Abstract
Cadmium (Cd) is an extremely toxic metal commonly found as an environmental contaminant from industrial and agricultural sources, posing severe risks to human health. In this study, the binding mechanism of Cd(II)–human serum albumin (HSA) complex and the effect of Cd(II) on the conformational stability and structural state of HSA were comprehensively investigated through a series of efficient and appropriate methods. X-ray photoelectron spectroscopy accurately described the microenvironmental changes around protein C, N, and O atoms in the presence of Cd(II). Fluorescence results indicated that the probable mechanism of Cd(II)–HSA interaction is a static quenching process. Fourier transform infrared spectroscopy and dynamic light scattering showed Cd(II) complexation altered HSA conformation and the microenvironments of Trp and Tyr residues, accompanied by the size increases of HSA aggregates. This research will be helpful for understanding the toxic effects of Cd(II) on protein function in vivo.

The detailed binding mechanism between Cd(II) and HSA accompanied with the conformational analysis of HSA was comprehensively investigated at the molecular level
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Cadmium (Cd), belonging to group IIB of the periodic table of elements, is a potent environmental toxin that is present in measurable amounts in almost everything that we eat, drink, and breathe due to the widespread nature of its occurrence (Flick et al. 1971; Godt et al. 2006). This heavy metal pollutant has been demonstrated to have numerous undesirable effects on health in experimental animals and humans, targeting the kidneys, liver, and vascular systems in particular (Friberg 1984; Godt et al. 2006; Yang et al. 2013). It has been reported that occupational exposure to Cd(II) compounds in a number of industries, such as electronic components, electroplating, metal mechanic processing, and mining (Haddam et al. 2011; Haq et al. 1999), has been associated with increased risk of pathological cancer of the lung, kidney, and liver (Feki-Tounsi et al. 2013; Reshetin et al. 2003). In the blood circulation system, about 90 % of Cd(II) is bound to serum albumin and α2-macroglobulin, and Cd(II) bound to albumin is readily exchangeable (Scott and Bradwell 1983). In addition, Cd(II) contamination also causes metabolism problems in microorganisms (Heo et al. 2010; Mclamore et al. 2010). Due to its toxic characteristics, wastes from industrial processing of cadmium products are usually requested to be displaced properly in order to avoid the environment contamination (Mockaitis et al. 2012). Plants may contain certain amounts Cd through taking up Cd-contaminated water (Dudka and Adriano 1997). For human beings, Cd(II) has probably no biological function and could infiltrate into organisms through the respiratory tract and skin and then damage tissues and organs owing to its interactions with biological macromolecules (Nursita et al. 2009). People are exposed to Cd upon consumption of Cd containing plants or animals (Hallenbeck 1984). As a widely used pigment, Cd can also be found in plastic toys and food containers (Kumar and Pastore 2007). Even though the deadly effects of Cd(II) on human health have been known for many years, the molecular mechanism of Cd(II) toxicity is poorly understood. Therefore, in order to better understand the toxic effects of Cd(II) on human health, it is necessary to elucidate the ion–protein interaction mechanism and the toxicological action of Cd(II) at molecular level.
Serum albumins (SAs) are the major soluble plasma proteins of the blood circulatory system in vivo, which is the most common media for transporting heavy metal ions and also has the capability to bind a wide range of endogenous and exogenous compounds (Carter and Ho 1994; Peters 1985). As a kind of serum albumin, human serum albumin (HSA) is a single-chain transporting protein of 585 amino acids with 17 tyrosine residues and only one tryptophanes located in position 214 (Curry et al. 1998; Sudlow et al. 1975), which is the ligand carrier’s protein in blood serum and serves to amplify the capacity of plasma for transporting small molecules, such as drugs, fatty acids, hormones, and environmental toxicants (He and Carter 1992; Kamal et al. 2004). On the other hand, it has been demonstrated that the conformation of plasma protein can be changed by interaction with exogenous toxicants, which may influence protein’s biological function as a carrier protein (Chi and Liu 2011; Ding et al. 2011). Therefore, the studies of binding of toxic Cd(II) to plasma proteins not only are helpful for understanding the metabolism and distribution of this toxicant in vivo but also can further elaborate the toxic effects of Cd(II) on protein function during the blood transportation process.
Because SA is one of the major binding proteins with cadmium in human body, several studies on the complexation of Cd(II) to SA have been reported in response to the increased demand for detection and study of adverse effect of cadmium ions in our body with various technologies, including equilibrium dialysis, radioactive-isotope dilution, isothermal microcalorimetry, 1H and 113Cd NMR, potentiometric titration, immunoadsorbent chromatography, and zone electrophoresis technologies (Perkins 1961; Qu et al. 2002; Rao and Lal 1958; Sadler and Viles 1996; Scott and Bradwell 1983). However, as one of widely used technologies for obtaining binding information between metal ions and macromolecules, spectroscopic methods have never been used to study the binding interaction of SA with Cd. Besides, since the range of symptoms of cadmium poisoning is so broad, no study on the conformational changes of SAs caused by interaction with Cd(II) has been reported. The exact molecular mechanism of cadmium poisoning is still unknown up to now.
In the present study, the binding characteristics of Cd(II) on HSA at the molecular level were systematically investigated by using X-ray photoelectron spectroscopy (XPS), fluorescence spectroscopy, Fourier transform infrared spectroscopy (FTIR), and dynamic light scattering (DLS) methods. The inner filter effect had been corrected before we estimated the Cd(II)–HSA binding information such as binding constants, quenching rate constants, binding modes, binding sites, and intermolecular distances. The effects of Cd(II) on the microenvironment and conformation of HSA were also discussed. These results clarify the binding mechanism between Cd(II) and HSA, which is not only helpful for understanding the metabolism and distribution of this toxic metal ion in vivo but also can further elaborate the toxicological actions of Cd(II) on protein function during the blood transportation process.
Experimental section
Reagents
Human serum albumin, lyophilized and stored at 2–8 °C, was purchased from Sigma Chemical Co. and used as received. The cadmium nitrate, GR grade, was purchased from Merck. All other reagents of analytical grade were used. Double-distilled water with a resistivity of 18.2 MΩ (Milli-Q water, Millipore, USA) at room temperature (25 °C) was used throughout experiments. HSA was dissolved in 0.05 M phosphate-buffered saline solution to form an aqueous protein solution and then preserved at 4 °C for later use.
Apparatus
XPS measurements were performed on an ESCALAB 250 X-ray photoelectron spectrometer (Thermo-VG Scientific Corporate, USA). Infrared spectra were recorded on a Spectrum One FTIR spectrometer (Perkin-Elmer Corporate, USA). Fluorescence data were obtained on a Cary Eclipse Spectrofluorometer (Varian Corporate, USA) equipped with 1.0 cm quartz cells. Dynamic light scattering measurements were done with a Nano ZS (model: ZEN3600, Malvern), employing a 5-mW, 633-nm He–Ne laser and equipped with a thermostated sample chamber. All pH measurements were conducted with a PHS-3C acidity meter.
Methods
Cd(II)–HSA complex formation
Required amounts of HSA and cadmium nitrate solutions with/without further dilution are mixed together and kept aside for half an hour. Cd(II) ions attach to the electroactive sites of HSA forming a metal ion–protein complex.
Spectroscopic characterizations
XPS measurements were performed on an X-ray photoelectron spectrometer (ESCALAB 250, Thermo-VG Scientific Corporation) with amonochromatic focused Al Kα X-ray source (1,486.6 eV) to determine C, N, and O on the sample surface. Two 250 μM of the sample were dropped on an aluminum substrate and dried in vacuum desiccator overnight to obtain a thin film. The dried sample was then transferred to the XPS sample holder. The pressure in the sample chamber was maintained at 10−9 mbar. The detection was performed at 45° with respect to the sample surfaces. Thermo Avantage software was used for background subtraction, peak integration, fitting, and quantitative chemical analysis. Surface elemental stoichiometry was determined from peak–area ratio. For each sample, the analyses were made at three different points on the slices, and atomic fractions were calculated from averages of the three.
For the fluorescence measurement, Cd(II) solution was added into the protein solutions in sequence. Both Cd(II) and HSA concentrations were calculated by weight for consistency. The systems were excited at 280 nm, and the emission wavelength was adjusted from 300 to 500 nm with a scanning speed of 600 nm min−1. The excitation and emission slit widths were both set at 5.0 nm. All the experiments were conducted in triplicate and further analyzed using Origin 8.0.
Time-resolved fluorescence spectra were executed in a time-correlated single photon counting system from FL920P spectrometer (Edinburgh Instruments, UK) with λ ex = 295 nm. The data are fitted to biexponential functions after deconvolution of the instrumental response function by an iterative reconvolution approach by the DAS6 decay analysis software utilizing reduced χ 2 and weighted residuals as parameters for goodness of fit.
Molecular theoretical calculations were carried out using ICM (v3.4–8) software. The native structure of HSA (PDB ID 4IW2) was taken from Protein Data Bank with a crystalline resolution at 2.41 Å (Wang et al. 2013). All water molecules were removed, and local search was used to search for the optimum binding site of small molecules to the protein. A grid size of 90 × 90 × 90 points with a grid spacing of 0.4 Å was applied. The docking conformation with the lowest binding free energy was further processed by Pymol software.
For infrared spectra, the cadmium nitrate solution was added dropwise to the protein solution with constant stirring to ensure the formation of homogeneous solution and reach the target Cd concentrations of 2, 5, and 10 mM with a final protein concentration of 0.25 mM. Then, the sample solutions were placed between CaF2 windows separated with a 25-μM spacer. Spectra were acquired at 4 cm−1 resolution and 100 scans.
The DLS experiments were carried out at 298 K with a Zetasizer Nano ZS (Malvern Instruments). The scattering intensity data were processed using the instrumental software to obtain the hydrodynamic diameter (d H) and the size distribution of the scatterer in each sample. The instrument measures the time-dependent fluctuation in the intensity of light scattered from the particles in solution at a fixed scattering angle. Hydrodynamic diameter, d H, is defined as,
where k B = Boltzmann constant, T = absolute temperature, η = viscosity coefficient of the solvent, and D = translational diffusion coefficient. Each measurement was performed on a freshly extracted aliquot of the protein sample and diluted with phosphate buffer to a final concentration of 10 μM at different time intervals. All the experiments were conducted in triplicate and further analyzed using Origin 8.0 software.
Results and discussion
XPS study for Cd(II)–HSA complexation
To accurately investigate the binding interaction between Cd(II) and HSA at atomic level, we firstly applied XPS study. Figure 1 shows that the XPS spectrum of HSA only contains signals for the elements of C, N, and O, which are the main constituents of the amino acid residues and occur at 285, 400, and 532 eV, respectively. Figure 2 shows that HSA before and after the treatment with Cd(II) have different XPS C 1s core level spectra. The C 1s spectra can be deconvoluted into three components, which are attributed to carbon atom bound to carbon and hydrogen (C–C and C–H), carbon singly bound to nitrogen and oxygen (C–N and C–O), and carbon involved in an amide bond (O=C–NH), respectively (Belatik et al. 2012; Wu et al. 2009). The binding energies and C atomic fractions at different chemical states with and without Cd(II) are listed in Table 1. The intensities of the component located at around 284.6 and 285.8 eV changed obviously, which were probably coming from the complexation of Cd with carbon atoms and induced changes in the skeletal structure of protein. However, the intensity of the component located at 287.6 originating from the peptide bond almost had no change, implying that the primary structure of HSA had not yet been destroyed at the Cd(II) concentration = 0.25 mM. Figures 3 and 4 show the XPS signals attributed for N and O. The nitrogen component that bound to carbon (C–N) is located at 399.6 eV, and the other nitrogen component bound to hydrogen (N–H2) is located at 400.1 eV. The oxygen signals represented C=O and O–H are positioned at 531.2 and 532.3 eV, respectively. After the addition of Cd(II), the binding energy and atomic fraction values were changed, indicating the direct interaction between Cd and nitrogen, oxygen atoms. The major fraction and intensity changes observed for N and O were coming from direct Cd coordination with nitrogen and oxygen atoms, which probably induced the formation of [Cd=O] and [Cd≡N] units in the Cd–HSA complex.
Moreover, the experimental atomic composition as determined from the XPS spectral analysis was also calculated for nitrogen to carbon (N/C) and oxygen to carbon (O/C) ratios in the absence and presence of Cd(II). The values of N/C and O/C are 18.25 and 24.44 % without Cd(II), while the corresponding values increased to 19.24 and 28.06 % in the presence of Cd(II). The major ratio changes observed for N and O were coming from direct Cd coordination with nitrogen and oxygen atoms, while the alterations of C ratios were related to the linkage of C atom to metal ion-bonded N and O atoms. The XPS results show that the microenvironment around protein C, N, and O changes in the presence of Cd(II).
Fluorescence spectra
Effect of Cd on the fluorescence spectra of HSA
Fluorescence quenching is a powerful tool to characterize the accessibility of fluorophores in the protein matrix to quenchers in solution. Detailed information of metal ions to protein such as the binding mode, binding constants, and conformational transformation can be obtained by the fluorescence technique (Lakowicz 2006). Herein, fluorescence quenching was utilized to study the interaction between Cd(II) and HSA.
Since the inner filter effect (IFE) may affect fluorescence measurements due to the formation of Cd(II)–HSA complex, we take advantage of some effective methods to correct the fluorescence of the complex and reduce the IFE. Firstly, each fluorescence spectrum of HSA in the presence of different concentrations of Cd(II) was corrected for any possible IFE using the following equation (Eq. 1) (Gu and Kenny 2009):
where F cor is the corrected fluorescence intensity obtained in the absence of inner-filter effect, F obs is the measured fluorescence, and A ex and A em are the sum of the absorbances of protein and ligand at the excitation and emission wavelengths, respectively, caused by ligand addition (in a 1 × 1-cm pathlength cuvette). Furthermore, in order to reduce as much as possible the inner filter effect caused by the absorption of both excitation radiation and emission radiation, we controlled the HSA concentration to be 2.0 × 10−6 mol L−1 and the maximum concentration of Cd(II) to be 1.0 × 10−4 mol L−1. Thus, any possible contribution of inner filter effect due to attenuation of the incident light by the quencher could be negligible. In addition, fluorescence backgrounds were also corrected for blank buffer solutions in each measurement.
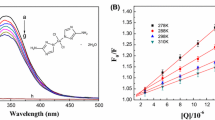
As shown in Fig. 5a, free HSA gave a strong fluorescence emission band at 345 nm when excited at 280 nm, and free Cd(II) did not show any fluorescence emission under the same condition. The addition of Cd(II) led to a concentration-dependent quenching of the intrinsic fluorescence of HSA, and the wavelength emission maximum λ max remained nearly unchanged in the HSA spectrum. HSA contains a single polypeptide of 585 amino acids with only one tryptophan (Trp-214) located in subdomain II A, and the Trp residue emission mainly dominates HSA fluorescence spectra in the UV region. The characteristic of the intrinsic fluorescence of HSA is very sensitive to its microenvironment, so changes in the intrinsic fluorescence of HSA can provide valuable information about its structure and dynamics. Therefore, the results imply that the binding of Cd(II) to HSA occurs and Cd(II) probably increases the exposure of the chromophore amino acids to the aqueous medium and therefore quench the intrinsic fluorescence of HSA.
a Effect of Cd(II) on HSA fluorescence at 293 K. The inset shows the dependence of the fluorescence intensities of HSA by Cd(II) at corresponding concentrations; b Stern–Volmer plots for the fluorescence quenching of HSA by the addition of Cd(II); c plots of log(F 0 − F)/F versus log{1/[Q] − (F 0 − F)[P t]/F 0)} for Cd(II)–HSA system at different temperatures. Conditions: [HSA] = 2.0 × 10−6 mol L−1; [Cd(II)] (a–q) = 0, 1.0, 2.0, 3.0, 4.0, 5.0, 6.0, 7.0, 8.0, 9.0, and 10.0 × 10−4 mol L−1; d The time-resolved fluorescence decay of 1.0 × 10−6 mol L−1 HSA in the presence of Cd(II) with different concentrations
The fluorescence quenching mechanism
Fluorescence quenching can occur in two different processes: static and dynamic quenchings (Lakowicz 2006). In this study, both static and dynamic processes are described by the Stern–Volmer Eq. 2.
where F 0 and F are the steady-state fluorescence intensities without and with quencher (ET), respectively; K q is the quenching rate constant; K SV is the Stern–Volmer quenching constant; and [Q] is the concentration of the quencher. Hence, Eq. 2 was applied to determine K SV by linear regression of a plot of F 0/F against [Q].
Figure 5b displays the Stern–Volmer plots of the quenching of HSA tryptophan residue fluorescence by Cd(II). The values of K SV and K q were further calculated and listed in Table 2. It shows that the Stern–Volmer plots are linear and the linearity does not affected by the increasing temperature. The linearity of the Stern–Volmer plots may indicate the existence of a single type of quenching, either static or dynamic. Table 2 showed that K SV and K q were inversely correlated with temperature, which indicated that the probable quenching mechanism of Cd(II)–HSA binding reaction might be initiated by complex formation rather than by dynamic collision (Liu et al. 2013a). Moreover, values for K q are much greater than the maximum diffusion collision quenching rate constant of the biomolecule (2.0 × 1010 M−1 s−1) (Lakowicz 2006); therefore, the static quenching could be the main mechanism of the fluorescence quenching of HSA by Cd(II).
In addition, static quenching can easily be distinguished from that of dynamic preferably by lifetime measurements (Ware 1975). In this study, we used the lifetime measurements to confirm the quenching mechanism as static. Figure 5d exhibits the fluorescence decay of HSA in the absence and presence of Cd(II). The results show that Cd(II) with increasing concentrations had no effect in the lifetime of HSA, which reveals that the quenching mentioned above is not initiated by dynamic collision but from the ground state complex formation of static quenching.
Binding constants and number of binding sites
The static mechanism is principally responsible for the observed fluorescence quenching when Cd(II) binds to HSA as discussed above. In order to investigate equilibrium between free and bound molecules and rationalize our experimental data on the Cd(II)–HSA systems, the following equation has been employed (Eq. 3) (Mandal and Ganguly 2009):
where F 0 and F are the fluorescence intensities before and after the addition of the quencher, K a is the apparent binding constant, and n is the average number of binding sites per Hb; [Q] is the total ligand concentration, and [P t] are the total protein concentration. By using Eq. 3, the values of K a and n at physiological pH 7.4 can be calculated. Figure 5c is the plots of log (F 0 − F)/F versus log (1/([Q] − (F 0 − F) × [P t]/F 0)) for the Cd(II)–HSA system at different temperatures, and the linearities are very well fitted with all R value >0.99. The binding constant is 7.692 × 103, 6.385 × 103, and 4.988 × 103 L mol−1, and the number of binding sites is 1.161, 1.142, and 1.114 at 293, 303, and 313 K, respectively. The results showed that the binding constants K a and binding sites n were decreased with increasing temperature, which may suggest that Cd(II) is partially dissociated from HSA as temperature increases. The number of binding sites n approximately equals 1, indicating that there is one binding site with higher binding affinity and selectivity in HSA for Cd(II) during their binding process.
Thermodynamic parameters
Generally, the binding forces between ligands and biomolecules may comprise hydrogen bonds, electrostatic forces, van der Waals forces, hydrophobic interactions, etc. (Gautier et al. 2008). The thermodynamic parameters, enthalpy change (∆H), free-energy change (∆G), and the entropy change (∆S) are the main evidences for confirming binding modes and could be calculated according to the van’t Hoff equation (Eq. 4) (Ross and Subramanian 1981):
where K a is analogous to the associative binding constants at the corresponding temperature and R is the gas constant. To elucidate the interaction between Cd(II) and HSA, the thermodynamic parameters can be calculated from the van’t Hoff plots. ∆G can then be estimated from the following relationship (Eq. 5):
The main binding forces can be judged by thermodynamic parameters. From the viewpoint of thermodynamics, ∆H > 0 and ∆S > 0 imply that hydrophobic interaction is the main force; ∆H < 0 and ∆S < 0 reflect van der Waals force or hydrogen bonding; ∆H < 0 and ∆S > 0 suggest that electrostatic forces play a key role (Kumar et al. 2002). On account of the little temperature effect, ∆H can be regarded as a constant if the temperature range is not too wide. The plots of lnK a versus 1/T are presented in Fig. 6 and show a good linear relationship. The calculated thermodynamic parameters and K a values for the binding interaction between Cd(II) and HSA are listed in Table 3. The corresponding negative ∆H and positive ∆S suggest that Cd(II) itself carries a positive charge and HSA carries negative charge in aqueous solution at physiological pH; therefore, the electrostatic forces play a major role in the formation of Cd(II)–HSA complex.
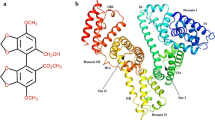
To further define the binding site, computational molecular docking was employed to improve the understanding of the interaction between Cd(II) and HSA. The best energy ranked result is shown in Fig. 7, and the geometry exhibits the lowest free energy. The docking result shows that Cd(II) mainly interacts with the negatively charged amino acid residues Asp451, Pro447, and Gln 221 predominately through electrostatic force, in agreement with the conclusion of thermodynamic analysis. The van der Waals interactions and some other forces might also exist between Cd(II) and HSA, but are not dominating. In addition, the formation of Cd(II)–HSA complex might give rise to a perturbation in the corresponding subdomain and then induce the structural alteration of HSA and microenvironment changes of Trp and Tyr, which could be reflected in the fluorescence intensity changes (Liu et al. 2013b).
Investigation on secondary structural changes of HSA
FTIR spectroscopy has been widely applied to monitor the conformational changes of proteins that spans different secondary structures such as α-helix (1,650–1,660 cm−1), β-sheet (1,610–1,640 cm−1), turn structure (1,660–1,680 cm-1), β-antiparallel (1,680–1,692 cm−1), and random coil (1,641–1,648 cm−1) (Mantsch and Chapman 1996). The infrared spectra of proteins are mainly comprised of amide I (1,600–1,700 cm−1) and amide II (1,500–1,600 cm−1), which are caused by the carbonyl stretching vibrations of the peptide backbone (amide I band) and the combination of N–H in-plane bending and C–N stretching vibrations of peptide groups (amide II band) (Kong and Yu 2007; Mantsch and Chapman 1996). Herein, the interaction between Cd(II) and HSA was characterized by infrared spectroscopy and its derivative methods. The difference spectra [(protein + Cd solution) − (buffer solution)] were obtained. Figure 8 shows that an intense band in the amide I region centered at 1,655 cm−1 could be observed in the original FTIR spectra of native HSA without Cd (II) at pH = 7.4, indicating that HSA is in an α-helix-rich conformation. As compared with that for the native HSA, the band in the Cd(II)–HSA complex system showed obvious changes in both shape and peak position (1,655–1,651 cm−1). The spectral shift for the amide I from 1,655 to 1,651 cm−1 in the spectra of Cd(II)–HSA complexes was due to the binding of Cd cation to protein C=O and C–N groups. In the presence of Cd(II), an increase in FTIR intensity was observed for the HSA amide I band, which was probably due to metal ion binding to protein C=O, C–N, and N–H groups. The results indicated that [Cd=O] and [Cd≡N] units were probably formed in the Cd–HSA complexes. Additional evidence to support the Cd interaction with C–N and N–H groups came from the shifting of the protein amide A band at 3,312 cm−1 (N=H stretching mode) in the free HSA to 3,304 upon metal ion–protein complexation (spectra not shown).
Moreover, since the secondary structures are stabilized by hydrogen bonds between amide C=O and N–H groups, the position and the area of the components could reflect the patterns and the strength of the hydrogen bonds (Byler and Susi 1986). Thus, the infrared self deconvolution with second derivative resolution enhancement and curve-fitting procedures was further applied to determine the protein secondary structure changes in the absence and presence of Cd(II). A quantitative analysis of the protein secondary structure for the free HSA and its Cd cation adducts in hydrated films were carried out, and the calculated results are listed in Table 4. According to Fig. 8a and Table 4, the free HSA has 58.12 % α-helix (1,651 cm−1), 13.35 % β-sheet (1,627 and 1,616 cm−1), 12.72 % turn structure (1,674 and 1,663 cm−1), 2.75 % β-antiparallel (1,684 cm−1), and 13.06 % random coil (1,640 cm−1). The results are consistent with the spectroscopic studies of human serum albumin previously reported (Beauchemin et al. 2007; Belatik et al. 2012). Upon Cd(II) interaction, an obvious decrease of α-helix from 58.12 % (free HSA) to 43.43 % (1 mM Cd(II)), 39.84 % (2 mM Cd(II)), and 32.14 % (10 mM Cd(II)) with an alteration in β-sheet from 13.35 % (free HSA) to 14.58 % (1 mM Cd(II)), 14.79 % (2 mM Cd(II)), and 10.39 % (10 mM Cd(II)) was observed. It was noted that an remarkable increase was observed for the random coil from 13.06 % (free HSA) to 19.86 % (1 mM Cd(II)), 20.44 % (2 mM Cd(II)), and 33.28 % (10 mM Cd(II)). The content of turn structure in HSA also increases from 12.72 % (free HSA) to approximately 20 % (1, 2, 10 mM Cd(II)) in the presence of Cd. Besides, a slight structural changes of β-antiparallel was also observed from 2.75 (free HSA) to 4.76 % (10 mM Cd(II)). Figure 9 clearly shows that some α-helix can be obviously converted into random coil when HSA is conjugated with Cd(II). The major decrease in α-helix structure and increase in random coil structures suggested a partial protein unfolding with the increase of Cd(II) concentration.
Besides, in order to examine the influence of the metal ion–protein binding interaction on the hydrophobic regions of HSA, the spectral changes of HSA antisymmetric and symmetric CH2 stretching vibrations in the region of 2,850–3,000 cm−1 were also monitored by infrared spectroscopy. As shown in Fig. 10, the CH2 bands of the free HSA located at 2,961, 2,938, and 2,876 cm−1 changed to 2,960, 2,939, 2,901, 2,875, 2,867, and 2,856 cm−1 in Cd(II)–HSA complex. The alterations of the protein antisymmetric and symmetric CH2 stretching vibrations suggested that the microenvironment of hydrophobic regions in HSA was changed through the metal ion–protein complexation.
Investigation on the aggregation structural changes of HSA
In order to further confirm the aggregation state of HSA due to the addition of Cd(II), we performed DLS experiments at pH = 7.4 with different molar ratios of Cd(II) to HSA to measure normalized hydrodynamic diameter (d H), which could give us an estimate regarding the average size of the total protein. Figure 11 represents d H distribution plot of HSA in the absence and presence of Cd(II). The d H of HSA alone in buffer was found to be around 5.16 nm (i.e., the hydrodynamic radius = 2.58 nm) at pH 7.4. The d H values for HSA increased to 5.67 and 6.56 nm at [Cd(II)]/[HSA] = 100:1 and 300:1, respectively. The increase in d H indicated that Cd(II) was assembled around HSA via the binding interaction between Cd(II) and HSA, leading the particle size of HSA aggregated to become larger. This result indicated that loss of charge from the binding site of albumin by the charged Cd(II) ions gave rise to dipole interaction which acted as the major contributor to promote protein aggregation. But the obvious reduce of d H value to 3.81 nm happened at [Cd(II)]/[HSA] = 1,000:1, which demonstrated that HSA suffered a maximum loss of its secondary structure and the conformation of HSA might be completely destroyed when Cd(II) was overdosed into the Cd(II)–HSA aqueous system. The results indicated that the presence of Cd(II) with low concentration could cause the formation of Cd(II)–HSA complex and further induce the aggregation structure changes of HSA, but Cd(II) with high concentration could result in structural damage of protein, leading to the size reduction of HSA aggregates.
Conclusion
In this work, the molecular mechanism involved in the interaction between Cd(II) and HSA was comprehensively investigated by a series of spectroscopic methods. Meanwhile, we also systematically studied the effect of food contaminant Cd(II) on the conformational stability of HSA in an aqueous environment. XPS analysis revealed the direct interaction of polypeptide C, N, and O atoms of HSA with Cd(II), which induced the microenvironmental changes around these protein atoms. The fluorescence results indicated that the probable mechanism of Cd–HSA interaction is a static quenching process and the binding reaction is spontaneous. Thermodynamic analysis combined with molecular docking results showed that Cd(II) mainly interacted with the negatively charged amino acid residues Asp 451 and Gln 221 predominately through electrostatic force. The structural analysis according to FTIR suggested that with the increased amount of Cd(II), the secondary structure of HSA underwent a decrease in α-helix and an increase in random coil structures. The aggregation states of HSA in the absence and presence of Cd(II) were further studied by DLS methods, and the results indicated that Cd(II) might interact with HSA and assembled around HSA, leading to the formation of HSA aggregates with larger size. However, Cd(II) with extremely high concentration could lead to the structural damage of the protein, leading to the size reduction of HSA aggregates. The basic information received from this work not only is helpful in clarifying the binding mechanism of Cd(II), the potential carcinogen, with HSA in vivo for further understanding its transportation, metabolism, elimination, and biodistribution during the blood transportation process but also will be of great importance in the toxicological evaluation of exogenous molecules at molecular level.
References
Beauchemin R, N'soukpoe-Kossi CN, Thomas TJ, Thomas T, Carpentier R, Tajmir-Riahi HA (2007) Polyamine analogues bind human serum albumin. Biomacromolecules 8:3177–3183
Belatik A, Hotchandani S, Carpentier R, Tajmir-Riahi HA (2012) Locating the binding sites of Pb(II) Ion with human and bovine serum albumins. Plos One 7:e36723
Byler DM, Susi H (1986) Examination of the secondary structure of proteins by deconvolved FTIR spectra. Biopolymers 25:469–487
Carter DC, Ho JX (1994) Structure of serum-albumin. Adv Protein Chem 45(45):153–203
Chi ZX, Liu RT (2011) Phenotypic characterization of the binding of tetracycline to human serum albumin. Biomacromolecules 12:203–209
Curry S, Mandelkow H, Brick P, Franks N (1998) Crystal structure of human serum albumin complexed with fatty acid reveals an asymmetric distribution of binding sites. Nat Struct Biol 5:827–835
Ding F, Wei L, Diao JX, Sun Y (2011) Characterization of alizarin Red S binding sites and structural changes on human serum albumin: a biophysical study. J Hazard Mater 186:352–359
Dudka S, Adriano DC (1997) Environmental impacts of metal ore mining and processing: a review. J Environ Qual 26:590–602
Feki-Tounsi M, Olmedo P, Gil F, Khlifi R, Mhiri MN, Rebai A, Hamza-Chaffai A (2013) Cadmium in blood of Tunisian men and risk of bladder cancer: interactions with arsenic exposure and smoking. Environ Sci Pollut R 20:7204–7213
Flick DF, Kraybill HF, Dimitrof JM (1971) Toxic effects of cadmium—review. Environ Res 4:71–85
Friberg L (1984) Cadmium and the kidney. Environ Health Persp 54:1–11
Gautier A, Kirkpatrick JP, Nietlispach D (2008) Solution-state NMR spectroscopy of a seven-helix transmembrane protein receptor: backbone assignment, secondary structure, and dynamics. Angewandte Chemie-International Edition 47:7297–7300
Godt J, Scheidig F, Grosse-Siestrup C, Esche V, Brandenburg P, Reich A, Groneberg DA (2006) The toxicity of cadmium and resulting hazards for human health. J Occup Med Toxicol 1:22
Gu Q, Kenny JE (2009) Improvement of inner filter effect correction based on determination of effective geometric parameters using a conventional fluorimeter. Anal Chem 81:420–426
Haddam N, Samira S, Dumont X, Taleb A, Lison D, Haufroid V, Bernard A (2011) Confounders in the assessment of the renal effects associated with low-level urinary cadmium: an analysis in industrial workers. Environ Health-Glob 10
Hallenbeck WH (1984) Human health-effects of exposure to cadmium. Experientia 40:136–142
Haq R, Zaidi SK, Shakoori AR (1999) Cadmium resistant Enterobacter cloacae and Klebsiella sp isolated from industrial effluents and their possible role in cadmium detoxification. World J Microb Biot 15:283–290
He XM, Carter DC (1992) Atomic-structure and chemistry of human serum-albumin. Nature 358:209–215
Heo DH, Baek IJ, Kang HJ, Kim JH, Chang M, Jeong MY, Kim TH, Choi ID, Yun CW (2010) Cadmium regulates copper homoeostasis by inhibiting the activity of Mac1, a transcriptional activator of the copper regulon, in Saccharomyces cerevisiae. Biochem J 431:257–265
Kamal JKA, Zhao L, Zewail AH (2004) Ultrafast hydration dynamics in protein unfolding: human serum albumin. Proc Natl Acad Sci U S A 101:13411–13416
Kong J, Yu S (2007) Fourier transform infrared spectroscopic analysis of protein secondary structures. Acta Bioch Bioph Sin 39:549–559
Kumar A, Pastore P (2007) Lead and cadmium in soft plastic toys. Curr Sci India 93:818–822
Kumar CV, Buranaprapuk A, Sze HC, Jockusch S, Turro NJ (2002) Chiral protein scissors: high enantiomeric selectivity for binding and its effect on protein photocleavage efficiency and specificity. Proc Natl Acad Sci U S A 99:5810–5815
Lakowicz JR (2006) Principles of fluorescence spectroscopy. Springer, New York, pp 278–290
Liu Y, Chen MM, Luo ZP, Lin JJ, Song L (2013a) Investigation on the site-selective binding of bovine serum albumin by erlotinib hydrochloride. J Biomol Struct Dyn 31:1160–1174
Liu Y, Lin JJ, Chen MM, Song L (2013b) Investigation on the interaction of the toxicant, gentian violet, with bovine hemoglobin. Food Chem Toxicol 58:264–272
Mandal P, Ganguly T (2009) Fluorescence spectroscopic characterization of the interaction of human adult hemoglobin and two isatins, 1-methylisatin and 1-phenylisatin: a comparative study. J Phys Chem B 113:14904–14913
Mantsch HH, Chapman D (1996) Infrared spectroscopy of biomolecules. Wiley-Liss, New York, 359pp
Mclamore ES, Zhang W, Porterfield DM, Banks MK (2010) Membrane-aerated biofilm proton and oxygen flux during chemical toxin exposure. Environ Sci Technol 44:7050–7057
Mockaitis G, Rodrigues JAD, Foresti E, Zaiat M (2012) Toxic effects of cadmium (Cd2+) on anaerobic biomass: kinetic and metabolic implications. J Environ Manage 106:75–84
Nursita AI, Singh B, Lees E (2009) Cadmium bioaccumulation in Proisotoma minuta in relation to bioavailability in soils. Ecotoxicol Environ Saf 72:1767–1773
Perkins DJ (1961) Studies on the interaction of zinc, cadmium and mercuric ions with native and chemically modified human serum albumin. Biochem J 80:668–672
Peters T (1985) Serum-albumin. Adv Protein Chem 37:161–245
Qu SS, Liu Y, Wang TZ, Gao WY (2002) Thermodynamics of binding of cadmium to bovine serum albumin. Chemosphere 46:1211–1214
Rao MSN, Lal H (1958) Metal protein interactions in buffer solutions. 2. A polarographic study of the interaction of Zn-II and Cd-II with bovine serum albumin. J Am Chem Soc 80:3222–3226
Reshetin VP, Kazazyan VI, Regens JL, Gunter JT (2003) Cancer risk assessment and ambient cadmium concentrations: mortality estimates from Moscow, Samara, and Saratov. Environ Sci Pollut R: 162–164
Ross PD, Subramanian S (1981) Thermodynamics of protein association reactions—forces contributing to stability. Biochemistry 20:3096–3102
Sadler PJ, Viles JH (1996) 1H and (113)Cd NMR investigations of Cd(2+) and Zn(2+) binding sites on serum albumin: competition with Ca(2+), Ni(2+), Cu(2+), and Zn(2+). Inorg Chem 35:4490–4496
Scott BJ, Bradwell AR (1983) Identification of the serum binding-proteins for iron, zinc, cadmium, nickel, and calcium. Clin Chem 29:629–633
Sudlow G, Birkett DJ, Wade DN (1975) The characterization of two specific drug binding sites on human serum albumin. Mol Pharmacol 11:824–832
Wang Y, Yu HY, Shi XL, Luo ZP, Lin DH, Huang MD (2013) Structural mechanism of ring-opening reaction of glucose by human serum albumin. J Biol Chem 288:15980–15987
Ware WR (1975) Photophysics of exciplexes—some kinetic aspects. Pure Appl Chem 41:635–660
Wu B, Mu CD, Zhang GZ, Lin W (2009) Effects of Cr3+ on the structure of collagen fiber. Langmuir 25:11905–11910
Yang H, Huo X, Yekeen TA, Zheng QJ, Zheng MH, Xu XJ (2013) Effects of lead and cadmium exposure from electronic waste on child physical growth. Environ Sci Pollut R 20:4441–4447
Acknowledgments
The authors gratefully acknowledge the financial supports from the National Natural Science Foundation of China (no. 20875055); the Natural Science Foundation of Fujian Province, China (no. 2013J01388); the State Key Lab of Structural Chemistry, Fujian Institute of Research on the Structure of Matter; and the Opening Research Foundation of Key Laboratory of Biomedical Material in Tianjin city.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Philippe Garrigues
Rights and permissions
About this article
Cite this article
Liu, Y., Chen, M., Jiang, L. et al. New insight into molecular interaction of heavy metal pollutant—cadmium(II) with human serum albumin. Environ Sci Pollut Res 21, 6994–7005 (2014). https://doi.org/10.1007/s11356-014-2610-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-014-2610-8