Abstract
The effect of copper oxide nanoparticles (CuONPs) on physiological and molecular level responses were studied in Arabidopsis thaliana. The seedlings were exposed to different concentrations of CuONPs (0, 0.5, 1, 2, 5, 10, 20, 50, and 100 mg/L) for 21 days in half strength Murashige and Skoog medium. The plant biomass significantly reduced under different concentrations (2, 5, 10, 20, 50, and 100 mg/L) of CuONPs stress. Exposure to 2, 5, 10, 20, 50, and 100 mg/L of CuONPs has resulted in significant reduction of total chlorophyll content. The anthocyanin content significantly increased upon exposure to 10, 20, 50, and 100 mg/L of CuONPs. Increased lipid peroxidation was observed upon exposure to 5, 10, and 20 mg/L of CuONPs and amino acid proline content was significantly high in plants exposed to 10 and 20 mg/L of CuONPs. Significant reduction in root elongation was observed upon exposure to 0.5–100 mg/L of CuONPs for 21 days. Exposure to CuONPs has resulted in retardation of primary root growth, enhanced lateral root formation, and also resulted in loss of root gravitropism. Staining with phloroglucionol detected the deposition of lignin in CuONPs-treated roots. Histochemical staining of leaves and roots of CuONPs-exposed plants with nitroblue tetrazolium and 3′3′-diaminobenzidine showed a concentration-dependant increase in superoxide and hydrogen peroxide formation in leaves and roots of CuONPs-exposed plants. Cytotoxicity was observed in root tips of CuONPs-exposed plants as evidenced by increased propidium iodide staining. Real-time PCR analysis showed significant induction of genes related to oxidative stress responses, sulfur assimilation, glutathione, and proline biosynthesis under CuONPs stress.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Engineered nanoparticles (ENPs) are widely used in several industrial, commercial, and medical products (www.nanotechproject.org). Due to their unique physicochemical properties, metal oxide nanoparticles (MONPs) are widely used in various consumer products such as in sunscreens, medicine, inks, food storage containers, and in diesel fuel (www.nanotechproject.org). Among the different MONPs, copper oxide nanoparticles (CuONPs) are used in various applications such as in electronics, air and liquid filtration, ceramics, wood preservation, bioactive coatings, films, textiles, skin products, inks, and in lubricant oils (Yang et al. 2006). The widespread use of CuONPs has raised concerns over their potential toxic impacts on ecosystem and human health due to their release from various products to the environment (Chen et al. 2012). Previous studies has shown that exposure to CuONPs caused toxic effects on aquatic organisms such as protozoa, crustaceans, algae, and zebrafish (Yoon et al. 2007; Griffitt et al. 2007; Aruoja et al. 2009; Gomes et al. 2010). Karlsson et al. (2008) reported that among the different MONPs tested, CuONPs caused maximum toxicity to human lung epithelial cells. Lee et al. (2008) reported that exposure to CuONPs has resulted in their uptake, bioaccumulation, and translocation in mung bean (Phaseolus radiates) and wheat (Triticum aestivum). However, apart from studying the uptake and bioaccumulation, detailed studies on the effect of CuONPs at the physiological and molecular level has not been undertaken in plants.
The toxicity of ENPs in plants is related to their size, concentration, surface chemistry, and dissolution to metal ions leading to interaction with different subcellular compartments of the cell resulting in excessive production of reactive oxygen species (ROS) causing oxidative stress, cytotoxicity, genotoxicity, necrosis, and apoptosis (Dietz and Herth 2011). In order to maintain the redox homeostasis, plants possess various enzymatic and nonenzymatic defense mechanisms in cells. The enzymatic antioxidant systems include superoxide dismutases (SODs, EC 1.15.1.1), catalases (CATs, EC 1.11.1.6), and ascorbate peroxidases (APXs, EC 1.11.1.11) (Mittler et al. 2004). The nonenzymatic antioxidant machinery in plants includes low molecular weight sulfur-containing compounds such as glutathione (GSH) and amino acid proline (Szabados and Savouré 2010; Gill and Tuteja 2010). In addition, sulfur assimilation plays an important role in the formation of sulfur-containing amino acids (Cys and Met), synthesis of proteins, vitamins, chlorophyll and GSH which are important for the growth and oxidative stress tolerance in plants (Saito 2000). Various genes are involved in the sulfur assimilation viz. ATP sulfurylase (ATPS), 3′-phosphoadenosine 5′-phosphosulfate reductase (APR), sulfite reductase (SiR) and cysteine synthase (CS). The final steps of GSH biosynthesis is catalyzed by glutamate-cysteine ligase (GCL), and glutathione synthetase (GS). In plants, proline is derived from glutamate or ornithine amino acid precursors through various enzymatic steps and the proline biosynthesis in the glutamate pathway is controlled by the bifunctional 1-pyrroline-5-carboxylate (P5CS) enzyme (Szabados and Savouré 2010).
In this study, the toxic effects of CuONPs at the physiological and molecular levels were studied in Arabidopsis thaliana which is currently used as a model plant in nanotoxicology studies (Ma et al. 2013). A. thaliana plants were grown in ½ MS agar medium containing different concentrations (i.e., 0, 0.5, 1, 2, 5, 10, 20, 50, and 100 mg/L) of CuONPs for 21 days under growth chamber conditions. At the physiological level, changes in plant growth, biomass, root elongation and root system architecture, ROS generation, cell death, root lignification, lipid peroxidation, total chlorophyll, anthocyanin, and proline content were studied. At the molecular level, transcriptional modulation of genes involved in various stress responses viz. MSD, CSD1, CSD2, CAT2, CAT3, APX1, APX2, ATPS, APR, SiR, CS, GCL, GS, P5CS1, and P5CS2 were studied. Characterization of CuONPs using transmission electron microscopy (TEM) and dynamic light scattering (DLS) were performed. The dissolution and plant uptake of CuONPs were analyzed using inductively coupled plasma-mass spectrometer (ICP-MS). Since it has been reported from previous studies that the biological effects of ENPs in aqueous solutions are also associated to the concentration of released metal ions (Wu et al. 2012), in this study, the toxic effects of CuONPs at different levels were compared with the toxicity of Cu2+ ions (0.1 and 0.2 mg/L), and medium without CuONPs was used as the negative control.
Materials and methods
Nanoparticles characterization
Copper oxide nanoparticles (purity of 99.9 %; surface area 29 m2/g) were purchased from Sigma-Aldrich, St. Louis, MO, USA. The morphology of CuONPs in deionised water and ½ strength MS medium was characterized using TEM LIBRA 120 at 80 kV (Carl Zeiss, Oberkochen, and Baden-Württemberg, Germany). TEM samples were prepared by placing 10 μL of CuONPs suspensions on carbon-coated copper grids (Tedpella Inc., Reading, USA) and dried O/N before imaging. Size distribution of CuONPs in deionized water was determined using photal dynamic light scattering spectrometer (DLS-7000, Otsuka Electronics Co., Inc., Osaka, Japan). The amount of cupric ion released from CuONPs in deionized water containing the highest concentration of CuONPs (i.e., 100 mg/L) was measured after sonication for 1 h and filtering through a 0.2-μm nylon membrane (Chromedisc® syringe filter, South Korea) using ICP-MS (Varian 820-MS, Palo Alto, CA, USA; detection limit for 65Cu <2 ng/L). The cupric ion released from three samples was analyzed using ICP-MS.
Plant exposure to CuONPs
A. thaliana (wild type, Columbia ecotype) seeds were surface sterilized with 70 % ethanol for 2 min and 1.5 % (w/v) sodium hypochlorite solution for 15 min. The seeds were washed with sterilized distilled water three times to remove the traces of the disinfectant. Half strength MS medium [pH 5.7; 20 g/L sucrose; 8 g/L agar (Duchefa Biochemie, Haarlem, The Netherlands)] was prepared and autoclaved at 121 °C for 15 min. Different concentrations of CuONPs (0, 0.5, 1, 2, 5, 10, 20, 50, and 100 mg/L) were added directly to the medium after autoclaving and shaken vigorously in order to break the agglomerates. Twenty-five milliliter of medium was poured on to 90 × 15 mm petri dishes (SPL Life Sciences, Gyeonggi-do, South Korea) and solidified immediately by keeping at 4 °C to avoid precipitation of CuONPs. After solidification, 15 A. thaliana seeds were sown in each plate (the floated seeds were removed to ensure that mature seeds were being used) and incubated under dark conditions at 4 °C O/N to break seed dormancy. The petri dishes were transferred to growth chamber (Hanbaek Scientific Co., Gyeonggi-do, South Korea) and incubated for 3 days horizontally and then kept vertically under conditions of 16 ± 8 h of light/dark, with light intensity 100 mE/m2/s at 26 ± 1 °C for 21 days. Cupric ion treatment of 0.1 and 0.2 mg/L (in the form of Cu(NO3)2) was used to test the effect of dissolved ions (concentration was selected based on the release of cupric ions from CuONPs). For each treatment and control, four replicate plates with 15 plants in each plate was maintained and the experiment was repeated twice.
Analysis of copper content in plants
The uptake of CuONPs was determined using A. thaliana plants grown in medium containing 20 mg/L of CuONPs for 21 days. The plants which were grown in medium containing 20 mg/L of CuONPs was selected for the analysis of Cu content in plant tissues, since sufficient plant material (shoot and root) was not available in plants which were exposed to 50 and 100 mg/L of CuONPs. The plants (30 mg, including shoot and roots) were thoroughly washed with sterilized distilled water several times and were dried at 75 °C for 24 h and subsequently digested with concentrated HNO3 at 115 °C for 1 h. The digests were diluted to 25 mL with deionized water and filtered through 0.2 μm nylon filters (Chromedisc® syringe filter) and analyzed using ICP-MS.
Biomass and root length
The root length of the control, CuONPs (0.5, 1, 2, 5, 10, 20, 50, and 100 mg/L) and Cu2+ ions (0.1 and 0.2 mg/L)-exposed A. thaliana plants were measured after 21 days of exposure using a ruler. For calculating the total plant biomass (Fresh weight, FW), the control, CuONPs- (0.5, 1, 2, 5, 10, 20, 50, and 100 mg/L) and Cu2+ ion (0.1 and 0.2 mg/L)-exposed plants were carefully removed after 21 days from medium and washed with demonized water. The root length and fresh weight of 15 seedlings from the control and CuONPs-exposed plants were calculated from two replications.
Total chlorophyll, anthocyanin, and proline contents
Total chlorophyll and anthocyanin content from the 21-day-old control, CuONPs- (0.5, 1, 2, 5, 10, 20, 50, and 100 mg/L), and Cu2+ ion (0.1 and 0.2 mg/L)-exposed A. thaliana plants were determined using modified method of Lichtenthaler (1987) and Rabino and Mancinelli (1986), respectively. The proline amino acid content of 21-day-old control, CuONPs- (0.5, 1, 2, 5, 10, and 20 mg/L), and Cu2+ ion (0.1 and 0.2 mg/L)-exposed plants were determined according to Bates (1973). The proline content of plant exposed to 50 and 100 mg/L of CuONPs was not checked since the root growth was very poor. The experiment was repeated four times using plants collected from four replications (Supplementary protocols 1, 2 and 3).
Measurement of lipid peroxidation
The level of lipid peroxidation was estimated using the method of Heath and Packer (1968) using 21-day-old control, CuONPs- (0.5, 1, 2, 5, 10, and 20 mg/L) and Cu2+ ion (0.1 and 0.2 mg/L)-exposed A. thaliana plants. The plants which were exposed to the abovementioned concentrations of CuONPs were selected to get enough plant material (shoots and roots) for the analysis since root elongation was affected at 50 and 100 mg/L of CuONPs. Thirty milligram of the plant samples were homogenized in 5 mL of 5 % trichloroacetic acid (TCA). The extracts were centrifuged at 7,000 × g for 10 min at room temperature and the supernatant (2 mL) was mixed with 2 mL of 0.67 % (w/v) thiobarbituric acid (TBA), boiled at 95 °C for 30 min and cooled immediately after centrifugation. Absorbance of the supernatant was read at 532 and 600 nm using a UV-Vis spectrophotometer (Optizen UV/Vis spectrophotometer, Mecasys Co. Ltd., Seoul, South Korea). The final malondialdehyde (MDA) concentration was calculated using the extinction coefficient 155 mM/cm. The experiment was repeated four times using plants collected from four replications.
In vivo detection of ROS, cell viability, and lignification
The effect of exposure to different concentrations of CuONPs in superoxide and H2O2 formation in leaves of plants exposed to 0, 0.5, 1, 2, 5, 10, 20, 50, and 100 mg/L of CuONPs and roots of plants exposed to 0, 1, 2, 5, 10, and 20 mg/L of CuONPs was determined as follows. In vivo detection of superoxide formation in leaves of A. thaliana plants was done by staining with nitroblue tetrazolium (NBT) which forms dark blue insoluble formazan when it reacts with superoxides (Fryer et al. 2002). The in vivo imaging of H2O2 formation in leaves and roots were checked using histochemical staining with 3′3′-diaminobenzidine (DAB) (Thordal-Christensen et al. 1997). The leaves were cleared from chlorophylls after NBT and DAB staining by boiling for 10 min in acetic acid-glycerol-ethanol (1:1:3[v/v/v]) solution in a dry bath.
Root cell viability of A. thaliana plants exposed to different concentrations of CuONPs (0, 1, 2, 5, 10, and 20 mg/L) was assayed by staining with 3 mg/L of propidium iodide (PI) solution for 15 min as described by Lequeux et al. (2010). The stained samples were mounted on 26 × 76 mm microscope slides (Citoglas®, Jiangsu, China) and imaged using a fluorescence microscope (BX61-32 FDIC, Olympus, Japan).
For checking lignin accumulation, the roots of the control and CuONPs (1, 2, 5, 10, and 20 mg/L)-exposed A. thaliana plants were incubated in 1 % (w/v) phloroglucinol-HCl 6 N solution for 5 min as described by Rogers et al. (2005). The stained samples were mounted on 26 × 76 mm microscope slides (Citoglas®, Jiangsu, China) and photographed using Nikon 80i (Nikon, Japan) microscope equipped with digital camera.
Total RNA isolation and cDNA synthesis
Total RNA was isolated from 21-day-old control, CuONPs- (2, 5, 10, and 20 mg/L), and Cu2+ ion (0.2 mg/L)-exposed A. thaliana plants using RNA isolation and cleaned up using RNA-Clean up kit (Qiagen, Hilden, Germany). The quality and quantity of RNA preparation was verified by agarose gel electrophoresis and absorbance spectrophotometry (A260/A280 > 1.8) (Optizen UV/Vis spectrophotometer). One microgram of total RNA was used for cDNA synthesis in 20 μL reaction volume using QuantiTect® reverse transcription kit integrated with genomic DNA contamination removal step (Qiagen) as per the manufacturer’s instructions. The primers for different genes viz. MSD, CSD1, CSD2, CAT2, CAT3, APX1, APX2, P5CS1, P5CS2, and ACT2 were designed using online Primer3 program (http://frodo.wi.mit.edu/primer3/). The primers for sulfur assimilation and GSH biosynthesis genes viz. ATPS, APR, SiR, CS, GCL, and GS were used as reported by previously by Ma et al. (2013) (Supplementary Table 1). The substrate specificity of the primers were verified by running PCR (T100 thermal cycler, Bio-Rad, Hercules, California, USA) using a representative cDNA preparation and by visualizing on a 1.5 % agarose gel stained with ethidium bromide.
Real-time RT-PCR
The real-time RT-PCR reaction included 1 μL of cDNA, 0.2 μM of corresponding forward and reverse primers, and 10 μL of 2X IQ SYBR Green Super Mix (Bio-Rad) in a final reaction volume of 20 μL. The reactions were run with an initial denaturing at 95 °C for 7 min followed by 45 cycles of 95 °C for 15 s, 55 °C for 1 min, and 72 °C for 15 s, and a melting curve analysis was done. Amplification and detection were performed using Chromo-4 opticon monitor RT-PCR detection system (Bio-Rad) and accompanying software according to the manufacturer’s instructions. The reactions were run for three biological replicates for each gene within each treatment and control and repeated twice. The expression level of each gene under different exposure conditions were calculated relative to the expression level of AtACT2 mRNA (Ma et al. 2013).
Data analysis
Cycle threshold (Ct) values were converted to relative gene expression levels by 2−ΔΔCt method (Livak and Schmittgen 2001) using the gene expression analysis software (Bio-Rad). One-way analysis of variance (ANOVA) was done with SPSS 12.0 KO (SPSS Inc., Chicago, IL, USA). Dunnett’s post-doc test was done to determine the effect of different exposures on various parameters. A probability level of p < 0.05, p < 0.01, and p < 0.001 was considered significant.
Results
CuONPs characterization
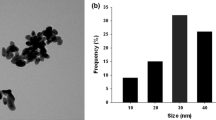
The TEM analysis showed dispersion of CuONPs in deionized water and based on the DLS analysis; the average size of the particles was 30 nm (Supplementary Fig. 1A, B, C). However, agglomeration of CuONPs was observed in growth medium (Supplementary Fig. 1D, E).
Effect of CuONPs on plant growth
The effect of exposure to different concentrations of CuONPs and Cu2+ ions on A. thaliana growth was evaluated after 21 days of exposure. As compared to control and Cu2+ ion-exposed plants, significant reduction in plant growth and plant biomass was observed with increasing concentrations of CuONPs exposure (Fig. 1a–i). The average fresh weight of seedlings exposed to 2, 5, 10, 20, 50, and 100 mg/L of CuONPs significantly reduced as compared to the controls (Fig. 1j). However, exposure to different concentrations of CuONPs was more evident in root growth. Significant reduction in root growth was observed upon exposure to 0.5, 1, 2, 5, 10, 20, 50, and 100 mg/L of CuONPs (Fig. 1h). More than 50 % reduction in average fresh weight and root growth was evident in plants which were exposed to 10 mg/L of CuONPs. However, the fresh weight and root length of plants exposed to 0.1 and 0.2 mg/L of Cu2+ ions was not significantly different from the control (Fig. 1j, h and Suppl. Fig. 2a). Exposure to CuONPs above 2 mg/L has reduced the primary root growth, increased the density of lateral root formation, and also resulted in loss of root gravitropism (Suppl. Fig. 2B, C, D, E).
a Phenotypes of A. thaliana seedlings grown in the presence of different concentrations of copper oxide nanoparticles for 21 days (left to right: control, 0.5, 1, 2, 5, 10, 20, 50, and 100 mg/L of CuONPs, respectively). b Average fresh weight and c root lengths of plants exposed to designated concentrations of Cu2+ ions and copper oxide nanoparticles for 21 days. Data are mean ± SE of four replicates. Asterisks indicate significant difference at p < 0.05*, p < 0.01**, and p < 0.001***
Analysis of copper content in plants
ICP-MS analysis showed dissolution of Cu2+ ions (0.08 μg/L) in deionized water after 24 h. Analysis of copper content in A. thaliana plants which were exposed to 20 mg/L CuONPs showed the presence of 82.6 ± 10.3 mg/Kg DW of Cu and the control plants also contained trace amounts of Cu.
Effect of CuONPs on chlorophyll, anthocyanin, and proline contents
The total chlorophyll content of A. thaliana plants did not change significantly upon exposure to 0.5 and 1 mg/L of CuONPs (Fig. 2a). However, significant reduction in the total chlorophyll content was observed upon exposure to 2, 5, 10 (p < 0.05), 20 (p < 0.01), 50, and 100 (p < 0.001) mg/L of CuONPs (Fig. 2a). Though not in a significant manner, an increase in the total chlorophyll content was observed in plants which were exposed to 0.1 and 0.2 mg/L of Cu2+ ions (Fig. 2a). The total anthocyanin content of the control, Cu2+ ions (0.1 and 0.2 mg/L) and 0.5, 1, 2, and 5 mg/L of CuONPs-exposed seedlings has not changed significantly as compared to the controls (Fig. 2b). However, exposure to 10, 20, 50, and 100 mg/L of CuONPs (p < 0.05 and p < 0.001) has resulted in significant increase in the total anthocyanin content of A. thaliana plants (Fig. 2b). The total proline content of A. thaliana plants which were exposed to 0.2 mg/L of Cu2+ ions and 2 and 5 mg/L of CuONPs did not change significantly (Fig. 2c). However, significant increase in proline content was observed after exposure to 10 and 20 mg/L (p < 0.05 and p < 0.01, respectively) of CuONPs (Fig. 2c).
a Total chlorophyll, b anthocyanin, c proline contents, and d lipid peroxidation of A. thaliana plants exposed to designated concentrations of Cu2+ ions and copper oxide nanoparticles for 21 days. Data are mean ± SE of four replicates. Asterisks indicate significant difference at p < 0.05*, p < 0.01**, and p < 0.001***
Effect of CuONPs on lipid peroxidation
The effect of exposure to different concentrations of CuONPs and Cu2+ ions on lipid peroxidation was measured as described in the “Materials and methods.” It was observed that exposure to 0.1, 0.2 mg/L of Cu2+ ions and 1–2 mg/L of CuONPs has not resulted in any significant change in lipid peroxidation as compared to the control (Fig. 2d). However, significant increase in lipid peroxidation was observed upon exposure to 5, 10, and 20 mg/L of CuONPs (p < 0.05, p < 0.001, and p < 0.01, respectively) (Fig. 2d).
Effect of CuONPs on ROS generation and root cell death
The effect of different concentrations of CuONPs on ROS generation in A. thaliana plants was assessed by in vivo histochemical staining with NBT and DAB. Staining with NBT showed a gradual increase in the formation of dark blue spots in leaves of seedlings exposed to different concentrations of CuONPs indicating superoxides accumulation (Fig. 3, panel 1A–H). Histochemical staining with DAB showed a gradual dose-dependant increase in the formation of dark brown coloration in leaves of CuONPs-exposed plants indicating excess H2O2 generation (Fig. 3, panel 2 A–H). In roots, DAB staining revealed a gradual increase in H2O2 accumulation with increasing concentrations of CuONPs, with highest accumulation of H2O2 in root tips (Fig. 3, panel 3 A–F).
Panel 1 Effect of different concentrations of copper oxide nanoparticles treatment on superoxides generation in leaves of A. thaliana plants after 21 days of exposure as revealed by NBT staining (A, control; B–H, plants exposed to 0.5, 1, 2, 5, 10, 20, 50, and 100 mg/L of CuONPs, respectively). Panel 2 Effect of different concentrations of copper oxide nanoparticles treatment on hydrogen peroxide generation in leaves of A. thaliana plants after 21 days of exposure as revealed by DAB staining (A, control; B–H, plants exposed to 0.5, 1, 2, 5, 10, 20, 50, and 100 mg/L of CuONPs, respectively). Panel 3 In vivo detection of hydrogen peroxide formation in roots of A. thaliana plants exposed to different concentrations of copper oxide nanoparticles (A, control; B–F, plants exposed to 1, 2, 5, 10, and 20 mg/L of CuONPs, respectively)
The effect of different concentrations of CuONPs on root cell death was assessed by checking the bright-red fluorescence as a result of penetration PI stain through damaged cell membranes and intercalation with genetic material of dead cells (Lequeux et al. 2010). Root cell death was not observed in control as well as in roots of plants exposed to 1 mg/L of CuONPs (Fig. 4, panel 1 A, B). However, PI staining showed a dose-dependant increase in cytotoxicity in lateral root tips of plants which were exposed to 2, 5, 10, and 20 mg/L of CuONPs as revealed by dark red fluorescence (Fig. 4, panel 1 C–F).
Panel 1 Effect of different concentrations of copper oxide nanoparticles on root apex cell death in A. thaliana plants. (A, control and B–F, plants exposed to 1, 2, 5, 10, and 20 mg/L of CuONPs, respectively). Panel 2 Effect of different concentrations of copper oxide nanoparticles on root lignifications in A. thaliana plants. (A, control and B–F, plants exposed to 1, 2, 5, 10, and 20 mg/L of CuONPs, respectively)
Accumulation of lignin in CuONPs-exposed roots
To understand the effect of exposure to different concentrations of CuONPs on lignin accumulation in roots, the roots of control as well as plants which were exposed to 1, 2, 5, 10, and 20 mg/L of CuONPs were incubated in phloroglucinol-HCl stain, which stains lignin in red color. It was observed that exposure to increasing concentrations of CuONPs resulted in a gradual increase in red-colored lignin accumulation in the middle part of the roots (Fig. 4, panel 2 a–f). However, lignin accumulation was not detected in control roots, showing that CuONPs exposure has resulted in lignin deposition in roots.
Changes in the expression of antioxidant genes
Exposure to 0.2 mg/L of Cu2+ ions and CuONPs (2 and 5 mg/L) has not resulted in any significant change in the expression of MSD gene (Fig. 5a). However, a 2.3- and 4.4-fold increase in MSD gene expression was observed upon exposure to 10 and 20 mg/L of CuONPs (p < 0.05 and p < 0.01, respectively) (Fig. 5a). The expression of CSD1 gene has not changed significantly upon exposure to 0.5 mg/L of Cu2+ ions and 20 mg/L of CuONPs (Fig. 5b). However, significant upregulation of CSD1 gene was observed upon exposure to 2, 5, and 10 mg/L of CuONPs (p < 0.05, p < 0.01, and p < 0.05, respectively) (Fig. 5b). Exposure to 2 and 20 mg/L of CuONPs has not changed the expression of CSD2 gene (Fig. 5c). However, significant upregulation of CSD2 gene was observed after exposure to 5 and 10 mg/L of CuONPs (p < 0.05 and p < 0.001, respectively) (Fig. 5c). The expression of CAT2 and CAT3 did not change significantly upon exposure to 0.2 mg/L of Cu2+ ions (Fig. 5d, e). However, significant upregulation of CAT2 and CAT3 genes were observed upon exposure 2, 5, 10, 20 mg/L and 5, 10, 20 mg/L of CuONPs, respectively. The highest expression of CAT2 (4.5-fold increase; p < 0.001) was observed upon exposure to 10 mg/L of CuONPs and that of CAT3 (5.3-fold increase; p < 0.01) was observed after exposure to 20 mg/L of CuONPs (Fig. 5d, e).
Effects of 21-day exposure to 0.2 mg/L of Cu2+ ions and 2, 5, 10, and 20 mg/L of copper oxide nanoparticles on the mRNA expression of a MSD, b CSD1, c CSD2, d CAT2, e CAT3, f APX1, and g APX2 genes in A. thaliana plants. Data are mean ± SE of three replicates. Asterisks indicate significant difference as compared to the control (control = 1) (p < 0.05*, p < 0.01**, and p < 0.001***)
The expression of APX1 gene was significantly upregulated upon exposure to 2, 5, 10, and 20 mg/L of CuONPs (Fig. 5f). However, no significant difference in the expression of APX1 gene was observed after exposure to 0.2 mg/L of Cu2+ ions (Fig. 5f). The expression of APX2 gene showed no significant change after exposure to 0.2 mg/L of Cu2+ ions and 2, 5, and 10 mg/L of CuONPs (Fig. 5g). However, significant upregulation of APX2 gene was observed upon exposure to 20 mg/L of CuONPs (p < 0.05) (Fig. 5g).
Changes in the expression of sulfur assimilation, GSH, and proline biosynthesis genes
The expression of ATPS gene did not change significantly upon exposure to 0.2 mg/L of Cu2+ ions and 2 and 5 mg/L of CuONPs (Fig. 6a). However, exposure to 10 and 20 mg/L of CuONPs has resulted in significant upregulation of ATPS gene (p < 0.05) (Fig. 6a). The APR gene expression has not changed after exposure to 0.2 mg/L of Cu2+ ions and 2 mg/L of CuONPs (Fig. 6b). However, significant upregulation of APR gene was observed upon exposure to 5, 10, and 20 mg/L of CuONPs (p < 0.01) (Fig. 6b). The expression of SiR gene was not significantly altered upon exposure to 0.2 mg/L of Cu2+ ions (Fig. 6c). However, exposure to 2, 5, 10, and 20 mg/L of CuONPs has significantly upregulated SiR gene expression with the highest expression being observed upon exposure to 10 mg/L of CuONPs (Fig. 6c). The CS mRNA expression has not significantly altered upon exposure to 0.2 mg/L of Cu2+ ions and 2 and 5 mg/L of CuONPs (Fig. 6d). However, significant increase of CS gene was observed after exposure to 10 and 20 mg/L of CuONPs (p < 0.05) (Fig. 6d).
Effects of 21 day exposure to 0.2 mg/L of Cu2+ ions and 2, 5, 10, and 20 mg/L of copper oxide nanoparticles on the expression of a sulfate adenylyltransferase (ATPS), b adenosine-5′-phosphosulfate reductase (APR), c sulfite reductase (SiR), d cysteine synthase (CS), e glutamyl-cystein ligase (GCL), f glutathione synthase (GS), g 1-pyrroline-5-carboxylate 1 (P5CS1), and h 1-pyrroline-5-carboxylate 2 (P5CS2) genes in A. thaliana. Data are mean ± SE of three replicates. Asterisks indicate significant difference as compared to the control (control = 1) (p < 0.05*, p < 0.01**, and p < 0.001***)
The expression of GCL gene was significantly increased upon exposure to 2, 5, and 10 mg/L of CuONPs (Fig. 6e). However, GCL gene has not changed significantly upon exposure to 0.2 mg/L of Cu2+ ions and 20 mg/L of CuONPs (Fig. 6e). Significant upregulation of GS gene was observed after exposure to 2, 5, 10, and 20 mg/L of CuONPs (Fig. 6f). However, no significant change in GS gene expression was observed after exposure to 0.2 mg/L of Cu2+ ions (Fig. 6f). The expression of P5CS1 and P5CS2 genes showed no significant change after exposure to 0.2 mg/L of Cu2+ ions and 2 mg/L of CuONPs (Fig. 6g, h). However, P5CS1 was significantly upregulated upon exposure to 10 and 20 mg/L of CuONPs (p < 0.05) (Fig. 6g). The expression of P5CS2 gene has significantly changed after exposure to 5 (p < 0.05), 10, and 20 mg/L of CuONPs (p < 0.01) (Fig. 6h).
Discussion
The present investigation was conducted to understand the toxic effects at the physiological and molecular levels in the model plant A. thaliana upon exposure to CuONPs under in vitro exposure conditions. As reported earlier by Lee et al. (2008), in the present study, we used ½ MS agar medium to avoid precipitation of CuONPs since they are water insoluble. Since there was no information on the phytotoxic concentration of CuONPs in A. thaliana plants, a wide range of CuONPs concentrations were tested in this study. It has been known from previous studies that the phytotoxic dose of different nanoparticles varies between different plants. For example, in the case of CuONPs, Wu et al. (2012) reported that the phytotoxic doses were different for lettuce (12.9 mg/L), radish (397.6 mg/L), and cucumber (175.4 mg/L). In yet another study, Lee et al. (2008) reported that the EC50 values for CuONPs in P. radiates and T. aestivum was 335 and 570 mg/L, respectively. In our studies, it was observed that seed germination was not affected even after exposure to 100 mg/L of CuONPs and this might be due to the protection offered by seed coat during germination. However, signs of phytotoxicity as evidenced by reduction in root growth were evident at concentrations as low as 1 mg/L of CuONPs. It was clear that exposure to 10 mg/L of CuONPs has resulted in more than 50 % reduction in the fresh weight as well as root growth of A. thaliana plants. Retardation of root growth was observed after exposure to 50 and 100 mg/L of CuONPs. In a recent study, Wang et al. (2012) has reported that exposure to even 100 mg/L of CuONPs had no effect on germination in maize but inhibited the growth of maize seedlings.
It was evident from the present study that A. thaliana seedling development was strongly inhibited under increasing concentrations of CuONPs exposure resulting in significant reduction of plant biomass. In a previous report, Wang et al. (2012) has also reported that exposure to CuONPs has affected the growth of maize seedlings. In a recent study, Shaw and Hossain (2013) reported significant reduction in shoot and root growth of rice seedlings exposed to CuONPs. In the present study, it was observed that exposure to 0.1 mg/L of Cu2+ ions has resulted in a nonsignificant increase in total plant biomass. In accordance with our results, Lee et al. (2008) has also reported an increase in plant biomass upon exposure to low concentrations of Cu2+ ions in P. radiates and T. aestivum. The possible reason for the increase in plant biomass is that Cu is an essential micronutrient for plant growth and low concentrations of Cu might have enhanced the plant growth.
In this study, significant reduction in total chlorophyll, an increase in anthocyanin and proline content was observed upon exposure to different concentrations of CuONPs. In addition, significant increase in MDA level, an indication of lipid peroxidation, was observed in CuONPs-stressed plants indicating membrane damage. It is known that, in plants, chloroplasts and mitochondria are the sources of ROS generation (Hernández et al. 1993) and exposure to ENPs could result in oxidative stress either through direct interaction or by dissolution into ions. The significant reduction in chlorophyll content might be due to the reduced shoot biomass upon exposure to higher concentrations of CuONPs or due to the membrane damage as a result of excess lipid peroxidation of chloroplast membranes under oxidative stress (Halliwell and Gutteridge 1989). An increased rate of lipid peroxidation has also been reported from plants and other biological models under CuONPs stress (Wang et al. 2012; Melegari et al. 2013). Recently, it has been reported that exposure to cerium oxide and indium oxide nanoparticles has resulted in significant reduction in plant biomass and total chlorophyll content in A. thaliana plants (Ma et al. 2013). The significant increase in anthocyanin, might be to protect the cells from oxidative stress caused by CuONPs exposure (Solfanelli et al. 2006). Anthocyanins are flavanoids (Tahara 2007) and could act as nonenzymatic antioxidants to protect the cells from oxidative stress by scavenging excess ROS (Gill and Tuteja 2010). The accumulation of proline, as observed in this study, might be to protect the plants from protein denaturation and also to protect the cellular structures from damage due to excess ROS generation (Chiang and Dandekar 1995). Accumulation of proline has been reported from different plants upon exposure to different metals (Tripathi and Gaur 2004; Choudhary et al. 2007).
In the present study, it was observed that the root growth of A. thaliana plants was adversely affected even at lowest concentration of CuONPs exposure. At concentrations of 50 and 100 mg/L of CuONPs, the root growth of A. thaliana plants were stunted and failed to penetrate the into the growth medium. Moreover, remodeling of root system architecture characterized by an inhibition of primary root growth and the stimulation of lateral root formation was also observed under CuONPs stress. The possible reason for the changes in root system architecture under CuONPs stress might be due to hormonal imbalances. For example, remodeling of root system architecture and enhanced lateral root formation upon exposure to excess Cu has been reported in A. thaliana plants and it was suggested that changes in hormonal status, especially the auxin concentrations, was responsible for the remodeling of root system architecture (Lequeux et al. 2010). In accordance with our results, previous studies has also shown that exposure to various MoNPs has resulted in the inhibition or reduction of root growth in plants (Lin and Xing 2008; Asli and Neumann 2009). In an earlier study, Liu et al. (2010) has reported loss of root gravitropism in A. thaliana plants exposed to water soluble fullerenes and it was speculated that the changes are caused due to the hormonal imbalances as a result of nanoparticles exposure (Liu et al. 2010). Moreover, in the present study, for the first time, we observed that CuONPs stress has resulted in lignin deposition in roots of A. thaliana roots as evidenced by red coloration after staining with phloroglucinol stain. Lignin deposition due to excess Cu exposure in A. thaliana roots has been reported earlier (Lequeux et al. 2010). The lignifications of roots under excess CuONPs stress, as observed in this study, might be due to the uptake and dissolution of CuONPs. For example, Wang et al. (2012) reported that CuONPs were transported from roots to shoots via xylem and translocated from shoots back to roots via phloem and they speculated that, during this translocation, CuONPs could be reduced to from Cu ions. Moreover, Shi et al. (2014) proposed that the dissolution of CuONPs could be promoted due to the pH reduction and the interaction with organic acids and proteins in plant tissues and cells. Since, in this study, lignification as evidenced by red coloration was only observed in the middle part of the root, the CuONPs might have translocated through the vascular tissues and the subsequent dissolution to Cu ions could have resulted in the lignifications of vascular tissues. Since the lignifications of root cells will limit the efflux of metals from the vascular cylinder and will result in the reduction of minerals in plants (Lequeux et al. 2010). Therefore, further studies are required to understand the lignification of root tissues under CuONPs stress, the translocation of the mineral through the roots to the shoots, and also on effect of CuONPs exposure on total mineral contents in the plant.
In vivo staining with DAB revealed the accumulation of H2O2 in lateral root tips of A. thaliana plants under CuONPs stress. Moreover, an increased amount of cell death was observed in lateral root apex as evidenced by PI staining. In an earlier study, Lequeux et al. (2010) reported that roots of A. thaliana plants under exposure to excess Cu has resulted in root apex cell death and they suggested that the excess generation of ROS as a result of higher concentrations of Cu exposure has resulted in increased cell death. In a recent study, Lee et al. (2013) reported that in buckwheat (Fagopyrum esculentum) exposure to CuONPs resulted in genotoxicity as a result of oxidative stress. Therefore, it is possible that the excess ROS generation under CuONPs stress could have resulted in cell death of lateral root apex cells of A. thaliana plants.
It is known that ROS production occurs in different compartments of the cell, such as mitochondria, chloroplasts, microsomes, glyoxysomes, peroxisomes, apoplasts, and the cytosol (Fridovich 1986), and different genes coding for enzymatic and nonenzymatic defense are activated in cells to protect from oxidative stress (Takahashi and Asada 1983). In the present study, it was observed that the expression levels of different SOD genes viz. MSD, CSD1, and CSD2 were upregulated upon exposure to different concentrations of CuONPs, showing that oxidative stress is caused in different cellular compartments of the cell as a result of exposure to CuONPs. The role of A. thaliana MSD gene in overcoming oxidative stress caused by different environmental pollutants has been reported from previous studies (Bowler et al. 1991; Slooten et al. 1995). Upregulation of A. thaliana CSD1 and CSD2 gene has been reported under oxidative stress conditions to maintain the redox homeostasis (Du et al. 2008). In addition, the SOD gene expression results were also supported by the histochemical detection of the formation of superoxides in leaves of A. thaliana plants exposed to CuONPs as evidenced by NBT staining.
In the present investigation, significant upregulation of CAT genes was also observed in A. thaliana plants upon exposure to CuONPs. Based on previous studies, it is well known that catalases are involved in maintaining cellular redox status in cells by scavenging excess H2O2 (Du et al. 2008). In vivo histochemical staining with DAB also revealed a dose-dependant increase in H2O2 generation upon exposure to CuONPs in leaves and roots of A. thaliana plants exposed to different doses of CuONPs. The upregulation of CAT genes has also been reported from rice and Arabidopsis as a result of oxidative stress induced by different environmental pollutants (Smeets et al. 2008; Fukao et al. 2011). Moreover, in this study, significant transcriptional upregulation of APX1 and APX2 genes were also observed in A. thaliana plants exposed to different concentration of CuONPs. It is known from earlier studies that the cells are protected from excess accumulation of H2O2 through the ascorbate-glutathione cycle, where APX reduces the H2O2 to H2O (Lu et al. 2010).
In this study, the transcriptional regulation of genes involved in sulfur assimilation, proline, and GSH biosynthesis was checked in A. thaliana under CuONPs stress. It is known that sulfur-containing compounds plays an important role in metal detoxification and also in the maintenance of cellular redox homeostasis (Schiavona et al. 2007). It has been reported earlier that enhanced consumption of cysteine for the synthesis of sulfur rich metal-chelating compounds has resulted in the upregulation of sulfur assimilation genes in plants (Dominguez-Solis et al. 2001; Nocito et al. 2002; Schiavona et al. 2007). In an earlier study, Queval et al. (2009) reported that exposure to H2O2 increased GSH content and upregulated genes coding for enzymes involved in cysteine synthesis in A. thaliana. Moreover, previous studies have shown that sulfur assimilation was involved in tolerance to oxidative stress caused as a result of exposure to Cd in A. thaliana (Harada et al. 2002). Recently, significant upregulation of genes involved in sulfur assimilation and GSH biosynthesis has also been reported upon exposure to MONPs in A. thaliana (Ma et al. 2013). In this study, the proline biosynthesis genes (P5CS1 and P5CS2) were significantly upregulated under CuONPs stress. It has been reported that knockout of the P5CS isoform1 in A. thaliana caused a reduction of stress-induced proline synthesis and showed an accumulation of ROS (Szekely et al. 2008). In addition, Krishnan et al. (2008) has reported that overexpression of the proline biosynthetic enzymes has resulted in proline accumulation and significantly lower ROS levels in mammalian cells. Moreover, different mammalian cell lines exposed to H2O2 has resulted in the upregulation of proline biosynthesis genes, indicating that upregulation of proline biosynthesis is an oxidative stress response (Krishnan et al. 2008). Therefore, the induction of P5CS genes and increase in the accumulation of proline, as observed in this study, might be to protect the plants from increased oxidative stress under CuONPs stress.
Conclusions
In conclusion, the results showed that exposure to CuONPs has resulted in significant reduction in the plant biomass and caused retardation of root growth in A. thaliana. Moreover, exposure to CuONPs has resulted in excess ROS generation, loss of root gravitropism, increased lateral root formation, decreased lateral root growth, caused lignification of root cells, and also resulted in root apex cell death. In vivo detection of ROS by histochemical staining with NBT and DAB as well as the upregulation of genes coding for enzymatic and nonenzymatic antioxidant defense mechanisms, sulfur assimilation pathways, and proline biosynthesis indicated an increased production of ROS as well as quick activation of plants defense mechanism to combat oxidative stress damage under CuONPs stress. Further studies are being conducted to understand the effect of lignification of roots and changes in mineral contents of plants upon exposure to CuONPs.
References
Aruoja V, Dubourguier HC, Kasemets K, Kahru A (2009) Toxicity of nanoparticles of CuO, ZnO and TiO2 to microalgae Pseudokirchneriella subcapitata. Sci Total Environ 407:1461–1468
Asli S, Neumann PM (2009) Colloidal suspensions of clay or titanium dioxide nanoparticles can inhibit leaf growth and transpiration via physical effects on root water transport. Plant Cell Environ 32:577–584
Bates LS (1973) Rapid determination of free proline for water-stress studies. Plant Soil 39:205–207
Bowler C, Slooten L, Vandenbranden S, De Rycke R, Botterman J, Sybesma C, Van Montagu M, Inzé D (1991) Manganese superoxide dismutase can reduce cellular damage mediated by oxygen radicals in transgenic plants. EMBO J 10:1723–1732
Chen Y, Wang D, Zhu X, Zheng X, Feng L (2012) Long-term effects of copper nanoparticles on wastewater biological nutrient removal and N2O generation in the activated sludge process. Environ Sci Technol 46:12452–12458
Chiang HH, Dandekar AM (1995) Regulation of proline accumulation in Arabidopsis during development and in response to desiccation. Plant Cell Environ 18:1280–1290
Choudhary M, Jetley UK, Abash KM, Zutshi S, Fatma T (2007) Effect of heavy metal stress on proline, malondialdehyde, and superoxide dismutase activity in the cyanobacterium Spirulina platensis. Ecotoxicol Environ Saf 66:204–209
Dietz KJ, Herth S (2011) Plant nanotoxicology. Trends Plant Sci 16:582–589
Dominguez-Solis JR, Gutierrez-Alcala G, Romero LC, Gotor C (2001) The cytosolic O-acetylserine (thiol) lyase gene is regulated by heavy-metals and can function in cadmium tolerance. J Biol Chem 276:9297–9302
Du YY, Wang PC, Chen J, Song CP (2008) Comprehensive functional analysis of the catalase gene family in Arabidopsis thaliana. J Integr Plant Biol 50:1318–1326
Fridovich I (1986) Superoxide dismutases. Adv Enzymol Relat Areas Mol Biol 58:61–97
Fryer MJ, Oxborough K, Mullineaux PM, Baker NR (2002) Imaging of photooxidative stress responses in leaves. J Ex Bot 53:1249–1254
Fukao T, Yeung E, Bailey-Serres J (2011) The submergence tolerance regulator SUB1A mediates crosstalk between submergence and drought tolerance in rice. Plant Cell 23:412–427
Gill SS, Tuteja N (2010) Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem 48:909–930
Gomes T, Pinheiro JP, Cancio I, Pereira CG, Cardoso C, Bebianno MJ (2010) Effects of copper nanoparticles exposure in the mussel Mytilus galloprovincialis. Environ Sci Technol 45:9356–9362
Griffitt RJ, Weil R, Hyndman KA, Denslow ND, Powers K, Taylor D, Barber DS (2007) Exposure to copper nanoparticles causes gill injury and acute lethality in zebrafish (Danio rerio). Environ Sci Technol 41:8178–8186
Halliwell B, Gutteridge JMC (1989) Free radicals in biology and medicine, 2nd edn. Clarendon Press, Oxford
Harada E, Yamaguchi Y, Koizumi N, Sano H (2002) Cadmium stress induces production of thiol compounds and transcripts for enzymes involved in sulfur assimilation pathway in Arabidopsis. J Plant Physiol 159:445–448
Heath RL, Packer L (1968) Photoperoxidation in isolated chloroplasts. I. Kinetics and stochiometry of fatty acid peroxidation. Arch Biochem Biophys 125:189–198
Hernández JA, Corpas FJ, Gómez M, del Río LA, Sevilla F (1993) Salt-induced oxidative stress mediated by activated oxygen species in pea leaf mitochondria. Physiol Plant 89:103–110
Karlsson HL, Cronholm P, Gustafsson J, Moller M (2008) Copper oxide nanoparticles are highly toxic: a comparison between metal oxide nanoparticles and carbon nanotubes. Chem Res Toxicol 21:1726–1732
Krishnan N, Dickman MB, Becker DF (2008) Proline modulates the intracellular redox environment and protects mammalian cells against oxidative stress. Free Radical Bio Med 44:671–681
Lee WM, An YJ, Yoon H, Kweon HS (2008) Toxicity and bioavailability of copper nanoparticles to the terrestrial plants Mung bean (Phaseolus radiatus) and wheat (Triticum aestivum): plant agar test for water-insoluble nanoparticles. Environ Toxicol Chem 27:1915–1921
Lee S, Chung H, Kim S, Lee I (2013) The genotoxic effect of ZnO and CuO Nanoparticles on early growth of Buckwheat, Fagopyrum esculentum. Water Air Soil Pollut 224:1668–1678
Lequeux H, Hermans C, Lutts S, Nathalie V (2010) Response to copper excess in Arabidopsis thaliana: impact on the root system architecture, hormone distribution, lignin accumulation and mineral profile. Plant Physiol Biochem 48:673–682
Lichtenthaler HK (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. In: Lester Packer RD (ed) Methods in enzymology, 148th edn. Academic Press, Waltham, pp 350–382
Lin D, Xing B (2008) Root uptake and phytotoxicity of ZnO nanoparticles. Environ Sci Technol 42:5580–5585
Liu Q, Zhao Y, Wan Y, Zheng J, Zhang X, Wang C, Fang X, Lin J (2010) Study of the inhibitory effect of water-soluble fullerenes on plant growth at the cellular level. ACS Nano 10:5743–5748
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt Method. Methods 25:402–408
Lu Y, Li X, He M, Zhao X, Liu Y, Cui Y, Pan Y, Tan H (2010) Seedlings growth and antioxidative enzyme activities in leaves under heavy metal stress differ between two desert plants: a perennial (Peganum harmala) and an annual (Halogeton glomeratus) grass. Acta Physiol Plant 32:583–590
Ma C, Chhikara S, Xing B, Musante C, White JC, Dhankher OP (2013) Physiological and molecular response of Arabidopsis thaliana (L.) to nanoparticle Cerium and Indium Oxide Exposure. ACS Sus Chem Eng 1:768–778
Melegari SP, Perreault F, Popovic RHRC, Radovan Matias WG (2013) Evaluation of toxicity and oxidative stress induced by copper oxide nanoparticles in the green alga Chlamydomonas reinhardtii. Aquat Toxicol 142–143:431–440
Mittler R, Vanderauwera S, Gollery M, Van Breusegem F (2004) Reactive oxygen gene network of plants. Trends Plant Sci 9:490–498
Nocito FF, Pirovano L, Cocucci M, Sacchi GA (2002) Cadmium-induced sulfate uptake in maize roots. Plant Physiol 129:1872–1879
Queval G, Thominet D, Vanacker H, Miginiac-Maslow M, Gakière B, Noctor G (2009) H2O2-activated up-regulation of glutathione in Arabidopsis involves induction of genes encoding enzymes involved in cysteine synthesis in the chloroplast. Mol Plant 2:344–356
Rabino I, Mancinelli AL (1986) Light, temperature and anthocyanin production. Plant Physiol 81:922–924
Rogers LA, Dubos C, Surman C, Willment J, Cullis IF, Mansfield SD, Campbell MM (2005) Comparison of lignin deposition in three ectopic lignification mutants. New Phytol 168:123–140
Saito K (2000) Regulation of sulfate transport and synthesis of sulfur containing amino acids. Curr Opin Plant Biol 3:188–195
Schiavona M, Zhang L, Abdel-Ghany SE, Pilon M, Malagoli M, Pilon-Smits EAH (2007) Variation in copper tolerance in Arabidopsis thaliana accessions Columbia, Landsberg erecta and Wassilewskija. Physiol Plant 129:342–350
Shaw AK, Hossain Z (2013) Impact of nano-CuO stress on rice (Oryza sativa L.) seedlings. Chemosphere 93:906–915
Shi J, Peng C, Yang Y, Yang J, Zhang H, Yuan X, Chen Y, Hu T (2014) Phytotoxicity and accumulation of copper oxide nanoparticles to the Cu-tolerant plant Elsholtzia splendens. Nanotoxicol 8:179–188
Slooten L, Capiau K, Van Camp W, Van Montagu M, Sybesma C, Inze D (1995) Overexpressing manganese superoxide dismutase in the chloroplasts. Plant Physiol 107:737–750
Smeets K, Ruytinx J, Semane B, Van Belleghem F, Remans T, Van Sanden S, Vangronsveld J, Cuypers A (2008) Cadmium-induced transcriptional and enzymatic alterations related to oxidative stress. Environ Exp Bot 63:1–8
Solfanelli C, Poggi A, Loreti E, Alpi A, Perata P (2006) Sucrose specific induction of the anthocyanin biosynthetic pathway in Arabidopsis. Plant Physiol 140:637–646
Szabados L, Savouré A (2010) Proline: a multifunctional amino acid. Trends Plant Sci 15:89–97
Szekely G, Abraham E, Cseplo A, Rigo G, Zsigmond L, Csiszar J, Ayaydin F, Strizhov N, Jasik J, Schmelzer E, Koncz C, Szabados L (2008) Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. Plant J 53:11–28
Tahara S (2007) A journey of twenty-five years through the ecological biochemistry of flavonoids. Biosci Biotechnol Biochem 71:1387–1404
Takahashi MA, Asada K (1983) Superoxide anion permeability of phospholipid membranes and chloroplast thylakoids. Arch Biochem Biophys 226:558–566
Thordal-Christensen H, Zhang Z, Wei Y, Collinge DB (1997) Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J 11:1187–1194
Tripathi BN, Gaur JP (2004) Relationship between copper- and zinc-induced oxidative stress and proline accumulation in Scenedesmus sp. Planta 219:397–404
Wang Z, Xie X, Zhao J, Liu X, Feng W, White JC, Xing B (2012) Xylem and phloem based transport of CuO nanoparticles in maize (Zea mays L.). Environ Sci Technol 46:4434–4441
Wu SG, Huang L, Head J, Chen DR, Kong IC, Tang YJ (2012) Phytotoxicity of metal oxide nanoparticles is related to both dissolved metals ions and adsorption of particles on seed surfaces. J Pet Environ Biotechnol 3:1000126
Yang JG, Okamoto T, Ichino R, Bessho T, Sarake S, Okido M (2006) A simple way for preparing antioxidation nano-copper powders. Chem Lett 35:648–649
Yoon KY, Byeon JH, Park JH, Hwang J (2007) Susceptibility constants of Escherichia coli and Bacillus subtilis to silver and copper nanoparticles. Sci Total Environ 373:572–575
Acknowledgments
This paper was supported by the SMART-Research Professor Program of Konkuk University, Seoul, South Korea to Dr. Prakash M. Gopalakrishnan Nair. This work was supported by a grant from the Next-Generation BioGreen 21 Program (Plant Molecular Center No. PJ009053), Rural Development Administration, Republic of Korea.
Conflict of interest
The authors declare no conflict of interest
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Elena Maestri
Highlights
• Studied the physiological and molecular level responses of CuONPs stress in A. thaliana.
• CuONPs stress significantly reduced plant biomass and root growth.
• CuONPs stress caused root growth modifications and enhanced lateral root formation.
• Histochemical staining revealed excess ROS generation.
• Propidium iodide staining indicated cell death in root apex.
• Induced antioxidant, sulfur assimilation, GSH biosynthesis genes.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Figure 1
Characterization of copper oxide nanoparticles using transmission electron microscopy (A, B) Morphology of CuONPs in deionized water, (C) Graph showing size distribution of copper oxide nanoparticles in deionized water based on dynamic light scattering analysis and (D, E) morphology of copper oxide nanoparticles in ½ MS medium. (GIF 735 kb)
Supplementary Figure 2
Phenotypes of A. thaliana plants grown in the presence of Cu2+ ions for 21 days (left to right: Control, 0.1 and 0.2 mg/L of Cu2+ ions) (B, C) Plants showing changes in root morphology and (D, E) loss of root gravitropism after exposure to 10 and 20 mg/L of copper oxide nanoparticles for 21 d. (GIF 1215 kb)
MOESM 3
(DOC 24 kb)
Table 1
(DOC 49 kb)
Rights and permissions
About this article
Cite this article
Nair, P.M.G., Chung, I.M. Impact of copper oxide nanoparticles exposure on Arabidopsis thaliana growth, root system development, root lignificaion, and molecular level changes. Environ Sci Pollut Res 21, 12709–12722 (2014). https://doi.org/10.1007/s11356-014-3210-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-014-3210-3