Abstract
Purpose
Bacterial community structure and the chemical components in aerosols caused by rotating brushes in an Orbal oxidation ditch were assessed in a Beijing municipal wastewater treatment plant.
Methods
Air samples were collected at different distances from the aerosol-generating rotating brushes. Molecular culture-independent methods were used to characterize the community structure of the airborne bacteria in each sample regardless of cell culturability. A clone library of 16S rDNA directly amplified from air DNA of each sample was constructed and sequenced to analyze the community composition and diversity. Insoluble particles and water-soluble ions emitted with microorganisms in aerosols were analysis by a scanning electron microscope together with energy dispersive X-ray spectroscopy and ion chromatogram analyzer.
Results
In total, most of the identified bacteria were Proteobacteria. The majority of sequences near the rotating brushes (the main source of the bioaerosols) were Proteobacteria (62.97 %) with β-(18.52 %) and γ-(44.45 %) subgroups and Bacteroidetes (29.63 %). Complex patterns were observed for each sampling location, suggesting a highly diverse community structure, comparable to that found in water in the Orbal oxidation ditch. Accompany with microorganisms, 46.36 μg/m3 of SO 2−4 , 29.35 μg/m3 of Cl−, 21.51 μg/m3 of NO −3 , 19.76 μg/m3 of NH +4 , 11.42 μg/m3 of PO 3−4 , 6.18 μg/m3 of NO −2 , and elements of Mg, Cl, K, Na, Fe, S, and P were detected from the air near the aerosols source.
Conclusions
Differences in the structure of the bacterial communities and chemical components in the aerosols observed between sampling sites indicated important site-related variability. The composition of microorganisms in water was one of the most important sources of bacterial communities in bioaerosols. Chemical components in bioaerosols may provide a media for airborne microorganism attachment, as well as a suitable microenvironment for their growth and survival in the air. This study will be benefit for the formulation of pollution standards, especially for aerosols, that take into account plant workers’ health.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Within wastewater treatment plant (WWTP), the sites that contain moving mechanical equipment for water aeration are the regions with the highest emission of bioaerosols (Orsini et al. 2002; Pascual et al. 2003; Sánchez-Monedero et al. 2008). As bioaerosols contain viruses as well as different bacteria and fungi, they may cause potential health hazards to plant workers and adjacent communities. Consequently, bioaerosol emissions have become of increasing concern and research interest in recent years.
Understanding bioaerosol production and transmission will help develop effective measures to minimize their generation. Previous research has investigated the concentrations, particle size distributions, and species diversity of airborne microorganisms at different stages in WWTPs by using different sampling and detection methods (Fannin et al. 1985; Buttner et al. 2001; Pillai and Ricke 2002; Grisoli et al. 2009). Major advances have been made in the understanding of bioaerosol characteristics and in identifying their hazards (Palmer et al. 1995; Carducci et al. 2000). These studies were based on culture-dependent methods, however, which do not necessarily reflect the real microflora in bioaerosol samples because only a small proportion (less than 10 %) of all microorganisms can be cultivated (Amann et al. 1995), especially in the atmospheric environment. In addition, the analyses used are time-consuming and laborious and lack sensitivity and specificity (Stetzenbach et al. 2004).
The development of various analysis techniques based on DNA extracted directly from natural samples has helped detect nonculturable airborne microorganisms and characterize their genetic structure and community diversity in complex environments. Polymerase chain reaction (PCR)-based methods have been used to detect and quantify aerosolized bacteria and fungus (Cruz-Perez et al. 2001; Williams et al. 2001; Rintala et al. 2004), as well as estimate airborne microorganisms present in WWTPs (Pascual et al. 2003) and rapidly detect and determine airborne Mycobacteria associated with whirlpools (Schafer et al. 2003). PCR-based methods have also been applied to distinguish seasonal microbial aerosol emissions from the surfaces of aeration tanks in industrial and municipal WWTPs (Ranalli et al. 2000), determine the presence of Varicella zoster virus in the atmosphere (Sawyer et al. 1994), and analyze environmental sample assays in response to bioterrorism attack (Higgins et al. 2003). Compared with culture-based techniques for the detection of aerosolized Escherichia coli DH1, results with solid-phase PCR showed an increase in detection sensitivity (Alvarez et al. 1994). Similar results have also been obtained for comparisons between real-time PCR and conventional quantized techniques for airborne microorganism quantification in environmental samples (An et al. 2006). The cloning/sequencing method has been used to examine the microbial quality of aerosols and determine the size distribution of microorganisms in the air over the eastern Mediterranean during a storm (Polymenakou et al. 2008), as well as to analyze community composition and diversity of air samples collected from northern France (Maron et al. 2005).
While such techniques are widely applied to analyze microbial diversity in the atmospheric environment, very few studies have reported their application in resolving the genetic diversity and structure of airborne bacterial communities emitted from WWTPs, especially those using oxidation ditch processes. The anaerobic-anoxic-oxic, sequencing batch reactor activated sludge, and oxidation ditch wastewater treatment processes are all applied widely in WWTPs. Among them, more than 60 % of WWTPs in China use the oxidation ditch treatment process in which rotating aeration brushes are installed to provide linear liquid motion and water aeration. Results of our previous investigation showed that high concentrations of airborne microorganisms were found near the rotating brush due to their mobilization, and the structure of bacterial communities in bioaerosols had a close relationship with that of microorganisms in the water (Li et al. 2011).
Unlike those released into the air from soil surfaces, bioaerosols generated from water sources are usually form with a thin layer of moisture surrounding the cell. In addition to microorganism zoogloea, sludge particles, and small water drops, inorganic compounds are also present in bioaerosols. Although they can significantly affect microorganism survival in the air, the chemical components emitted with microorganisms in aerosols have seldom been reported in previous research.
The main objectives of the present study were to analyze the microbial structure of bioaerosols caused by rotating brushes and their relationship with water in the Orbal oxidation ditch by using a cloning/sequencing method. Bioaerosol samples were collected at different distances from the rotating brushes. The production and transmission of aerosols were described and determined by their distributions and variations in species diversity and chemical components at different distances from the aerosol source in the treatment plant.
2 Materials and methods
2.1 Sampling locations
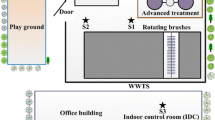
The study was conducted at an Orbal oxidation ditch in a municipal WWTP, located in southern Beijing (Li et al. 2011). Five outdoor sampling sites were established in July 2010 at the WWTP Orbal oxidation ditch for microbiological and chemical assays (Fig. 1 and Table 1). The sampling sites monitored airflow from the aerosol source (rotating brushes in oxidation ditch) and were situated 0.5 m before (PBA), 0.5 m after (BBA), and 40 m downwind (BFBA) from the rotating brushes. Water (W) in the Orbal oxidation ditch was collected 10 m before the rotating brushes for use as the control measurement (Table 1). Parallel to the microbiological assays, observations of select meteorological parameters were conducted. During air sampling, the temperature and relative humidity were recorded by a Dewpoint Thermohygrometer (WD-35612, OAKTON, Germany). The wind speed and irradiance were measured by a portable anemometer (HD2303, Delta OHM, Italy) and irradiance meter (HD2302.0, Delta OHM, Italy), respectively (Table 1).
2.2 Air sample collection
All air samples were collected 1.5 m above the water surface. An impingement airborne microorganism sampler (SKC BioSampler, SKC Inc., USA) was used for airborne bacteria collection. The sampler was made of glass and consisted of an inlet, a nozzle section with three tangential sonic nozzles, and a collection vessel filled with 20 ml of sterile phosphate-buffered saline as the collection medium. The SKC BioSampler was designed for sampling airborne particulates of biological origin (e.g., bacteria, fungi, pollen, and viruses) and their fragments and by-products. All samplers were sterilized in advance in an autoclave for 120 min, and the inside surfaces were maintained in a sterile condition until sampling. Air samples were transferred into the collection medium by drawing the gas at a flow rate of 12.5 l/min. Once the required volume of air had been drawn through, the sampler was removed and sealed to prevent contamination. Cells in liquid were concentrated according to previous research (Moletta et al. 2007). Parallel to the microbiological samples, chemicals including insoluble particles and water-soluble ions in the aerosols were analyzed. They were collected with the same sampler with the collection vessel filled with 20 ml of sterile distilled water. The flow rate and corresponding volume of the collected samples were 12.5 l/min and 2.0 m3, respectively. The control measurement (SO) was performed 100 m upwind of the Orbal oxidation ditch. After sampling, the liquor was then separated from the insoluble solid residue using a 0.1-μm membrane filter. The solution was used for the determination of water-soluble ions, and the residue together with filter was completely dried and observed through a scanning electron microscope together with energy dispersive X-ray spectroscopy (SEM/EDAX) (HITACHI S-3000 N /EDAX Inc., Japan).
2.3 Insoluble particles and water-soluble ions analysis
Concentrations of Cl−, NO −2 , NO −3 , SO 2−4 and PO 3−4 ions in each sample were determined with an ion chromatogram analyzer (ICS-1000, Dionex ion chromatography system, USA). Concentrations of NH +4 were analyzed by a UV–VIS spectrophotometer (UV-1700 PharmaSpec, Shimadzu, Japan) according to standard methods (China Bureau of Environmental Protection 2002). Elements in the insoluble components were determined by the SEM/EDAX.
2.4 DNA extraction and PCR amplification
Total DNA isolation was accomplished with a DNA Autoplate (TanBead, Taiwan) by Automatic Plateform for Magnetic System-16 (TanBead, Taiwan). The DNA bands were observed by 1.0 % agarose gel electrophoresis. All samples were assayed in duplicate. Universal primers F16S-27 (5′-AGAGTTTGATCCTGGCTCAG-3′) and R16S-1492 (5′-CGGTTACCTTGTTACGACTTC-3′) were used to amplify the eubacterial 16S rDNA segment. The PCR amplification reaction was performed using the Gene AmpR PCR System (9700, AB, USA) at a final volume of 50 μl. The reaction mixture contained 2.0 μl of both primers, 3.0 μl of each dNTPs, 5 μl of 10× PCR buffer (Takara, China), and 2.0 μl of Taq DNA polymerase (TaKaRa, China). The temperature cycling conditions were 94°C for 5 min, followed by 25 cycles of 94 °C for 30 s, 56 °C for 30 s, and 72 °C for 90 s. A final extension at 72 °C for 10 min was used. After purification by an Agarose Gel Extraction Kit (Dingguo, China), the PCR products were ligated to the 1.0-μl pMD19-T vector (Takara, China), and transformed into E. coli DH5α competent cells. The clones were cultured on LB medium with X-Gal, IPTG, and Amp. A total of 60 clones were selected for PCR detection, and their positive clones were submitted for sequencing using the ABI 3730DXL DNA sequencer (AB, USA).
2.5 Clone library construction and phylogenetic analysis of sequences in clone libraries
All sequences obtained from the clone libraries were manually checked and trimmed to exclude vector sequences, then checked for chimeras using Bellerophon on the Greengenes website (http://greengenes.lbl.gov/). These sequences were aligned using the DNAMAN software. Aligned sequences were grouped into universal operational taxonomic units (OTUs) at a threshold of 97 % minimum similarity; individual sequences were also grouped into OTUs to identify bacterial distribution in bioaerosols from different sampling sites. The representative sequence of each OTU in the group-specific libraries was aligned to the NCBI database by the BLAST website. To identify the phylogenetic affiliation of all OTUs, a phylogenetic tree including representative sequences of each OTU and related sequences from the previous NCBI database was constructed using the neighbor joining algorithm by MEGA version 3.1.
2.6 Diversity and richness estimation of clone libraries
To determine whether the size of a clone library was large enough to represent the diversity of an original community, the coverage of each clone library was calculated according to the equation C = 1 − (n/N), where n is the number of unique clones and N is the total number of sequences examined (Good 1953). The diversity of each sequence set was estimated using the Shannon–Wiener index (H) (Ding et al. 2008). Rarefaction curves were created using the species diversity function of the Analytic Rarefaction 1.3 statistical software (Holland 2003).
2.7 Accession numbers for nucleotide sequences
The representative OTU sequences obtained in this study were deposited in the NCBI database under the accession numbers shown in Table 2.
3 Results
3.1 Microbial characterizations
3.1.1 Clone libraries coverage and phylogenetic analysis
The diversity and coverage of the 16S rDNA-based phylotypes were determined by analysis of clones from each of the clone libraries. A total of 199 clones were compared. There were 39 different OTUs among the 51 screened clones from the W clone library, where 43 OTUs of 47 screened clones, 53 OTUs of 54 screened clones, and 41 OTUs of 47 screened clones were identified from the PBA, BBA, and BFBA clone libraries, respectively. Coverage analysis indicated that the bioaerosol libraries represented approximately 76.47–98.11 % of the total number of clones examined, providing a dependable inventory of the bacterial 16S rRNA gene sequences present in the bioaerosols. The C values observed in all the samples were larger than 0.70, which indicates that the clone number analyzed in each sample in the present study is accepted as valid in microbial diversity analysis (Pace 1997; Han et al. 2010). Rarefaction curves are generated for the 16S rDNA clone libraries (Fig. 2). Both coverage (C) and rarefaction curves can reflect the uniqueness of the microbial in air and water environment.
Rarefaction curves from 16S rDNA clone libraries of the bioaerosols and water samples. Rarefaction curves were created using the species diversity function of the Analytic Rarefaction 1.3 statistical software (Holland 2003)
Figure 3 showed the phylogenetic position of the detected bacteria in all samples. Bacterial groups of Proteobacteria with α-, β-, γ- and δ-subgroups, Bacteroidetes, Actinobacteria, Spirochaetes, Verrucomicrobia, Planctomycetes, and unclassified bacterium were widespread, indicating that bioaerosols and water samples from WWTP presented with high species richness. The bacterial communities of W, PBA, BBA, and BFBA were exhibited in Figs. 4, 5, 6, and 7, respectively.
3.1.2 Genetic structure of airborne bacterial communities
Sequences were assigned to a bacterial phylum according to their position in the phylogenetic tree. The bacteria that most closely represented the OTUs microorganisms were those isolated from activated sludge in the WWTP and soil ecosystems.
The majority of sequences in PBA were assigned to uncultured bacteria (74.48 %) and Proteobacteria with β-(4.26 %) and γ-(6.38 %) subgroups (Fig. 8). The minor groups were the Verrucomicrobia, Planctomycetes, and Spirochaetes. For BBA, the most represented bacterial groups were Proteobacteria (62.97 %) and Bacteroidetes (29.63 %). The Proteobacteria were distributed in the β-(18.52 %) and γ-(44.45 %) subgroups, with the α- and δ-subgroups not found. The other OTUs were either related to uncultured environmental bacteria (7.41 %) or not closely related to any known sequence present in the databases. The represented bacterial groups in BFBA were Proteobacteria (48.95 %) with α-(19.16 %), β-(27.66 %), and δ-(2.13 %) subgroups; uncultured bacteria (36.20 %); and Actinobacteria (14.9 %). These sequence results suggest that Bacteroidetes OTUs were widespread within BBA and W, and that the Proteobacteria OTUs were clustered in all sampling sites. These findings are in agreement with previous research on the genetic structure and diversity of airborne bacterial communities from northern France as determined by DNA fingerprinting and 16S rDNA clone library, which found that 60 % of bacteria were Proteobacteria belonging to the α- (19 %), β-(21 %), γ-(12 %), and δ-(8 %) subdivisions (Maron et al. 2005).
Multiplicity of microorganisms appeared in water. Over 70 % of the bacterial groups in the W clone library were uncultured bacteria. Other identified groups were Proteobacteria (13.72 %) with β-(5.88 %) and γ-(7.84 %) subgroups and Bacteroidetes (5.88 %). Actinobacteria and Spirochaetes were minor groups.
The diversities of bacterial species were 3.26 for W, 2.88 for PBA, 2.20 for BBA, and 2.73 for BFBA. Water in the oxidation ditch presented the highest species richness among the four clone libraries. It was also confirmed by rarefaction curves (Fig. 2). The diversity of bacteria collected after the rotating brushes (BBA) dropped dramatically compared to that sampled before the rotating brushes (PBA). Diversity increased to approach the lever of PBA when the distance from the rotating brushes reached 40 m (BFBA). The distribution of bioaerosol diversity was uneven and exhibited a site-related relationship.
3.1.3 Site-related variation of OTU distribution
The distribution and diversity of bacteria in the bioaerosols demonstrated site-related variation. A similar phenomenon occurred in the proportions of Proteobacteria, the major phylum of identified bacteria, in each sampling site. Obvious increases were observed in β-Proteobacteria percentages, from 4.26 % in PBA to 18.52 % in BBA, and γ-Proteobacteria percentages, from 6.38 % in PBA to 44.45 % in BBA (Fig. 8). Seven OTUs (24 clones in total, JN981922–JN981925 and JN981927–JN981929) formed γ-Proteobacteria in BBA. They were closed to cloned 16S rDNA isolated from aquatic and soil environments. Two OTUs (JN981922 and JN981928) belonged to Proteus sp. The same observation was made for OTUs belonging to a similar group detected in soil (Pearson et al. 2008). Two other OTUs (10 clones, JN981921 and JN981931) that formed β-Proteobacteria obtained from aerobic sludge and distantly related to Alcaligenes faecalis fell into the same subgroup described previously (Ansede et al. 2001). The other dominating bacteria (29.63 %) belonging to Bacteroidetes emerged in BBA. Three OTUs (JN981871 and JN981893 in W and JN981926 in BBA) related to one species of Bacteroides and four OTUs (JN981891 in W, JN981933 in BBA, JN981889 in W, and JN981930 in BBA) representing two species of uncultured bacterium (F11 and A5-814) were detected simultaneously in W and BBA but were not found in the sampling site before the rotating brushes (PBA). The phylogenetic affiliation of the OTUs revealed that there were two groups of OTUs (JN981881, JN981913, and JN981937; and JN981894, JN981901, and JN981948) that were originally in W and also appeared in PBA and BFBA. Compared to that collected from the sampling site before the rotating brushes (PBA), the enhancement in distribution and proportion of Bacteroidetes and β- and γ-Proteobacteria after the rotating brushes (BBA) indicated that these microorganisms might be prone to emit from water into the air. No OTU related to γ-Proteobacteria and Bacteroidetes was present in BFBA, located 40 m from the rotating brushes. The bacteria present in BBA may have succumbed to gravitational settling by the time they reached the sampling site 40 m from the aerosol source.
Most bacteria present in PBA, BBA, and BFBA were detected in W. The composition of microorganisms in the water was one of the most important sources of bacterial communities in bioaerosols. β-Proteobacteria was present at all sampling sites; γ-Proteobacteria was detected in PBA, BBA, and W; Bacteroidetes existed in BBA and W; Actinobacteria was found in BFBA and W; and Spirochetes was present in PBA and W. Almost all identified bacteria in W were homologously related to corresponding bacteria in PBA, BBA, and BFBA.
3.2 Chemical characterizations
3.2.1 Elements of insoluble components
The concentrations of insoluble components were 4.5 μg/m3 for SO, 3.5 μg/m3 for PBA, 4.0 μg/m3 for BBA, and 3.5 μg/m3 for BFBA, respectively. Samples collected from SO had the largest concentration of insoluble components, while those obtained from PBA and BFBA had the smallest.
Analysis by SEM/EDAX provided information on the chemical elements present in the insoluble components collected from each sampling site. The distribution of elements at the individual sampling sites was not homogeneous. As shown in Table 3, only C, O, Al, and Si emerged in the atmosphere (SO). The maximum number of elements in the insoluble components was recorded at W from collected water. Those elements, e.g., N, Mg, Cl, Ca, K, Na, Fe, S, and P, present in W were not present in the SO sample. The species of elements in the insoluble airborne particulate matter collected at BBA rose significantly compared to PBA, reaching up to 12 species. The major increase was observed for Mg, Cl, K, Na, Fe, S, and P, which were also detected in water. All samples had C and O present. For the SO, PBA, and BFBA samples, the ratio of C and O were almost equal to the filter. Significantly, more C was detected in W and BBA compared to other samples, showing that they possessed some organic matter in the insoluble components. The elemental components occurred in BBA were similar to those in W and most of the elemental components did not appear in SO at the same time, indicating that those elements originally in the water were transferred into the atmosphere after passing through the rotating brushes and presented in BBA. When the sampling site was 40 m from the rotating brushes, the percentages of detected elements dropped dramatically compared to that of BBA. The decrease in elements occurred mostly for Mg, Ca, K, Na, and P, which had higher percentages in water.
3.2.2 Ions species of soluble components
Ions collected from individual sampling sites are shown in Fig. 9. The highest and lowest concentrations of total detected ion species were achieved in BBA and SO, respectively. The determined concentrations of ions were 6.79 μg/m3 of NH +4 , 5.24 μg/m3 of NO −3 , and 19.41 μg/m3 of SO 2−4 in the control sample (SO). The main ion in SO was SO 2−4 , which is similar to previous reports on Beijing atmosphere (Yao et al. 2003; Mao et al. 2011). Peaks at BBA were observed in the concentrations of all ions except that of NO −2 . The concentration order of the ions species in BBA was SO 2−4 (46.36 μg/m3) > Cl− (29.35 μg/m3) > NO −3 (21.51 μg/m3) > NH +4 (19.76 μg/m3) > PO 3−4 (11.42 μg/m3) > NO −2 (6.18 μg/m3). Table 4 shows that SO 2−4 , NO −3 , and NH +4 were the three main ion species found in water. In addition, Cl−, NO −2 , and PO 3−4 were detected in water but not in SO. Compared with SO, the composition of ion species in BBA was similar to that in W. The increased and newly emerged ions found in BBA probably came from the water, and the increase was consistent with the SEM/EDAX results.
The concentration of Cl−, SO 2−4 , NH +4 , NO −3 , and PO 3−4 in air samples collected at BBA increased dramatically compared with those collected before the rotating brushes (PBA). The concentrations then decreased at BFBA, when the distance from the rotating brushes increased to 40 m. The significant decrease occurred in SO 2−4 and NO −3 , with Cl− and SO 2−4 dominating at the sampling site of BFBA.
4 Discussion
The transfer of microorganisms from the water to the air occurs mainly during aeration, and the mechanical agitation of water will accelerate this transformation (Fannin et al. 1985; Carducci et al. 2000; Sánchez-Monedero et al. 2008). Rotating brushes were used for aeration in the oxidation ditch to provide linear liquid motion and aerate wastewater by revolving a number of paddles mounted on a large diameter horizontal shaft. The rotating speeds of brush aerators were maintained at 40 to 60 rpm. The paddles agitated the water, causing a spray of water which was broken into fine droplets as it passed through the brush bristles. The surface-active particles in the water, e.g., zoogloea containing microorganisms and activated sludge particles, were dispersed with the water spray. Morphological analysis of the resultant fine droplets revealed that mycelioids clustered with activated sludge particles to form zoogloea surrounded by water (Fig. 10). The fine droplets that contained zoogloea with microorganisms acted as a media for bioaerosol generation from water and transmission into the air, which, in turn, resulted in the close relationship of microbial structure with water and the surrounding atmosphere.
The microbial structure and chemical components comprising the aerosol varied with distance from the aerosol source, atmospheric dispersal mechanisms, and environmental conditions at a particular site. The PBA and BBA sampling sites were situated before and after the rotating brushes, respectively, and significant differences in microbial structure and chemical concentrations were observed between them. Compared to those present in the site before the rotating brushes (PBA), significant increases in Proteobacteria and Bacteroidetes occurred in BBA. The migration of bacteria from water to air was mainly caused by the mechanical aeration of the brush in the oxidation ditch. Different to those emitted from soil or non-aqueous surfaces, bioaerosols generated from water sources (such as during splashing and wave action) usually form with a thin layer of moisture surrounding the microorganisms (Wickman 1994). Therefore, the percentages of elements and concentrations of soluble ions in the air were significantly increased at BBA, just after the rotating brushes.
Meteorological conditions including wind speed and wind direction were related to the compositions and concentrations of the microbial populations and chemical elements. Diffusion by wind from regions of higher concentration to regions of lower concentration is a significant factor for treatment facilities in outdoor environments, such as the oxidation ditch in WWTP. During the sampling day, the average wind speed was 1.5 m/s. Open air diffusion was an important consideration for the decreasing levels of microorganisms and chemicals during their transport downwind. Elements and water-soluble ions showed concentration reductions at BFBA, 40 m downwind from the rotating brushes. Some species, e.g., γ-Proteobacteria and Bacteroidetes, were not present at BFBA not only because of biological inactivation but also due to wind dilution.
Gravitational settling is also associated with decreasing microorganisms and chemicals. For aerosols greater than 1 μm, gravitational settling is a much more influential factor than other forces such as Brownian motion (Pillai and Ricke 2002). The gravitational effect on an aerosol particle is countered by the drag or frictional force exerted on that particle. When the two forces are equal, the particle reaches its final or terminal velocity. Thus, in our study, large particles tended to fall rather quickly due to gravitational settling, whereas smaller particles were more likely detected at sampling sites farther downwind from the rotating brushes (e.g., 40 m).
The risks associated with exposure to aerosols are not only related to concentration but also to the size of aerosolized particles. Small particles are very easily carried to large distances by the wind, ranging from a few hundred meters to several kilometers. Consequently, small-sized aerosols can have a greater influence on urban air quality as well as human health.
Some studies indicate that even though transport through the environment can be detrimental to the viability of aerosolized microbial cells, cell growth can occur in airborne particles (Dimmick et al. 1979). Insoluble components can serve as attachment media for microorganism adhesion, and water-soluble ions might provide nutrition for the growth of cells in aerosols. In addition, the layer of moisture surrounding the microorganisms may provide protection against UV damage. The chemical components in bioaerosols may also provide suitable microenvironments for airborne microorganism growth and survival in the air. Some bacteria can survive long distances based on their characteristics. For example, the microorganism affiliated to Rhodocyclales found in this study has bacteria chlorophyll and carotenoids. Photoautotrophic growth with molecular hydrogen may be possible if growth factors are supplied. In addition, the acid proof characteristics of Burkholderia also ensured activity over long-distance transmission, which is concerning as these bacteria are a potential health hazard in humans.
Most wastewater treatment processes are conducted by automatic systems without human labor. However, some WWTP workers still remain on-site for facilities maintenance. Many OTUs (13 OTUs of 47 screened clones) that appeared in water still present in BFBA, 40 m from the aerosol source. In addition to the potentially pathogenic β-Proteobacteria, most of bacteria in BFBA are related to uncultured environmental microorganisms, which cannot be detected by conventional culturing methods. As expected, the risk of bioaerosol exposure was greater at closer to the aerosol source. The exposure level of airborne microorganisms should be assessed at aerosols sources as the surrounding air at WWTPs is a major exposure route to potential pathogens in bioaerosols for on-site workers. Until recently, there is no standard to regulate the level of bioaerosols in WWTPs. This study will be benefit for the formulation of pollution standards, especially for aerosols, that take into account plant workers’ health.
References
Alvarez AJ, Buttner MP, Toranzos GA, Dvorsky EA, Toro A, Heikes TB, Mertikas-Pifer LE, Stetzenbach LD (1994) Use of solid-phase PCR for enhanced detection of airborne microorganisms. Appl Environ Microbiol 60:374–376
Amann RI, Ludwig W, Schleifer KH (1995) Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Mol Biol R 59:143–169
An HR, Mainelis G, White L (2006) Development and calibration of real-time PCR for quantification of airborne microorganisms in air samples. Atmos Environ 40:7924–7939
Ansede JH, Friedman R, Yoch DC (2001) Phylogenetic analysis of culturable dimethyl sulfide-producing bacteria from a Spartina-dominated salt marsh and estuarine water. Appl Environ Microbiol 67:1210–1217
Buttner MP, Cruz-Perez P, Stetzenbach LD (2001) Enhanced detection of surface-associated bacteria in indoor environments by quantitative PCR. Appl Environ Microbiol 67:2564–2570
Carducci A, Tozzi E, Rubulotta E, Casini B, Cantiani L, Rovini E, Muscillo M, Pacini R (2000) Assessing airborne biological hazard from urban wastewater treatment. Water Res 34:1173–1178
China Bureau of Environmental Protection (2002) Water and wastewater monitor and analysis method (4th). China Environmental Science, Beijing, pp 279–281
Cruz-Perez P, Buttner MP, Stetzenbach LD (2001) Detection and quantitation of Aspergillus fumigatus in pure culture using polymerase chain reaction. Mol Cell Probes 15:81–88
Dimmick RL, Wolochow H, Chatigny MA (1979) Evidence that bacteria can form new cells in airborne particles. Appl Environ Microbiol 37:924–927
Ding Y, Wu WX, Han ZY, Chen YX (2008) Correlation of reactor performance and bacterial community composition during the removal of trimethylamine in three-stage biofilters. Biochem Eng J 38:248–258
Fannin KF, Vana SC, Jakubowski W (1985) Effect of an activated sludge wastewater treatment plant on ambient air densities of aerosols containing bacteria and viruses. Appl Environ Microbiol 49:1191–1196
Good IJ (1953) The population frequencies of species and the estimation of population parameters. Biometrika 40:237–264
Grisoli P, Rodolfi M, Villani S, Grignani E, Cottica D, Berri A, Picco AM, Dacarro C (2009) Assessment of airborne microorganism contamination in an industrial area characterized by an open composting facility and a wastewater treatment plant. Environ Res 109:135–142
Han SF, Liu YC, Zhou ZG, He SX, Cao YN, Shi PJ, YaoB RE (2010) Analysis of bacterial diversity in the intestine of grass carp (Ctenopharyngodon idellus) based on 16S rDNA gene sequences. Aquac Res 42:47–56
Higgins JA, Cooper M, Schroeder-Tucker L, Black S, Miller D, Karns JS, Manthey E, Breeze R, Perdue ML (2003) A field investigation of Bacillus anthracis contamination of US Department of Agriculture and other Washington, D.C., buildings during the anthrax attack of October 2001. Appl Environ Microbiol 69:593–599
Holland SM (2003) Analytic rarefaction 1.3. Available at http://www.uga.edu/strata/software/ anRareReadme.html.
Li L, Gao M, Liu JX (2011) Distribution characterization of microbial aerosols emitted from a wastewater treatment plant using the Orbal oxidation ditch process. Process Biochem 46:910–915
Mao HY, Tian G, Huang YH, Li G, Song GW (2011) Mass size distributions and existing forms of sulfate and nitrate at atmospheric environment in Beijing (in Chinese). Environ Sci 32:1237–1241
Maron P-A, Lejon DPH, Carvalho E, Bizet K, Lemanceau P, Ranjard L, Mougel C (2005) Assessing genetic structure and diversity of airborne bacterial communities by DNA fingerprinting and 16S rDNA clone library. Atmos Environ 39:3687–3695
Moletta M, Delgenes JP, Godon JJ (2007) Differences in the aerosolization behavior of microorganisms as revealed through their transport by biogas. Sci Total Environ 379:75–88
Orsini M, Laurenti P, Boninti F, Arzani D, Ianni A, Romano-Spica V (2002) Research note. A molecular typing approach for evaluating bioaerosol exposure in wastewater treatment plant workers. Water Res 36:1375–1378
Pace NR (1997) A molecular view of microbial diversity and the biosphere. Science 276:734–740
Palmer CJ, Bonilla GF, Roll B, Paszko-Kolva C, Sangermano LR, Fujioka RH (1995) Detection of Legionella species in reclaimed water and air with the environAmp Legionella PCR kit and direct fluorescent antibody staining. Appl Environ Microbiol 61:407–412
Pascual L, Pérez-Luz S, Yáñez MA, Santamaría A, Gibert K, Salgot M, Apraiz D, Catalán V (2003) Bioaerosol emission from wastewater treatment plants. Aerobiologia 19:261–270
Pearson MM, Sebaihia M, Churcher C, Quail MA, Seshasayee AS, Luscombe NM, Abdellah Z, Arrosmith C, Atkin B, Chillingworth T, Hauser H, Jagels K, Moule S, Mungall K, Norbertczak H, Rabbinowitsch E, Walker D, Whithead S, Thomson NR, Rather P, Parkhill NJ, Mobley HLT (2008) Complete genome sequence of uropathogenic Proteus mirabilis, a master of both adherence and motility. J Bacteriol 190:4027–4037
Pillai SD, Ricke SC (2002) Bioaerosols from municipal and animal wastes: background and contemporary issues. Can J Microbiol 48:681–696
Polymenakou PN, Mandalakis M, Stephanou EG, Tselepides A (2008) Particle size distribution of airborne microorganisms and pathogens during an intense African dust event in the eastern Mediterranean. Environ Health Perspect 116:292–296
Ranalli G, Principi P, Sorlini C (2000) Bacterial aerosol emission from wastewater treatment plants: culture methods and bio-molecular tools. Aerobiologia 16:39–46
Rintala H, Hyvärinen A, Paulin L, Nevalainen A (2004) Detection of Streptomycetes in house dust—comparison of culture and PCR methods. Indoor Air 14:112–119
Sánchez-Monedero MA, Aguilar MI, Fenoll R, Roig A (2008) Effect of the aeration system on the levels of airborne microorganisms generated at wastewater treatment plants. Water Res 42:3739–3744
Sawyer MH, Chamberlin CJ, Wu YN, Aintablian N, Wallace MR (1994) Detection of Varicella-zoster virus DNA in air samples from hospital rooms. J Infect Dis 169:91–94
Schafer MP, Martinez KF, Mathews ES (2003) Rapid detection and determination of the aerodynamic size range of airborne Mycobacteria associated with whirlpools. Appl Occup Environ Hyg 18:41–50
Stetzenbach LD, Buttner MP, Cruz P (2004) Detection and enumeration of airborne biocontaminants. Curr Opin Biotechnol 15:170–174
Wickman HH (1994) Deposition, adhesion and release of bioaerosols. In: Lighthart B, Mohr AJ (eds) Atmospheric microbial aerosols, theory and applications. Chapman & Hall, New York, pp 99–165
Williams RH, Ward E, McCartney HA (2001) Methods for integrated air sampling and DNA analysis for detection of airborne fungal spores. Appl Environ Microbiol 67:2453–2459
Yao XH, Lau APS, Fang M et al (2003) Size distributions and formation of ionic species in atmospheric particulate pollutants in Beijing, China: 1-inorganic ions. Atmos Environ 37:2991–3000
Acknowledgments
We appreciate Wenjie Ding for his help in sequence alignment and Dr. Watts for the English revision of the manuscript. We also appreciate the University of Georgia Stratigraphy Lab for providing the software (Analytic Rarefaction 1.3) in rarefaction calculations. The work was financially supported by the National Natural Science Foundation of China (no. 51178451 and 51138009).
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Philippe Garrigues
Rights and permissions
About this article
Cite this article
Han, Y., Li, L., Liu, J. et al. Microbial structure and chemical components of aerosols caused by rotating brushes in a wastewater treatment plant. Environ Sci Pollut Res 19, 4097–4108 (2012). https://doi.org/10.1007/s11356-012-0885-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-012-0885-1