Abstract
Two strains capable of degrading cyclohexane were isolated from the soil and sludge of the wastewater treatment plant of the University of Stuttgart and a biotrickling filter system. The strains were classified as gram negative and identified as Acidovorax sp. CHX100 and Chelatococcus sp. CHX1100. Both strains have demonstrated the capability to degrade cycloalkanes (C5–C8), while only strain CHX1100 used as well short linear n-alkanes (C5–C8) as the sole source of carbon and energy. The growth of Acidovorax sp. CHX100 using cyclohexane was much faster compared to Chelatococcus sp. CHX1100. Degenerated primers were optimized from a set sequences of cyclohexanol dehydrogenase genes (chnA) as well as cyclohexanone monooxygenases (chnB) and used to amplify the gene cluster, which encodes the conversion of cyclohexanol to caprolactone. Phylogenetic analysis has indicated that the two gene clusters belong to different groups. The cyclohexane monooxygenase-induced activity which oxidizes also indole to 5-hydroxyindole has indicated the presence of a CYP-type system monooxygenase involved in the transformation of cyclohexane to cyclohexanol.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Cyclohexane is a highly volatile and colorless cyclic alkane originated from petroleum derivates. Its production in the European Union (EU) is 950,000 t/a, worldwide around 4,400,000 t/a (European Union 2004). Cyclohexane is mainly used for the synthesis of adipic acid and caprolactam, precursors in nylon manufacturing. Also, it is used as a solvent in craft industries and as an auxiliary in chemical production processes, such as a precipitating and extraction agent. Cyclohexane can be widespread to the environment through petroleum and fuel spills as well as through its usage and production processes. The European Union (2004) has estimated that cyclohexane emissions on a continental level are as follows: by production, 544 t/a; used as solvent, 15,400 t/a; and as intermediate, 864 t/a. The US Environmental Protection Agency (EPA) (1994) has reported environmental issues due to gas emissions containing cyclohexane. Moreover, the toxicity of cyclohexane is related to its octanol-water partition coefficient (Log K ow) with a value of 3.2 causing a high cell damage as reported by Sardessai and Bhosle (2004).
The saturated cyclic structure of cyclohexane makes it more recalcitrant compared to alkanes or monoaromatic compounds. The degradation of cyclohexane using bacterial strains has first been reported by de Klerk and van der Linden (1974), who described the co-metabolism of cyclohexane using heptane as the main substrate by Pseudomonas spp. and a symbiosis with another Pseudomonas spp. for further degradation of cyclohexanol. The strains were unable to use cyclohexane as the sole source of carbon and energy. Stirling and Watkinson (1977) have isolated a strain identified as Nocardia sp. capable of using cyclohexane as carbon source requiring only biotin as a complement in the media. Rouverie and Chen (2003) reported Brachymonas petroleovorans CHX capable to degrade cyclohexane under the presence of yeast extract in the mineral medium. Strains able to degrade cyclohexane without any complement in the medium were reported by Anderson et al. (1980) and Trower et al. (1985), who studied the degradation with Pseudomonas sp. and Xanthobacter sp. Moreover, two gram-positive strains Rhodococcus sp. EC1 and Bacillus lentus LP32 have been reported as cyclohexane-degrading strains by Lee and Cho (2008) and Opere et al. (2013), respectively. Although some strains able to use cyclohexane as the sole carbon source have been already reported, the potential in bioremediation is still limited by several factors, e.g., the low solubility of cyclohexane in water and difficulty in biofilm formation.
The purpose of this study is to isolate novel strains that are capable to use cyclohexane as the sole source of carbon and energy and to provide an understanding of the microbiology and genetics of the degradation process of cyclohexane.
Materials and methods
Growth media and enrichment of bacterial strains
For the enrichment of strains able to use cyclohexane as carbon source, two approaches were designed: batch growth and a setup of biotrickling filter (BTF). The bacterial strains were cultivated on a liquid mineral medium (MM) with the following: KH2PO4 1.0 g, Na2HPO4 2.79 g, (NH4)2SO4 1.0 g, Ca(NO3)2 · 7H2O 0.01 g, C6H8O7 · Fe · NH4 0.01 g, 1 mL of trace mineral solution, and distilled water to reach 1 L. The trace mineral solution contained the following: H3BO3 0.3 g, CoCl2 · 6H2O 0.2 g, ZnSO4 · 7H2O 0.1 g, Na2MoO4 · 2H2O 0.03 g, MnCl2 · 4H2O 0.03 g, NiCl2 · 6H2O 0.02 g, and CuCl2 · 2H2O 0.01 g.
For batch growth, inoculum was obtained from soil samples and sludge from the wastewater treatment plant of the Institute for Sanitary Engineering, Water Quality and Solid Waste Management, University of Stuttgart. The inoculum samples were processed as described by Dobslaw and Engesser (2012) and Salamanca et al. (2013). Cyclohexane was added directly (5.5 mM) to the medium, and the flasks were sealed air tight and incubated at 30 °C, 150 rpm.
For the BTF, inoculum from a wastewater treatment plant of a chemical company was used. The BTF was set up with a total length of 1 m and an internal diameter of 0.314 m. The BTF was filled with a packing material of polyurethane foam (PU) with a specific area of 620 m2/m3 and porosity of 96–98 %. The recirculation system was programmed to intervals of 4 min and a pumping period of 1 min in a counter-current mode at a constant empty bed retention time (EBRT) of 37 s. Nutrients were supplied to the recirculation tank. The initial cyclohexane concentration was 61.21 ± 1.053 mg C/m3 and monitored using a micro flame ionization detector (FID) from Hartmann & Braun. After a running period of 3 weeks, packing material containing biofilm was extracted, and the strains were grown on MM agar plates using cyclohexane as carbon source. The bacterial strains were isolated according to the procedure described for batch growth experiments.
Genetic characterization of the isolated strains and sequence analysis
The total DNA was isolated using the DNA Mini-Prep Kit (Qiagen). For 16S ribosomal RNA (rRNA) PCR, gene amplification standard oligonucleotides 27f (AGAGTTTGATCMTGGCTCAG) and 1492r (TACGGYTACCTTGTTACGACTT) were used. The nucleotide sequencing was done by GATC Biotech (Konstanz, Germany), and the results were compared to those of the GenBank Database at the National Center for Biotechnology Information. The phylogenetic tree was estimated and plotted using the neighbor-joining tree method by a bootstrap analysis of 1,000 replications.
Growth rate
The pure strains were precultured for a period of 3 days in 1 L of MM medium containing 5.5 mM of cyclohexane. Subsequently, the precultures were centrifuged at 9,600g for 15 min. Then, the bacterial pellet was washed twice with distilled water, resuspended in 50 mL of fresh MM, and used as inoculum, distributing it into flasks containing MM in order to reach an initial optical density (OD) of 0.05 to 0.2. For the strain CHX100, an OD of 0.05 is equivalent to 1.83 × 107 colony formation units (CFU)/mL, and for CHX1100, an OD of 0.2 is equivalent to 1.68 × 108 CFU/mL. The strains were cultivated with 7.5 mM of cyclohexane and incubated at 30 °C, 150 rpm. Samples were taken every hour, and the growth rates of the strains were determined measuring the optical density at 546 nm. Cyclohexane was measured in the water and gas phase through gas chromatography–mass spectrometry (GC-MS).
Metabolites and lag phase experiments
The main metabolites in the degradation pathway of cyclohexane are described to be cyclohexanol and cyclohexanone (Perry 1984; Cheng et al. 2000). The growth rate of each strain was estimated using cyclohexanol or cyclohexanone. In order to determine a possible lag phase caused by the metabolites, three precultures with cyclohexane, cyclohexanol, and cyclohexanone in 1 L MM were prepared. The precultures were centrifuged twice at 9,600 rpm for 10 min and resuspended on 300 mL MM. From each preculture, the volume was split into three flasks (100 mL MM) and fed with cyclohexane and the metabolites separately (initial concentration of 7.5 mM each). The growth was determined by measuring the OD at 546 nm.
Degradation of other carbon sources
The ability to degrade other carbon sources by the isolated strains was evaluated using compounds such as primary and secondary alcohols (methanol, ethanol, 2-dodecanol, 2-hexadecanol, 3-hexanol, 1-hexanol, 2-hexanol, 1-octanol), ketones and lactones (acetone, dibenzophenone, caprolactone, cyclohexanone, and cyclododecanone), chlorinated compounds (chlorocyclohexane, dichloromethane, tetrachloroethylene), ethers (octyl ether, butyl ether, tert-butyl ether, methyl tert-butyl ether), cyclic alkanes (cyclopentane, cycloheptane, cyclooctane, cyclodecane, cyclododecane, methylcyclohexane), monoaromatic (benzene, toluene, ethylbenzene, m-xylene, p-xylene, and o-xylene), and alkanes (butane, pentane, hexane, heptane, octane, nonane, decane). The strains were precultivated on MM with cyclohexane until reaching at an OD of 1.5. Subsequently, the samples were washed twice in 20 mL sterile minimal medium for being inoculated with each substrate at 30 °C, 150 rpm. The cultures were incubated for a period of 7 days. To define the mechanism of degradation, co-oxidation experiments were only carried out in a substrate such as 1-chlorocyclohexane and methylcyclohexane using 5 mM cyclohexane as base substrate.
GC-MS
GC-MS analysis was carried out using an Agilent GC 7890A gas chromatograph with a 30 m × 0.25 mm column (Agilent Technologies 190915–433) and a MS 5975 analysis detector. A splitless injection was performed with the injector at 250 °C, the transfer line at 280 °C, and the detector at 230 °C using the following program: 80 °C (1 min) to 180 °C (1 min) at 7 °C min−1 to 240 °C at 12 °C min−1 and, finally, to 300 °C (8 min) at 20 °C min−1. The metabolite formation and cyclohexane degradation were studied by taking 1 mL of the liquid medium and acidifying it with 100 μL of phosphoric acid. The extraction was carried out in 1 mL ethylacetate. The organic phase was separated and analyzed by GC-MS.
Detection of genes involved in the degradation pathway of cyclohexane
In order to identify the genes involved in the degradation pathway of cyclohexane, degenerated primers were designed using the alignment of multiple sequences from cyclohexanol dehydrogenase and cyclohexanone monooxygenase genes from different bacterial strains, such as Pseudomonas mendocina (YMP CP000680.1), Burkholderia xenovorans LB400 (CP000270.1), Sphingomonas sp. SKA58 (NZ_AAQG01000028.1), Acidovorax sp. JS42 (NC_008782.1), B. petroleovorans (AY437859.1), Arthrobacter sp. L661 (EF538713.1), Xanthobacter flavus (AJ418061.1), Comamonas testosteroni (AJ418060.1), Acinetobacter sp. EB104 (NC_002760.1), Nocardiopsis dassonvillei (NR_076750.1), Polaromonas sp. JS666 (NC_007950.1), Pseudonocardia dioxanivorans CB1190 (CP002593.1), Rhodococcus erythropolis PR4 (NC_012490.1), and Rhodococcus sp. TK6 (AY486161.1). The resulting degenerated oligonucleotides were the following: for the cyclohexanone monooxygenase, ChnB-1_F (CCACCGGYGTTCAGGTKAT) and ChnB-1_R (TCGSHGATCCABTCGACCTG) and ChnB-2_F (CCGGCCTSGGCATGGCSATCC) and ChnB-2_R (CCGCARTACAGCGTGCCG), and for the cyclohexanol dehydrogenase, ChnA-1_F (GTCACYGGCGGYGCCATGG) and ChnA-1_R (CCATGCACGAACGAKGMYTC). Amplification was carried out through a PCR program consisting of initial denaturation at 95 °C for 240 s, 30 cycles at 94 °C for 30 s, 60.5 °C for 60 s, 72 °C for 60 s, and a final elongation step at 72 °C for 600 s. The sequence analysis was carried out as specified in the 16S rRNA PCR procedure.
Indole test
An indole test was used to detect the presence of monooxygenases. Indole is hydrolyzed by monooxygenases and oxygenases forming compounds which can be monitored colorimetrically (Sugimori et al. 2004; Qing-Shan et al. 2000; McClay et al. 2005). The procedure described by McClay et al. (2005) was modified to evaluate the presence of monooxygenases directly in the liquid media. The isolated strains were pregrown until the exponential phase on different substrates was achieved, and then, 2.5 mM of indole was added. The formation of metabolites in the samples was analyzed using HPLC analysis by taking 1 mL of the culture and centrifuged for 5 min at 15,000g. The supernatant was transferred into HPLC vials and analyzed using a Beckman Coulter System Gold HPLC device with a mix of methanol/water (70:30) at a flow rate of 1 mL/min. Moreover, the cell pellet was resolved using dimethylformamide in order to analyze the insoluble metabolites such as indigo. Indole and its metabolites were analyzed at a wavelength of 270 nm. Indole was detected at 2.5 min, 5-hydroxyindole at 1.5 min, and indigo at 6 min; 7.5 mM of the following substrates served as carbon and energy source: cyclohexane, cyclohexanol, and cyclohexanone. For control of the substrates acetate, n-hexane and caprolactone were used.
Results
Isolation and identification of the strains
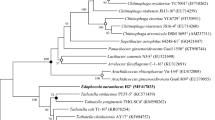
An isolation of bacteria able to degrade cyclohexane was carried out under batch growth conditions from an enrichment using sludge and soil as an inoculum from the wastewater treatment plant of the Institute for Sanitary Engineering, Water Quality and Solid Waste Management, University of Stuttgart. A cyclohexane strain designed as CHX1100 was identified by 16S rRNA as Chelatococcus sp. The isolation of a strain using biotrickling filter as a system for cleaning air containing cyclohexane as contaminant was inoculated from a sludge of a chemical company, and after a period of adaptation of approximately 15 days, degradation of cyclohexane was detected, indicating a biological activity towards cyclohexane (see Fig. 1). The biotrickling filter had an elimination efficiency of 97.3 ± 1.7 % and elimination capacity of 5.4 kg/m3. Biofilm formation was observed on the packing material, and a gram-negative strain was isolated and identified as Acidovorax sp. (CHX100). The phylogenetic tree of the cyclohexane degraders shows that the strains belong to separate groups (see Fig. 2).
Performance of a biotrickling filter used as an enrichment system of bacterial strains able to degrade cyclohexane as a contaminant model. The BTF was inoculated from sludge of a chemical company. The cyclohexane concentration was monitored using micro-FID. The BTF was filled with a packing material of polyurethane foam (PU) to enhance biofilm production. Biofilm samples were taken after a running period of 3 weeks and inoculated as described in batch growth experiments
Growth rates of the strain CHX100 and CHX1100
The strains CHX100 and CHX1100 were able to grow in the presence of cyclohexane by an increase of the OD at 546 nm in the liquid medium (see Fig. 3). The growth rates at 30 °C of the strains CHX100 and CHX1100 were determined in a minimal medium using cyclohexane (7.5 mM) as the sole source of carbon and energy. The growth rate of CHX100 was μ = 0.199 h−1, and for CHX1100, it was μ = 0.031 h−1, with a generation time of 3.5 and 22 h, respectively (see Fig. 3). The strains were able to degrade 99 % of the initially cyclohexane added. The strain CHX1100 requires a higher amount of bacterial inoculum compared to CHX100 in order to start growing in the minimal medium containing cyclohexane. The strain CHX100 was able to use cyclohexane concentrations up to 14 mM and the strain CHX1100 up to 9.2 mM (data not shown).
Growth of the strains CHX100 and CHX1100 on cyclohexane as the sole source of carbon and energy. The strains were inoculated on a minimal medium using an initial cyclohexane concentration of 7.5 mM. The growth was determined by measuring the optical density at 546 nm. The degradation of cyclohexane was determined using GC-MS analysis. Filled circle indicates growth of CHX100 on cyclohexane, empty circle degradation of cyclohexane by strain CHX100, filled square growth of CHX1100 on cyclohexane, and empty square degradation of cyclohexane by the strain CHX1100. Discontinuous line indicates sample containing only minimal medium and cyclohexane
Genes involved in the degradation of cyclohexane
Degenerated primers targeting the cyclohexanol and cyclohexanone sequence were designed in this study, amplifying the genes involved in the transformation of cyclohexanol to caprolactone. The primers ChnA-1 and ChnB-1 have amplified partial genes for the strain CHX100. A 712-bp DNA fragment was amplified and found to be homologous to a cyclohexanone monooxygenase within a similarity to the following: 98.13 % Acidovorax sp. JS42, 80.57 % to Polaromonas sp. J666, 74.23 % Rhodococcus sp. HI-31, and Rhodococcus sp. TK6. For the cyclohexanol dehydrogenase, a 401-bp fragment was amplified and was homologous to the following: 98.21 % Acidovorax sp. JS42 (short-chain dehydrogenase), 81.39 % to Polaromonas sp. J666, 72.56 % C. testosteroni, and 68.23 % X. flavus. The primers ChnA-1_R and ChnB-1_F were used to determine whether the genes belong to the same gene cluster, and a fragment of 1,781 bp has been amplified in the strain CHX100. For the strain CHX1100, the sequence obtained from the primers ChnB-2 was homologous to the following: 98.05 % Beijerinckia indica ATCC 9039, 70.51 % Rhodococcus jostii RH4, 70.12 % Rhodococcus sp. DK17, and 70.11 % Gordonia sp. TY-5. The degenerated primers ChnA-1_F and ChnA-1_R have not provided any gene amplification for determining the cyclohexanol dehydrogenase in the strain CHX1100.
Degradation of other hydrocarbons
The strains CHX100 and CHX1100 are able to degrade cycloalkanes from C5 to C8 (see Table 1). CHX1100 degrades other recalcitrant compounds as short n-alkanes (C5–C8) including some aromatic hydrocarbons (benzene and toluene). For chlorinated compounds, only strain CHX100 was able to transform chlorocyclohexane in co-oxidation mode with cyclohexane as base substrate. Metabolites identified as cis- and trans-4-chlorocyclohexanol were detected using GC-MS (see Fig. 4). The metabolites were not further degraded by the strain CHX100. The strains were not able to co-oxidize methylcyclohexane.
GC-MS co-oxidation of 1-chlorocyclohexane. For co-oxidation process, the samples were inoculated using 5.5 mM cyclohexane and 1 mM of 1-chlorocyclohexane. The metabolites were extracted using ethyl acetate as solvent and analyzed using GC-MS. The metabolites were identified as cis- and trans-4-chlorocyclohexanol. The strain CHX100 was unable to degrade these metabolites further
Induction and lag phase
The results suggest that cyclohexane and its proposed degradation intermediates (cyclohexanol and cyclohexanone) produce an inhibition on the growth rate, and their behaviors differ in each strain: strain CHX100 has a growth rate of 0.075 h−1 on cyclohexanol and 0.201 h−1 on cyclohexanone, and strain CHX1100 has a growth rate of 0.083 h−1 on cyclohexanol and 0.178 h−1 on cyclohexanone. To ensure the identical conditions of the experiments, each substrate has an initial concentration of 7.5 mM and was added directly to the minimal medium. Extended lag phases were detected when the strains fed with cyclohexane and cyclohexanone were exposed to cyclohexanol as carbon source and energy (see Fig. 5).
Lag phase and induction by cyclohexane, cyclohexanol, and cyclohexanone. The experiments were carried out using 7.5 mM of each substrate for reaching exponential growth. Subsequently, the samples were washed and inoculated with fresh media and substrate. a Initial growth using cyclohexane as carbon source. b Initial growth using cyclohexanol as carbon source. c Initial growth using cyclohexanone as carbon source. Filled square indicates strain CHX100 using cyclohexane, filled triangle strain CHX100 using cyclohexanol, filled circle strain CHX100 using cyclohexanone, empty square strain CHX1100 using cyclohexane, empty triangle strain CHX1100 using cyclohexanol, and empty circle strain CHX1100 using cyclohexanone
Indole test
The formation of blue color during the co-oxidation of indole has shown the presence of the cyclohexanone monooxygenase in the strains CHX100 and CHX1100. Blue color was produced due to the formation of indigo by a combined action of cyclohexanone as base substrate. A yellow color was formed when the strains were using cyclohexane as carbon source and the metabolite produced by the co-oxidation of indole was 5-hydroxyindole. Nevertheless, if the sample was incubated for a longer period, a formation of greenish color was observed, possibly indicating the induction of the cyclohexanone monooxygenases involved in the degradation pathway of cyclohexane (see Fig. 6). In the samples containing acetate, n-hexane (only CHX1100) and caprolactone, color formation, and metabolites were not detected. A variation in the product distribution resulting from indole co-oxidation has been estimated through HPLC data analysis (see Table 2). The cyclohexane strains were unable to use indole as carbon source and energy. Toluene and hexane as base substrate used by the strain CHX1100 did not induce the oxidation of indole, and metabolite formation was not observed.
Identification of indole co-oxidation products during the degradation cyclohexane and its metabolites. The strains were grown until reaching exponential phase using cyclohexane and cyclohexanone as carbon source. Subsequently, 1 mM indole was added. The indole metabolites were detected using HPLC. a Formation of 5-hydroxyindole using cyclohexane as main substrate (yellow color). b Formation of indigo after further incubation of the sample containing cyclohexane and indole (greenish color). c Formation of indigo during the co-oxidation of indole using cyclohexanone as main substrate (blue color)
Discussion
In this study, two novel strains were isolated for being capable to degrade cyclohexane using two different methods: batch growth and a continuous system (biotrickling filter). The analysis of 16S rRNA gene sequences reveals that the strain CHX100 is related to Acidovorax sp. and has a similarity of 99.72 % with Acidovorax temperans PHL which belongs to the Comamonadaceae family, and the strain CHX1100 is phylogenetically related to Chelatococcus sp., and its nearest neighbors are Chelatococcus daeguensis K106 and Chelatococcus asaccharovorans TE2, with values of 94.96 and 94.71 %, respectively. Chelatococcus sp. belongs to the family Beijerinckiaceae. This is the first report dealing with two different strains using cyclohexane as the sole source of carbon and energy.
Acidovorax sp. CHX100 and Chelatococcus sp. CHX1100 are gram negative and oxidase and catalase positive. The strains show high capability to grow on short-chain cycloalkanes (C5–C8). The strains are unable to use medium-chain cycloalkanes (C10–C12), suggesting that the degradation of short cycloalkanes is carried out by a specific enzyme. Acidovorax sp. CHX100 has a generation time (3.5 h) about six times faster than that of CHX1100 (22 h), exhibiting the fastest generation time ever reported by a cyclohexane-degrading strain. Trower et al. (1985) has reported 6 h of generation time of the strain Xanthobacter sp. Another study (Anderson et al. 1980) reported a Pseudomonas sp. having a generation time of 8 h. Moreover, Acidovorax sp. CHX100 has a remarkable short lag phase of approximately 9 h in the presence of cyclohexane as carbon source, indicating that the strain has a specialized system for uptaking the substrate and using it for its metabolism. Xanthobacter sp. has a lag phase of approximately 15 h before starting to grow on cyclohexane (Trower et al. 1985), and Anderson et al. (1980) reported a lag phase between 16 and 20 h in Pseudomonas sp. In contrast, the strain CHX1100 has a lag phase similar to one reported in the strain Rhodococcus sp. EC1 of approximately 20 h (Lee and Cho 2008).
The most important aspect of the strains isolated in this research compared to bacterial strains from other studies is the remarkable ability to degrade cyclohexane completely in concentrations up to 14 mM strain CHX100 and 9.2 mM strain CHX1100. Lee and Cho (2008) has reported a degradation of 92.3 % of cyclohexane at an initial concentration of 7.5 mM for the strain Rhodococcus sp. EC1, but at higher concentrations (9.2 mM), the degradation and growth were inhibited. Opere et al. (2013) has reported a similar behavior by strain B. lentus LP32 degrading 67.71 % of cyclohexane in a period of 10 days, and further degradation (92.43 %) was reached in a total period of 18 days. Other bacillus strains have been reported to degrade 25.9 % at an initial concentration of 2.4 mM within a period of 7 days (Lee et al. 2013). Rouviere and Chen (2003) has reported a Brachymonas sp. with a 70 % degradation of 1 mM cyclohexane using yeast extract for supporting the growth. In the studies with Xanthobacter sp. (Trower et al. 1985) and Pseudomonas sp., (Anderson et al. 1980), the cyclohexane concentration was estimated to be around 9.24 mM; however, there was a lack of information about the degradation process.
A significant difference between the isolated strains is the capability of degrading other recalcitrant compounds. Chelatococcus sp. CHX1100 is degrading short linear alkanes (C5–8) and some aromatic compounds, suggesting that the strain could be useful for implementation in bioremediation with different hydrocarbon contaminations.
The co-oxidation of 1-chlorocyclohexane by the strain CHX100 is accomplished by the induced monooxygenase during the degradation of cyclohexane to cyclohexanol. Nevertheless, the main metabolites cis- and trans-4-chlorocyclohexanol were not further degraded by the strain CHX100. Such co-oxidation has not been reported for the degradation of 1-chlorocyclohexane, and it is related to the activity of haloalkane dehalogenases in Xanthobacter autotrophicus GJ10 (Schanstra and Janssen 1996), Rhodococcus rhodochrous NCIMB 13064 (Kulakova et al. 1995), and Sphingomonas paucimobilis UT26 (Prokop et al. 2003).
The strain CHX100 isolated from a biotrickling filter demonstrates a novel approach to cleaning waste gas containing cyclohexane. In the BTF, the degradation of cyclohexane has been detected after an operational time of almost 15 days which suggests that strains initially need an adaptation time. Cox and Deshusses (2002) and Yang et al. (2011) have reported start-up periods from days to weeks during the degradation of volatile compounds in BTFs. Under the operational conditions of the biotrickling filter, a high removal rate of cyclohexane (97.3 ± 1.7 %) was obtained using a load of 306.1 mg/h. Biofilm formation on PU foam shows the high ability of the strain CHX100 for adaptation to different environments.
The degradation of cyclohexane by the strains CHX100 and CHX1100 follows the typical lactone formation pathway (Perry 1984; Cheng et al. 2000) and does not involve aromatization as it was reported by Yi et al. (2011). The gene cluster organization of cyclohexanol dehydrogenase and cyclohexanone monooxygenase from Acidovorax sp. CHX100 is different from that of the strain Chelatococcus sp. CHX1100, due to the fact that the two genes (cyclohexanol and cyclohexanone) belong to the same gene cluster in the strain CHX100. The variety of cyclohexanol and cyclohexanone gene cluster has been studied in detail by Cheng et al. (2000). The structure of the genes from Acidovorax sp. CHX100 is similar to the previously characterized from Acidovorax sp. JS42 and Polaromonas sp J666. The strain CHX1100 has a cyclohexanone monooxygenase homologous to Rhodococcus jostii RH4; however, in the strain CHX1100, the cyclohexanol dehydrogenase was not found in the same gene cluster of cyclohexanone monooxygenase. A possible explanation for this phenomena is that the cyclohexanol dehydrogenase gene could be encoded in another part of the chromosome of the strain Chelatococcus sp. CHX1100 or the transformation to cyclohexanol is carried out by an unspecific dehydrogenase.
A monooxygenase specifically active for cyclohexane is still unknown, and co-oxidation of indole to its different derivates could be used as a method for detecting such system. The yellow color is produced by the formation of 5-hydroxyindole during the co-oxidation of indole. The formation of 5-hydroxyindole could indicate the presence of a CYP-type system as the enzyme involved in the transformation of cyclohexane to cyclohexanol (Park et al. 2010). Warburton et al. (1990) proposed CYP450 as a possible system for the hydroxylation of cyclohexane. In contrast, co-oxidation of indole has not been detected when the strain CHX1100 was inoculated using hexane as carbon source and energy, indicating that the degradation of hexane is carried out by another system such as alkane hydroxylases which have not been reported to oxidize indole.
The monooxygenase systems involved in the degradation of cyclohexane and cyclohexanone could be used as novel advance in the green chemistry for the biosynthesis of pigments such as 5-hydroxyindole and indigo.
In conclusion, the results demonstrate the potential applicability of Acidovorax sp. CHX100 and Chelatococcus sp. CHX1100 for the treatment of waste gas containing cycloalkanes (C5–C8) and other recalcitrant compounds. The complete identification and isolation of the enzyme involved in the transformation of cyclohexane to cyclohexanol is subject of further research.
References
Anderson MS, Hall R, Griffin M (1980) Microbial metabolism of alicyclic hydrocarbons: cyclohexane catabolism by a pure strain of Pseudomonas sp. J Gen Microbiol 120:89–94
Cheng Q, Thomas SM, Kostichka K, Valentine JR, Nagarajan V (2000) Genetic analysis of a gene cluster for cyclohexanol oxidation in Acinetobacter sp. strain SE19 by in vitro transposition. J Bacteriol 17:4744–4751
Cox HHJ, Deshusses MA (2002) Co-treatment of H2S and toluene in a biotrickling filter. Chem Eng J 87(1):101–110
de Klerk H, van der Linden AC (1974) Bacterial degradation of cyclohexane participation of a co-oxidation reaction. Ant van Leeuw 40:7–15
Dobslaw D, Engesser KH (2012) Degradation of 2-chlorotoluene by Rhodococcus sp. OCT 10. Appl Microbiol Biotechnol 93:2205–2214
European Union (EU) (2004) Risk assessment report cyclohexane, CAS no: 110-82-7
Kulakova AN, Stafford TM, Larkin MJ, Kulakov LA (1995) Plasmid pRTL1 controlling 1-chloroalkane degradation by Rhodococcus rhodochrous NCIMB13064. Plasmid 33:208–217
Lee EH, Cho KS (2008) Characterization of cyclohexane and hexane degradation by Rhodococcus sp. EC1. Chemosphere 71:1738–1744
Lee S, Oh H, Kim O (2013) Isolation and characterisation of volatile organic compounds-degrading Bacillus strains from Loess. J Environ Protect 4:50–57
McClay K, Boss C, Keresztes I, Steffan RJ (2005) Mutations of toluene-4-monooxygenase that alter regiospecificity of indole oxidation and lead to production of novel indigoid pigments. Appl Environ Microbiol 71:5476–5483
Opere BO, Obayori OS, Raji AA (2013) Degradation of cyclohexane and cyclohexanone by Bacillus lentus strain LP32. Afr J Biotechnol 12(47):6632–6635
Park SH, Kim DH, Kim D, Kim DH, Jung HC, Pan JG, Ahn T, Kim D, Yun CH (2010) Engineering bacterial cytochrome P450 (P450) BM3 into a prototype with human P450 enzyme activity using indigo formation. Drug Metab Dispos 38(5):732–739
Perry JJ (1984) Microbial metabolism of cyclic alkanes. R.M. Atlas (Ed.), Petroleum. Microbiology 61–67
Prokop Z, Monincová M, Chaloupková R, Klvana M, Nagata Y, Janssen DB, Damborský J (2003) Catalytic mechanism of the haloalkane dehalogenase LinB from Sphingomonas paucimobilis UT26. J Biol Chem 278:45094–45100
Qing-Shan L, Schwaneberg U, Fischer P, Schmid RD (2000) Directed evolution of the fatty-acid hydroxylase P450 BM-3 into an indole-hydroxylation catalyst. Chem Eur J 6:1531–1536
Rouviere PE, Chen MW (2003) Isolation of Brachymonas petroleovorans CHX, a novel cyclohexane-degrading β-proteobacterium. FEMS Microbiol Lett 227:101–106
Salamanca D, Strunk N, Engesser KH (2013) Chromate reduction in anaerobic systems by bacterial strain Pseudomonas aeruginosa CRM100. Chemie Ingenieur Technik 85:1575–1580
Sardessai YN, Bhosle S (2004) Industrial potential of organic solvent tolerant bacteria. Biotechnol Progress 20:655–660
Schanstra JP, Janssen DB (1996) Kinetics of halide release of haloalkane dehalogenase: evidence for a slow conformational change. Biochemistry 35:5624–5632
Stirling LA, Watkinson RJ (1977) Microbial metabolism of alicyclic hydrocarbons: isolation and properties of a cyclohexane-degrading bacterium. J Gen Microbiol 99:119–125
Sugimori D, Sekiguchi T, Hasumi F, Kubo M, Shirasaka N, Ikunaka M (2004) Microbial hydroxylation of indole to 7-hydroxyindole by Acinetobacter calcoaceticus strain 4-1-5. Biosci Biotechnol Biochem 68(5):1167–1169
Trower MK, Buckland MB, Higgins R, Griffin M (1985) Isolation and characterization of a cyclohexane-metabolizing Xanthobacter sp. Appl Environ Microbiol 49:1282–1289
US Environmental Protection Agency (EPA) (1994) Chemical summary for cyclohexane. Washington D.C
Warburton EJ, Magor AM, Trower MK, Griffin M (1990) Characterization of cyclohexane hydroxylase; involvement of a cytochrome P-450 system from a cyclohexane grown Xanthobacter sp. FEMS Microbiol Lett 66:5–9
Yang C, Yu G, Zeng G, Yang H, Chen F, Jin C (2011) Performance of biotrickling filters packed with structured or cubic polyurethane sponges for VOC removal. J Environ Sci 23:1325–1333
Yi T, Lee EH, Ahn YG, Hwang GS, Cho KS (2011) Novel biodegradation pathways of cyclohexane by Rhodococcus sp. EC1. J Hazard Mater 191(1–3):393–396
Acknowledgements
We thank Prof. Dr. Andreas Schmidt, PD Dr. Katja Bühler and Rohan Karande from the Laboratory of Chemical Biotechnology, TU Dortmund University, for sharing their expertise in the field of biodegradation process.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Robert Duran
Rights and permissions
About this article
Cite this article
Salamanca, D., Engesser, KH. Isolation and characterization of two novel strains capable of using cyclohexane as carbon source. Environ Sci Pollut Res 21, 12757–12766 (2014). https://doi.org/10.1007/s11356-014-3206-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-014-3206-z