Abstract
Introduction
Processing delays after blood collection is a common pre-analytical condition in large epidemiologic studies. It is critical to evaluate the suitability of blood samples with processing delays for metabolomics analysis as it is a potential source of variation that could attenuate associations between metabolites and disease outcomes.
Objectives
We aimed to evaluate the reproducibility of metabolites over extended processing delays up to 48 h. We also aimed to test the reproducibility of the metabolomics platform.
Methods
Blood samples were collected from 18 healthy volunteers. Blood was stored in the refrigerator and processed for plasma at 0, 15, 30, and 48 h after collection. Plasma samples were metabolically profiled using an untargeted, ultrahigh performance liquid chromatography–tandem mass spectrometry (UPLC–MS/MS) platform. Reproducibility of 1012 metabolites over processing delays and reproducibility of the platform were determined by intraclass correlation coefficients (ICCs) with variance components estimated from mixed-effects models.
Results
The majority of metabolites (approximately 70% of 1012) were highly reproducible (ICCs ≥ 0.75) over 15-, 30- or 48-h processing delays. Nucleotides, energy-related metabolites, peptides, and carbohydrates were most affected by processing delays. The platform was highly reproducible with a median technical ICC of 0.84 (interquartile range 0.68–0.93).
Conclusion
Most metabolites measured by the UPLC–MS/MS platform show acceptable reproducibility up to 48-h processing delays. Metabolites of certain pathways need to be interpreted cautiously in relation to outcomes in epidemiologic studies with prolonged processing delays.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Metabolomics involves the study of small molecules (< 1500 Da) related to metabolism in cells, biofluids, and tissues using high-throughput analytical technologies (Wishart 2008). Metabolomics is increasingly being used in epidemiologic studies to identify metabolic variations related to disease development (Shah and Newgard 2015; Mayers et al. 2014; Sreekumar et al. 2009; Holmes et al. 2008), early detection biomarkers (Gaul et al. 2015; Ishikawa et al. 2016) and lifestyle biomarkers (Cross et al. 2014; Moore et al. 2014; Playdon et al. 2017, 2016). Because pre-analytical factors during sample collection and storage can introduce measurement error to metabolite levels and attenuate the associations with disease outcomes, evaluating the reproducibility of metabolites given the existing pre-analytical factors is critical before applying the technology in epidemiologic studies.
A potential source of pre-analytic variation in many epidemiologic studies is the delay in sample processing that often occurs when blood is collected outside of facilities with appropriate processing laboratories, and thus must be transported to a different facility for fractionation and subsequent storage and the delay could go beyond 24 h. During prolonged processing delays, certain metabolite levels can change primarily due to active metabolism of red blood cells (Bernini et al. 2011; Bervoets et al. 2015; Fliniaux et al. 2011; Jain et al. 2017; Jobard et al. 2016; Malm et al. 2016; Trezzi et al. 2016).
Several studies have evaluated the effect of processing delays on serum or plasma metabolic profiles (Bernini et al. 2011; Bervoets et al. 2015; Breier et al. 2014; Dunn et al. 2008; Fliniaux et al. 2011; Jain et al. 2017; Jobard et al. 2016; Kamlage et al. 2014; Malm et al. 2016; Midttun et al. 2014; Townsend et al. 2013; Trezzi et al. 2016; Yin et al. 2013; Shurubor et al. 2007). However, only three of these studies evaluated processing delays longer than 24 h (Jain et al. 2017; Midttun et al. 2014; Shurubor et al. 2007), and among those studies either a limited number of metabolites was evaluated (Midttun et al. 2014; Shurubor et al. 2007) or no quantitative individual metabolite results were provided (Shurubor et al. 2007; Jain et al. 2017). Thus, information regarding the reproducibility of individual metabolites over longer processing delays is lacking.
The statistical method used to quantify the influence of processing delay is important when informing calculation of statistical power or sample size in epidemiologic studies of metabolomics and diseases. In epidemiologic studies that examine relationships with disease outcomes, metabolite reproducibility, assessed by intraclass correlation coefficient (ICC, the ratio of between-person variance to total variance), is more meaningful to consider than absolute level change. When between-person variation is much larger than variation due to processing delay, the impact of processing delay on the metabolite-disease association will be minimal (Bervoets et al. 2015). ICC has been used to calculate attenuated risk estimates given the true risk estimates (Platz et al. 2010; Rosner et al. 1992). The attenuated risk estimate determines the effect size and statistical power of the study. Some previous studies reported significant changes in metabolite levels due to processing delay but didn’t calculate variation due to processing delay (Breier et al. 2014; Bernini et al. 2011; Jobard et al. 2016; Kamlage et al. 2014; Malm et al. 2016; Trezzi et al. 2016; Yin et al. 2013). Several other studies reported systematic differences in blood metabolic profiles using multivariate analysis such as principal component analysis and suggested that between-person variation was greater than processing delay variation but didn’t calculate ICC (Bervoets et al. 2015; Fliniaux et al. 2011; Jain et al. 2017; Teahan et al. 2006). Only two of the previous studies quantified reproducibility over processing delays using ICC (Townsend et al. 2013; Midttun et al. 2014).
Therefore, we conducted a study to test the reproducibility of metabolites measured by an untargeted, ultrahigh performance liquid chromatography-tandem mass spectrometry (UPLC–MS/MS) platform over extended processing delays up to 48 h, mimicking the conditions during sample transportation and processing in large population studies. We aimed to examine the reproducibility of the metabolites and identify a list of metabolites that are poorly reproduced during processing delays to inform studies with similar pre-analytic conditions. We also aimed to evaluate the reproducibility of the platform using sample replicates.
2 Methods
2.1 Participants and study design
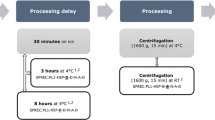
A convenience sample of 18 healthy volunteers donated non-fasting blood for this study. A total of 240 ml blood was collected using 10 ml sodium EDTA vacutainers. Blood samples were placed in a rack and kept in the refrigerator until being processed by centrifugation for 10 min at 1000×g after 0, 15, 30, and 48 h. Plasma was transferred into the 1.8 ml cryovials using a sterile, disposable plastic pipette and stored at − 80 °C until needed for analysis. For each participant, the samples collected at the 15- and 48-h time points were analyzed in duplicate whereas those for the 0- and 30-h time point were analyzed in triplicate. Thus, ten samples were analyzed from each volunteer.
The 180 samples (18 × 10) were analyzed in two batches (108 and 72 samples on each batch) on two consecutive days. Triplicates of 12 samples were distributed among the study samples to allow for the calculation of within and between batch variation.
2.2 Metabolomics analysis
Metabolomic profiling was conducted by Metabolon, Inc. (Durham, NC) using UPLC–MS/MS described elsewhere (Evans et al. 2009). Briefly, methanol was added to precipitate protein, followed by centrifugation. The resulting extract was divided into five fractions: two for analysis by two separate reversed phase (RP)/UPLC–MS/MS methods with positive ion mode electrospray ionization (ESI), one for analysis by RP/UPLC–MS/MS with negative ion mode ESI, one for analysis by HILIC/UPLC–MS/MS with negative ion mode ESI, and one sample was stored as backup. Individual metabolites were identified by comparison with the Metabolon chemical spectral library of more than 3300 authentic purified standards based on retention time, mass to charge ratio, and fragment ion spectra. The majority of the named metabolites are validated identification according to the metabolomics standards initiative (Sumner et al. 2007; Schrimpe-Rutledge et al. 2016). Metabolites that are structurally similar but have a side group that could not be definitively placed in the molecule were given the same chemical name followed by a number in parentheses to differentiate them from each other.
2.3 Statistical analysis
Before any statistical analysis, we excluded metabolites that were below the detection limit in ≥ 90% samples (n = 14 of 1026) to reduce noise. Missing values were assigned the minimum detection value for the included metabolites. To correct for day-to-day variation from the platform, each metabolite was divided by its daily median.
To evaluate the reproducibility of metabolites over processing delays and reproducibility of the platform, two types of ICCs were calculated (Sampson et al. 2013). ICCs representing reproducibility of processing delay denote the proportion of total variance attributable to between-person variance (Eq. 1). Samples collected at two time points (15 vs 0 h, 30 vs 0 h, 48 vs 0 h) were used to calculate ICCt (ICC15, ICC30, ICC48). Total variance is the sum of between-person variance \((\sigma _{B}^{2})\), within-person variance or variance due to processing delay \((\sigma _{W}^{2})\), and variance of random error \((\sigma _{\varepsilon }^{2})\). These variance components were estimated from a mixed-effects model with subject (Si), interaction term of subject and processing delay (SiTi), and random error being random variables, and processing delay (Ti) being a fixed variable (Model 1).
Model 1
Model 2 further separated batch as a random effect (Bi). The reproducibility of the platform—technical ICC (ICCtech) was calculated as the proportion of the total variance that is attributable to biological variance (sum of between-person and processing delay variance), as opposed to random laboratory error (Eq. 2). We further calculated percentage of variance due to batch effect (Varbatch %) using Eq. 3.
Model 2
We considered ICCs < 0.4 to be poor, 0.4–0.59 to be fair, 0.6–0.75 to be good and > 0.75 to be excellent (Cicchetti 1994). All statistical analyses were performed using the computing environment R 3.4.2.
3 Results
3.1 Reproducibility over processing delays
In the analysis, the 1012 metabolites included 31% unknown metabolites, 30% lipids, 17% amino acids, 10% xenobiotics, 3% nucleotides, 3% peptides, 2% carbohydrates, 2% vitamins and cofactors, and 1% energy-related metabolites. At 15-h processing delay, 7.1% (n = 72) metabolites had poor reproducibility (ICC15 < 0.4), and 71.4% (n = 723) had excellent reproducibility (ICC15 > 0.75) (Table 1). For 30- and 48-h processing delay, the number of metabolites with poor reproducibility were slightly higher [85 (8.4%) and 83 (8.2%), respectively] and metabolites with excellent reproducibility were slightly fewer [696 (68.8%) and 697 (68.9%), respectively] compared to 15 h. When stratified by super-pathway, processing delays primarily affected nucleotides (median ICCs ≤ 0.58), approximately 30% with ICCs < 0.4; smaller proportions of energy-related metabolites (11–22%), peptides (15%), and carbohydrates (10%) had ICCs < 0.4 over processing delays, however the median ICCs for the latter three pathways were considered good or excellent. Reproducibility across all processing delay times was excellent for approximately 70% of amino acids, cofactors and vitamins, lipids, unknowns and xenobiotics.
The ICCs for individual metabolites are shown in Supplemental Table 1. The metabolites with poor reproducibility at any time point (n = 102) included those involved in sub-pathways such as glutathione metabolism, histidine metabolism, glycolysis, TCA cycle, lysolipids, γ-glutamyl amino acid, purines, pyrimidines and several subtypes of fatty acids. Of these metabolites, 30 had acceptable reproducibility at 15 h (ICC15 ≥ 0.4) but poor reproducibility at either 30 or 48 h (ICCs < 0.4).
We further calculated the fold-change of metabolite levels from 0 to 15, 0 to 30, and 0 to 48 h (complete results of fold-change are shown in Supplemental Table 1). We evaluated the relationship between ICC and absolute value of percent change in all metabolites, and found statistically significant but low inverse correlation between them (r = − 0.24, − 0.14, − 0.14 for 15, 30, and 48 h, respectively; P values < 0.0001). For the majority of metabolites, there was no clear pattern between fold-change and ICC. Fold-change and ICCs were shown side by side in Fig. 1 for 75 known metabolites with poor reproducibility at either 15-, 30- or 48-h processing delay.
3.2 Technical reproducibility
The contribution of batch variance to total variance was very low with a median of 3% which is not unexpected because the two batches were measured on consecutive days (Supplemental Table 1). Sixty-six percent of metabolites (n = 665) had excellent technical reproducibility (ICCtech > 0.75). The overall high technical reproducibility (median ICCtech = 0.84, IQR = 0.68–0.93) indicated that the platform is highly reproducible (Table 2). Correlation analyses indicated a high correlation between ICC15 and ICCtech (r = 0.81, P < 0.0001), ICC30 and ICCtech (r = 0.79, P < 0.0001) and between ICC48 and ICCtech (r = 0.81, P < 0.0001). Most metabolites with high ICC30 and ICC48 have high ICCtech because between-person variance which is in the numerator of ICCtech equation is relatively large compared with total variance. However, metabolites with poor ICC over processing delays could also have high ICCtech due to the large processing delay variance which is also in the numerator of the equation (Fig. 2).
4 Discussion
In this study that evaluated reproducibility of metabolites over up to 48-h processing delays, the majority of the metabolites were highly reproducible over 15-, 30- or 48-h processing delays. The untargeted UPLC–MS/MS platform used for the metabolomics analysis was highly reliable.
The human metabolome is a dynamic and complex matrix with a high inter-individual variability. A series of reactions occur in vitro after blood collection, such as erythrocyte metabolism, enzymatic activities, degradation and synthesis of certain metabolites. Erythrocytes account for 40–45% of the volume of whole blood, and active metabolism of erythrocytes is a major event causing metabolism perturbation during processing delays (Jain et al. 2017). Several studies suggest that metabolites related to erythrocytes can be used as indicators of prolonged processing delays when documentation of sample processing delay is missing (Breier et al. 2014; Jain et al. 2017; Kamlage et al. 2014; Malm et al. 2016; Trezzi et al. 2016). Examples of these metabolites include carbohydrates involved in glycolysis such as glucose, lactate, and pyruvate (Breier et al. 2014; Kamlage et al. 2014; Malm et al. 2016), 5-oxoproline, ornithine/arginine ratio (Jain et al. 2017), and ascorbic acid/lactic acid ratio (Trezzi et al. 2016). Importantly, among these studies, the longest processing delay was up to 4 days after collection (Jain et al. 2017). Similarly, we found significant fold-change for 30- and 48-h processing delays in glucose, lactate, pyruvate, and 5-oxoproline. In addition, the ornithine/arginine ratio also increased over processing delay time in our study (fold-change = 1.18., 1.27, 1.30 for 15, 30, and 48 h, respectively). Ascorbic acid was not measured thus the ratio of ascorbic acid to lactate was not investigated. ICCs for the above metabolites at 30 and 48 h were also low, however ornithine/arginine ratio has excellent reproducibility at both times (ICC30 = 0.87, ICC48 = 0.81). Although the absolute value of percent change and ICC are inversely correlated, the correlation coefficients are very low. Both ICC and fold-change are important when assessing the stability of metabolites over processing delay. Only ICC is informative when calculating effect size and statistical power in epidemiologic studies of metabolomics and outcomes.
In addition to the metabolites involved in glycolysis, TCA cycle, and glutathione metabolism, poor reproducibility was also found for metabolites involved in γ-glutamyl amino acid, and for several fatty acid subtypes. The low ICCs of purines and pyrimidines observed in this study are similar to the findings of a study conducted in the Nurses’ Health Study and Health Professionals Follow-up Study, and could be due to the ongoing enzymatic reactions in whole blood (Townsend et al. 2013).
There are 30 metabolites with acceptable reproducibility at 15 h but poor reproducibility at 30 or 48 h, such as 5-oxoproline and cysteinylglycine involved in glutathione metabolism, succinate in TCA cycle, DHA, sphingomyelin, and sphingosine 1-phosphate. These metabolites might be reliable when processing delay is within 24 h but need to be interpreted cautiously when processing delay goes beyond 1 day.
The present study has several advantages. It evaluated reproducibility across prolonged processing delays for a larger number of metabolites than had been examined in previous studies. It quantified reproducibility using the ICC which is directly linked to effect size and statistical power in epidemiologic studies. There are also limitations in the present study. Time of day of blood collection and fasting status were not controlled. Previous studies show that several lipid species and amino acids vary across time during the day, partially due to the effect of circadian rhythmicity (Ang et al. 2012; Begum et al. 2016). Therefore the between-person variation and total variation might be larger for these metabolites than those in a study where these conditions are controlled. Only three processing delay times were evaluated in this study. However, large cohort studies typically experience a range of distribution of processing delays. Therefore the ICC obtained from this study may not be accurate when the samples are a mix of different processing delays, which can contribute to measurement error in effect size and power calculations. If the majority of the samples experience similar processing delays, this impact may be minimal.
5 Conclusions
In this study designed to evaluate metabolite reproducibility over extended processing delays, most metabolites measured by the untargeted UPLC–MS/MS platform were shown to be highly reproducible over 15-, 30- or 48-h processing delays. The results demonstrated that pre-analytical constraints commonly imposed on sample processing in large cohort studies do not preclude metabolomic analyses to identify meaningful metabolite-disease associations. Metabolites of certain pathways need to be interpreted cautiously in relation to disease outcomes in future etiological analyses when prolonged processing delay is a common condition.
References
Ang, J. E., Revell, V., Mann, A., Mäntele, S., Otway, D. T., Johnston, J. D., et al. (2012). Identification of human plasma metabolites exhibiting time-of-day variation using an untargeted liquid chromatography–mass spectrometry metabolomic approach. Chronobiology International, 29(7), 868–881. https://doi.org/10.3109/07420528.2012.699122.
Begum, H., Li, B., Shui, G., Cazenave-Gassiot, A., Soong, R., Ong, R. T.-H., et al. (2016). Discovering and validating between-subject variations in plasma lipids in healthy subjects. Scientific Reports, 6, 19139. https://doi.org/10.1038/srep19139.
Bernini, P., Bertini, I., Luchinat, C., Nincheri, P., Staderini, S., & Turano, P. (2011). Standard operating procedures for pre-analytical handling of blood and urine for metabolomic studies and biobanks. Journal of Biomolecular NMR, 49(3–4), 231–243. https://doi.org/10.1007/s10858-011-9489-1.
Bervoets, L., Louis, E., Reekmans, G., Mesotten, L., Thomeer, M., Adriaensens, P., et al. (2015). Influence of preanalytical sampling conditions on the 1H NMR metabolic profile of human blood plasma and introduction of the Standard PREanalytical Code used in biobanking. Metabolomics, 11(5), 1197–1207. https://doi.org/10.1007/s11306-015-0774-y.
Breier, M., Wahl, S., Prehn, C., Fugmann, M., Ferrari, U., Weise, M., et al. (2014). Targeted metabolomics identifies reliable and stable metabolites in human serum and plasma samples. PLoS ONE, 9(2), e89728. https://doi.org/10.1371/journal.pone.0089728.
Cicchetti, D. V. (1994). Guidelines, criteria, and rules of thumb for evaluating normed and standardized assessment instruments in psychology. Psychological Assessment, 6(4), 284–290.
Cross, A. J., Boca, S., Freedman, N. D., Caporaso, N. E., Huang, W. Y., Sinha, R., et al. (2014). Metabolites of tobacco smoking and colorectal cancer risk. Carcinogenesis, 35(7), 1516–1522. https://doi.org/10.1093/carcin/bgu071.
Dunn, W. B., Broadhurst, D., Ellis, D. I., Brown, M., Halsall, A., O’Hagan, S., et al. (2008). A GC-TOF-MS study of the stability of serum and urine metabolomes during the UK Biobank sample collection and preparation protocols. International Journal of Epidemiology, 37(Suppl 1), i23–i30. https://doi.org/10.1093/ije/dym281.
Evans, A. M., DeHaven, C. D., Barrett, T., Mitchell, M., & Milgram, E. (2009). Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Analytical Chemistry, 81(16), 6656–6667. https://doi.org/10.1021/ac901536h.
Fliniaux, O., Gaillard, G., Lion, A., Cailleu, D., Mesnard, F., & Betsou, F. (2011). Influence of common preanalytical variations on the metabolic profile of serum samples in biobanks. Journal of Biomolecular NMR, 51(4), 457–465. https://doi.org/10.1007/s10858-011-9574-5.
Gaul, D. A., Mezencev, R., Long, T. Q., Jones, C. M., Benigno, B. B., Gray, A., et al. (2015). Highly-accurate metabolomic detection of early-stage ovarian cancer. Scientific Reports, 5, 16351. https://doi.org/10.1038/srep16351.
Holmes, E., Loo, R. L., Stamler, J., Bictash, M., Yap, I. K., Chan, Q., et al. (2008). Human metabolic phenotype diversity and its association with diet and blood pressure. Nature, 453(7193), 396–400. https://doi.org/10.1038/nature06882.
Ishikawa, S., Sugimoto, M., Kitabatake, K., Sugano, A., Nakamura, M., Kaneko, M., et al. (2016). Identification of salivary metabolomic biomarkers for oral cancer screening. Scientific Reports, 6, 31520. https://doi.org/10.1038/srep31520.
Jain, M., Kennedy, A. D., Elsea, S. H., & Miller, M. J. (2017). Analytes related to erythrocyte metabolism are reliable biomarkers for preanalytical error due to delayed plasma processing in metabolomics studies. Clinica Chimica Acta, 466, 105–111. https://doi.org/10.1016/j.cca.2017.01.005.
Jobard, E., Tredan, O., Postoly, D., Andre, F., Martin, A. L., Elena-Herrmann, B., et al. (2016). A systematic evaluation of blood serum and plasma pre-analytics for metabolomics cohort studies. International Journal of Molecular Sciences. https://doi.org/10.3390/ijms17122035.
Kamlage, B., Maldonado, S. G., Bethan, B., Peter, E., Schmitz, O., Liebenberg, V., et al. (2014). Quality markers addressing preanalytical variations of blood and plasma processing identified by broad and targeted metabolite profiling. Clinical Chemistry, 60(2), 399–412. https://doi.org/10.1373/clinchem.2013.211979.
Malm, L., Tybring, G., Moritz, T., Landin, B., & Galli, J. (2016). Metabolomic quality assessment of EDTA plasma and serum samples. Biopreservation and Biobanking, 14(5), 416–423. https://doi.org/10.1089/bio.2015.0092.
Mayers, J. R., Wu, C., Clish, C. B., Kraft, P., Torrence, M. E., Fiske, B. P., et al. (2014). Elevation of circulating branched-chain amino acids is an early event in human pancreatic adenocarcinoma development. Nature Medicine, 20(10), 1193–1198. https://doi.org/10.1038/nm.3686.
Midttun, O., Townsend, M. K., Nygard, O., Tworoger, S. S., Brennan, P., Johansson, M., et al. (2014). Most blood biomarkers related to vitamin status, one-carbon metabolism, and the kynurenine pathway show adequate preanalytical stability and within-person reproducibility to allow assessment of exposure or nutritional status in healthy women and cardiovascular patients. The Journal of Nutrition, 144(5), 784–790. https://doi.org/10.3945/jn.113.189738.
Moore, S. C., Matthews, C. E., Sampson, J. N., Stolzenberg-Solomon, R. Z., Zheng, W., Cai, Q., et al. (2014). Human metabolic correlates of body mass index. Metabolomics, 10(2), 259–269. https://doi.org/10.1007/s11306-013-0574-1.
Platz, E. A., Sutcliffe, S., De Marzo, A. M., Drake, C. G., Rifai, N., Hsing, A. W., et al. (2010). Intra-individual variation in serum C-reactive protein over 4 years: An implication for epidemiologic studies. Cancer Causes and Control, 21(6), 847–851. https://doi.org/10.1007/s10552-010-9511-z.
Playdon, M. C., Moore, S. C., Derkach, A., Reedy, J., Subar, A. F., Sampson, J. N., et al. (2017). Identifying biomarkers of dietary patterns by using metabolomics. The American Journal of Clinical Nutrition, 105(2), 450–465. https://doi.org/10.3945/ajcn.116.144501.
Playdon, M. C., Sampson, J. N., Cross, A. J., Sinha, R., Guertin, K. A., Moy, K. A., et al. (2016). Comparing metabolite profiles of habitual diet in serum and urine. The American Journal of Clinical Nutrition, 104(3), 776–789. https://doi.org/10.3945/ajcn.116.135301.
Rosner, B., Spiegelman, D., & Willett, W. C. (1992). Correction of logistic regression relative risk estimates and confidence intervals for random within-person measurement error. American Journal of Epidemiology, 136(11), 1400–1413.
Sampson, J. N., Boca, S. M., Shu, X. O., Stolzenberg-Solomon, R. Z., Matthews, C. E., Hsing, A. W., et al. (2013). Metabolomics in epidemiology: Sources of variability in metabolite measurements and implications. Cancer Epidemiology and Prevention Biomarkers, 22(4), 631–640. https://doi.org/10.1158/1055-9965.EPI-12-1109.
Schrimpe-Rutledge, A. C., Codreanu, S. G., Sherrod, S. D., & McLean, J. A. (2016). Untargeted metabolomics strategies-challenges and emerging directions. Journal of the American Society for Mass Spectrometry, 27(12), 1897–1905. https://doi.org/10.1007/s13361-016-1469-y.
Shah, S. H., & Newgard, C. B. (2015). Integrated metabolomics and genomics: Systems approaches to biomarkers and mechanisms of cardiovascular disease. Circulation: Genomic and Precision Medicine, 8(2), 410–419. https://doi.org/10.1161/CIRCGENETICS.114.000223.
Shurubor, Y. I., Matson, W. R., Willett, W. C., Hankinson, S. E., & Kristal, B. S. (2007). Biological variability dominates and influences analytical variance in HPLC-ECD studies of the human plasma metabolome. BMC Clinical Pathology, 7, 9. https://doi.org/10.1186/1472-6890-7-9.
Sreekumar, A., Poisson, L. M., Rajendiran, T. M., Khan, A. P., Cao, Q., Yu, J., et al. (2009). Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature, 457(7231), 910–914. https://doi.org/10.1038/nature07762.
Sumner, L. W., Amberg, A., Barrett, D., Beale, M. H., Beger, R., Daykin, C. A., et al. (2007). Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics, 3(3), 211–221. https://doi.org/10.1007/s11306-007-0082-2.
Teahan, O., Gamble, S., Holmes, E., Waxman, J., Nicholson, J. K., Bevan, C., et al. (2006). Impact of analytical bias in metabonomic studies of human blood serum and plasma. Analytical Chemistry, 78(13), 4307–4318. https://doi.org/10.1021/ac051972y.
Townsend, M. K., Clish, C. B., Kraft, P., Wu, C., Souza, A. L., Deik, A. A., et al. (2013). Reproducibility of metabolomic profiles among men and women in 2 large cohort studies. Clinical Chemistry, 59(11), 1657–1667. https://doi.org/10.1373/clinchem.2012.199133.
Trezzi, J. P., Bulla, A., Bellora, C., Rose, M., Lescuyer, P., Kiehntopf, M., et al. (2016). LacaScore: A novel plasma sample quality control tool based on ascorbic acid and lactic acid levels. Metabolomics, 12, 96. https://doi.org/10.1007/s11306-016-1038-1.
Wishart, D. S. (2008). Metabolomics: Applications to food science and nutrition research. Trends in Food Science & Technology, 19(9), 482–493.
Yin, P., Peter, A., Franken, H., Zhao, X., Neukamm, S. S., Rosenbaum, L., et al. (2013). Preanalytical aspects and sample quality assessment in metabolomics studies of human blood. Clinical Chemistry, 59(5), 833–845. https://doi.org/10.1373/clinchem.2012.199257.
Author information
Authors and Affiliations
Contributions
YW and VLS conceived and designed research. YW and BDC conducted data analysis. YW wrote the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
All authors declared no conflicts of interest.
Ethical approval
All aspects of this study were approved by the Emory University (Atlanta, GA, USA) Institutional Review Board.
Additional information
The views expressed here are those of the authors and do not necessarily represent the American Cancer Society or the American Cancer Society – Cancer Action Network.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, Y., Carter, B.D., Gapstur, S.M. et al. Reproducibility of non-fasting plasma metabolomics measurements across processing delays. Metabolomics 14, 129 (2018). https://doi.org/10.1007/s11306-018-1429-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11306-018-1429-6