic resistance bacteria, pathogens, and mobile genetic elements composed a complex microbial matrix hospital wastewater. Intricate relationship among the different components will lead to diversity of antibiotic resistance genes and pathogens. Multiple antibiotic resistance pathogens and superbugs may be emerged in hospital wastewater, which perhaps it is butterfly effect. Antibiotic resistance has been making an unprecedented trouble for human. Methicilin-resistant Staphylococcus aureus were absolutely resistant against β-lactams and cephalosporins and have different degrees of resistance to aminoglycosides, macrolides, tetracyclines, fluoroquinolones, sulfonamides, and lifestyle.
However, the diverse foodborne bacteria and pathogenic bacteria in hospital wastewater were capable to parasitize in animals and humans. Once these bacteria acquired antibiotic resistance, it would pose a serious threat to the treatment and cure of bacteria infectious diseases for human. Resende et al. reported that the resistance of various micrococcus species was investigated in anaerobic digestion matter of cattle manure; the results revealed Escherichia coli, Enterococcus faecium, S. aureus, Enterococcus faecalis, and Enterococcus hirae were resistant to the different classes of antibiotics (Resende et al. 2014; Tahrani et al. 2015). Worryingly, the potentially pathogenic and antibiotic-resistant bacteria were prevented in foodstuffs by entering the food chain via contaminated crops or groundwater. S. aureus and Shigella isolated from fresh vegetables exhibited resistance against sulfamethoxazole, tetracycline, and fluoroquinolones (Hassan et al. 2011). Ruimy et al. (2010) reported the gram-negative bacteria expressing resistance to the antibiotics observed in the organic and conventional fruits and vegetables. In the conditions that absence of antibiotic contaminations, ARGs were still detected in deep rock terrestrial subsurface (Brown and Balkwill 2009), Antarctic marine waters (De Souza et al. 2006), and deep Greenland glacier ice core (Miteva et al. 2004), which was associated with earth material circulation system. Material circulation system played an important role in large-scale spread of antibiotics and antibiotic resistance. When pathogens were released into the natural environment system through the wastewater treatment processes, they can be irrigated into agricultural soils and then permeated into the groundwater system. Treated hospital wastewater may be directly released into rivers and lakes or even as drinking water after further treatment. These potential routes of spread lead to ARGs were detected anywhere contained vital signs. However, the dissemination behavior of pathogens and ARGs just were negative relationship with treatment pathogen infectious diseases. Meanwhile, the potentiality of foodborne pathogens and commensal bacteria to humans and animals resistant to antibiotics was increasing. These behaviors were serious threats to the environmental microbial community structure and effectively treat bacterial infectious diseases in human medicine. The diverse dissemination mechanism of ARGs contributed to sustained presence persistence, transfer, and propagation in the microbial communities. As the role of material recycling systems, ARB and ARGs can be introduced into a wide variety of ecological systems for microbial growth.
Conclusions
This study investigated the occurrence of 11 antibiotics and 12 ARGs in the wastewater samples from different hospitals in Xinjiang. The high concentrations of antibiotics and the relative concentrations of ARGs were observed. The findings revealed that qnrB, qepA, sulA, and their corresponding antibiotics exhibited strong correlations. However, the relationships between TC and several noncorresponding antibiotic genes also showed significance. The relationships between the contamination level of ARGs and the concentrations of antibiotics should be further explored because the majority of ARGs showed weakly correlated levels of antibiotics. Furthermore, integrons played a significant role in influencing the dissemination and abundance of ARGs in environment systems. On account of the removal efficiencies of antibiotics and the steady dynamics of ARGs in the wastewater treatment process, they were released into environment systems, including rivers, lakes, and soils. This study suggested that the WWTPs of hospitals were a major reservoir for the evolution and dissemination of antibiotics and ARGs. The spread of antibiotics and ARGs in environment systems would pose a serious risk to ecosystems and human health. Further research should improve the removal efficiencies of antibiotics and ARGs and the propagation mechanisms of ARGs in the microbial community.
References
Adelowo OO, Fagade OE (2009) The tetracycline resistance gene tet39 is present in both Gram-negative and Gram-positive bacteria from a polluted river, Southwestern Nigeria. Lett Appl Microbiol 48:167–172. doi:10.1111/j.1472-765X.2008.02523.x
Akinbowale OL, Peng H, Barton MD (2007) Diversity of tetracycline resistance genes in bacteria from aquaculture sources in Australia. J Appl Microbiol 103:2016–2025. doi:10.1111/j.1365-2672.2007.03445.x
Aktan Y, Tan S, Icgen B (2013) Characterization of lead-resistant river isolate Enterococcus faecalis and assessment of its multiple metal and antibiotic resistance. Environ Monit Assess 185(6):5285–5293. doi:10.1007/s10661-012-2945-x
Aminov RI, Garrigues JN, Mackie RI (2001) Molecular ecology of tetracycline resistance: development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protection proteins. Appl Environ Microbiol 67:22–32. doi:10.1128/AEM.67.1.22-32.2001
Belinda H, Elisabet M, Meritxell G, Pilar L, Marcelo P, Joan A, Damià B, Jose LB, Sara RM, Rafael M (2013) Exploring the links between antibiotic occurrence, antibiotic resistance, and bacterial communities in water supply reservoirs. Sci Total Environ 456–457:161–170. doi:10.1016/j.scitotenv.2013.03.071
Berglund B, Fick J, Lindgren PE (2015) Urban wastewater effluent increases antibiotic resistance gene concentrations in a receiving northern European river. Environ Toxicol Chem 32:192–196. doi:10.1002/etc.2784
Brown MG, Balkwill DL (2009) Antibiotic resistance in bacteria isolated from the deep terrestrial subsurface. Microb Ecol 57(3):484–93. doi:10.1007/s00248-008-9431-6
Cattoir V, Poirel L, Rotimi V, Soussy CJ, Nordmann P (2007) Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J Antimicrob Chemother 60:394–397. doi:10.1093/jac/dkm204
Chen B, Liang X, Nie X, Huang X, Zou S, Li X (2015) The role of class I integrons in the dissemination of sulfonamide resistance genes in the Pearl River and Pearl River Estuary, South China. J Hazard Mater 282:61–67. doi:10.1016/j.jhazmat.2014.06.010
Cheng W, Chen H, Su C, Yan S (2013) Abundance and persistence of antibiotic resistance genes in livestock farms: a comprehensive investigation in eastern China. Environ Int 61C:1–7. doi:10.1016/j.envint.2013.08.023
Dahmen S, Poirel L, Mansour W, Bouallègue O, Nordmann P (2010) Prevalence of plasmid-mediated quinolone resistance determinants in Enterobacteriaceae from Tunisia. Clin Microbiol Infect 16:1019–1023. doi:10.1111/j.1469-0691.2009.03010.x
De Souza MJ, Nair S, Loka Bharathi PA, Chan DD (2006) Metal and antibiotic-resistance in psychrotrophic bacteria from Antarctic Marine waters. Ecotoxicology 15(4):379–384
Dorival-García N, Zafra-Gómez A, Navalón A, González J, Vílchez JL (2013) Removal of quinolone antibiotics from wastewaters by sorption and biological degradation in laboratory-scale membrane bioreactors. Sci Total Environ 442:317–328. doi:10.1016/j.scitotenv.2012.10.026
Gao P, Mao D, Luo Y, Wang L, Xu B, Xu L (2012) Occurrence of sulfonamide and tetracycline resistant bacteria and resistance genes in aquaculture environment. Water Res 46:2355–2364. doi:10.1016/j.watres.2012.02.004
Goldstein C, Lee MD, Sanchez S, Hudson C, Phillips B, Register B, Grady M, Liebert C, Summers AO, White DG, Maurer JJ (2001) Incidence of class 1 and 2 integrases in clinical and commensal bacteria from livestock, companion animals, and exotics. Antimicrob Agents Chemother 45:723–726. doi:10.1128/AAC.45.3.723-726.2001
Guillard T, Moret H, Brasme L, Carlier A, Vernet-Garnier V, Cambau E, de Champs C (2011) Rapid detection of qnr and qepA plasmid-mediated quinolone resistance genes using real-time PCR. Diagn Microbiol Infect Dis 70:253–259. doi:10.1016/j.diagmicrobio.2011.01.004
Hassan SA, Altalhi AD, Gherbawy YA, EI-Deeb BA (2011) Bacterial load of fresh vegetables and their resistance to the currently used antibiotics in Saudi Arabia. Foodborne Pathog Dis 8(9):1011–1018. doi:10.1089/fpd.2010.0805
Huang MH, Zhang W, Liu C, Hu HY (2015) Fate of trace tetracycline with resistant bacteria and resistance genes in an improved AAO wastewater treatment plant. Process Saf Environ Prot 93:68–74. doi:10.1016/j.psep.2014.04.004
Jiang H, Zhang D, Xiao S, Geng C, Zhang X (2013a) Occurrence and sources of antibiotics and their metabolites in river water, WWTPs, and swine wastewater in Jiulongjiang River basin, south China. Environ Sci Pollut Res 20:9075–9083. doi:10.1007/s11356-013-1924-2
Jiang L, Hu X, Xu T, Zhang H, Sheng D, Yin D (2013b) Prevalence of antibiotic resistance genes and their relationship with antibiotics in the Huangpu River and the drinking water sources, Shanghai, China. Sci Total Environ 458–460:267–272. doi:10.1016/j.scitotenv.2013.04.038
Karthikeyan KG, Meyer MT (2006) Occurrence of antibiotics in wastewater treatment facilities in Wisconsin, USA. Sci Total Environ 361:196–207. doi:10.1016/j.scitotenv.2005.06.030
Kim SR, Nonaka L, Suzuki S (2004) Occurrence of tetracycline resistance genes tet(M) and tet(S) in bacteria from marine aquaculture sites. FEMS Microbiol Lett 237(1):147–156. doi:10.1016/j.femsle.2004.06.026
Li J, Wang T, Shao B, Shen J, Wang S, Wu Y (2012) Plasmid-mediated quinolone resistance genes and antibiotic residues in wastewater and soil adjacent to swine feedlots: potential transfer to agricultural lands. Environ Health Perspect 120:1144–1149. doi:10.1289/ehp.1104776
Li W, Shi Y, Gao L, Liu J, Cai Y (2013) Occurrence and removal of antibiotics in a municipal wastewater reclamation plant in Beijing, China. Chemosphere 92:435–444. doi:10.1016/j.chemosphere.2013.01.040
Liu M, Zhang Y, Yang M, Tian Z, Ren L, Zhang S (2012) Abundance and distribution of tetracycline resistance genes and mobile elements in an oxytetracycline production wastewater treatment system. Environ Sci Technol 46(14):7551–7557. doi:10.1021/es301145m
Liu L, Liu YH, Wang Z, Liu CX, Huang X, Zhu GF (2014) Behavior of tetracycline and sulfamethazine with corresponding resistance genes from swine wastewater in pilot-scale constructed wetlands. J Hazard Mater 278:304–310. doi:10.1016/j.jhazmat.2014.06.015
Luo Y, Mao D, Rysz M, Zhou Q, Zhang H, Xu L, J J Alvarez P (2010) Trends in antibiotic resistance genes occurrence in the Haihe River, China. Environ Sci Technol 44:7720–7225. doi:10.1021/es100233w
Luo Y, Xu L, Rysz M, Wang Y, Zhang H, Alvarez PJ (2011) Occurrence and transport of tetracycline, sulfonamide, quinolone, and macrolide antibiotics in the Haihe River Basin, China. Environ Sci Technol 45:1827–1833. doi:10.1021/es104009s
MacDougall C, Powell JP, Johnson CK, Edmond MB, Polk RE (2005) Hospital and community fluoroquinolone use and resistance in Staphylococcus aureus and Escherichia coli in 17 US hospitals. Clin Infect Dis 41:435–440. doi:10.1086/432056
Mazel D (2006) Integrons: agents of bacterial evolution. Nat Rev Microbiol 4:608–620. doi:10.1038/nrmicro1462
McKinney CW, Loftin KA, Meyer MT, Davis JG, Pruden A (2010) tet and sul antibiotic resistance genes in livestock lagoons of various operation type, configuration, and antibiotic occurrence. Environ Sci Technol 44:6102–6109. doi:10.1021/es9038165
Miteva VI, Sheridan PP, Brenchley JE (2004) Phylogenetic and physiological diversity of microorganisms isolated from a deep Greenland glacier ice core. Appl Environ Microbiol 70(1):202–213
Nõlvak H, Truu M, Tiirik K, Oopkaup K, Sildvee T, Kaasik A, Mander Ü, Truu J (2013) Dynamics of antibiotic resistance genes and their relationships with system treatment efficiency in a horizontal subsurface flow constructed wetland. Sci Total Environ 461-462C:636–644. doi:10.1016/j.scitotenv.2013.05.052
Pei R, Kim SC, Carlson KH, Pruden A (2006) Effect of river landscape on the sediment concentrations of antibiotics and corresponding antibiotic resistance genes (ARG). Water Res 40:2427–2435. doi:10.1016/j.watres.2006.04.017
Resende JA, Silva VL, de Oliveira TL, de Oliveira FS, da Costa CJ, Otenio MH, Diniz CG (2014) Prevalence and persistence of potentially pathogenic and antibiotic resistant bacteria during anaerobic digestion treatment of cattle manure. Bioresour Technol 153:284–91. doi:10.1016/j.biortech.2013.12.007
Robicsek A, Jacoby GA, Hooper DC (2006) The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect Dis 6:629–640. doi:10.1016/S1473-3099(06)70599-0
Rodriguez-Mozaz S, Chamorro S, Marti E, Huerta B, Gros M, Sànchez-Melsió A, Borrego CM, Barceló D, Balcázar JL (2015) Occurrence of antibiotics and antibiotic resistance genes in hospital and urban wastewaters and their impact on the receiving river. Water Res 69:234–242. doi:10.1016/j.watres.2014.11.021
Ruimy R, Brisabois A, Bernede C, Skurnk D, Barnat S, Arlet G, Momcilovic S, Elbaz S, Moury F, Vibet MA, Courvalin P, Guillemot D, Andremont A (2010) Organic and conventional fruits and vegetables contain equivalent counts of Gram-negative bacteria expressing resistance to antibacterial agents. Environ Microbiol 12(3):608–615. doi:10.1111/j.1462-2920.2009.02100.x
Sarmah AK, Meyer MT, Boxall AB (2006) A global perspective on the use, sales, exposure pathways, occurrence, fate and effects of veterinary antibiotics (VAs) in the environment. Chemosphere 65:725–759. doi:10.1016/j.chemosphere.2006.03.026
Sayah RS, Kaneene JB, Johnson Y, Miller R (2005) Patterns of antimicrobial resistance observed in Escherichia coli isolates obtained from domestic-and wild-animal fecal samples, human septage, and surface water. Appl Environ Microbiol 71:1394–1404. doi:10.1128/AEM.71.3.1394-1404.2005
Su HC, Ying GG, Tao R, Zhang RQ, Zhao JL, Liu YS (2012) Class 1 and 2 integrons, sul resistance genes and antibiotic resistance in Escherichia coli isolated from Dongjiang River, South China. Environ Pollut 169:42–49. doi:10.1016/j.envpol.2012.05.007
Tahrani L, Soufi L, Mehri I, Najjari A, Hassan A, Van Loco J, Reyns T, Cherif A, Mansour HB (2015) Isolation and characterization of antibiotic-resistant bacteria from pharmaceutical industrial wastewaters. Microb Pathog 89:54–61. doi:10.1016/j.micpath.2015.09.001
Tao CW, Hsu BM, Ji WT, Hsu TK, Kao PM, Hsu CP, Shen SM, Shen TY, Wan TJ, Huang YL (2014) Evaluation of five antibiotic resistance genes in wastewater treatment systems of swine farms by real-time PCR. Sci Total Environ 496:116–121. doi:10.1016/j.scitotenv.2014.07.024
Udikovic-KoLic N, Wichmann F, Broderick NA, Handelsman J (2014) Bloom of resident antibiotic-resistant bacteria in soil following manure fertilization. Proc Natl Acad Sci U S A 111(42):15202–7. doi:10.1073/pnas.1409836111
Wu C, Spongberg AL, Witter JD (2009) Sorption and biodegradation of selected antibiotics in biosolids. J Environ Sci Health 44:454–461. doi:10.1080/10934520902719779
Xu J, Xu Y, Wang H, Guo C, Qiu H, He Y, Zhang Y, Li X, Meng W (2015) Occurrence of antibiotics and antibiotic resistance genes in a sewage treatment plant and its effluent-receiving river. Chemosphere 119:1379–1385. doi:10.1016/j.chemosphere.2014.02.040
Yang SF, Lin CF, Wu CJ, Ng KK, Lin AY, Hong PK (2012) Fate of sulfonamide antibiotics in contact with activated sludge—sorption and biodegradation. Water Res 46:1301–1308. doi:10.1016/j.watres.2011.12.035
Zhang XX, Zhang T (2011) Occurrence, abundance, and diversity of tetracycline resistance genes in 15 sewage treatment plants across China and other global locations. Environ Sci Technol 45(7):2598–2604. doi:10.1021/es103672x
Zhang XX, Zhang T, Zhang M, Fang HH, Cheng SP (2009) Characterization and quantification of class 1 integrons and associated gene cassettes in sewage treatment plants. Appl Microbiol Biotechnol 182:1169–1177. doi:10.1007/s00253-009-1886-y
Zhou T, Lu J, Tong Y, Li S, Wang X (2014) Distribution of antibiotic resistance genes in Bosten Lake, Xinjiang, China. Water Sci Technol 70(5):925–931. doi:10.2166/wst.2014.321
Acknowledgments
The authors are grateful to the Key Laboratory for Green Processing of Chemical Engineering of Xinjiang Bingtuan, School of Chemistry and Chemical Engineering, Shihezi University. This research was supported by the National Science Foundation of China (No. 21267019).
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Philippe Garrigues
Rights and permissions
About this article
Cite this article
Li, C., Lu, J., Liu, J. et al. Exploring the correlations between antibiotics and antibiotic resistance genes in the wastewater treatment plants of hospitals in Xinjiang, China. Environ Sci Pollut Res 23, 15111–15121 (2016). https://doi.org/10.1007/s11356-016-6688-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-016-6688-z