Abstract
Inflammation plays a prominent role in the pathogenesis of Alzheimer’s disease, affecting both brain and the peripheral system. Thus, modulation of inflammation in animal models of this neurodegenerative disorder may be of great interest to elucidate the pathological mechanisms underlying this inflammatory component. To this end, a metabolomic investigation on a triple transgenic mouse model obtained by crossing the APP/PS1 mice with interleukin-4 knockout mice (a model of impaired immune function) was performed for the first time. Serum samples from transgenic mice and wild type animals were analyzed by direct infusion mass spectrometry followed by multivariate statistics in order to identify altered metabolites. Subsequently, metabolic pathway analysis allowed the elucidation of potential pathological mechanisms associated with the development of Alzheimer-type disorders in response to interleukin-4 deficiency, such as impaired homeostasis of histamine, altered metabolism of amino acids (threonine, aspartate and tyrosine), deregulated urea cycle and increased production of eicosanoids. Therefore, this work demonstrates the potential of this triple transgenic model with modulated immunity for the study of pathological mechanisms associated with inflammation in Alzheimer’s disease.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Inflammation is a primary pathological hallmark of Alzheimer’s disease (AD), generated in response to the deposition of senile plaques containing β-amyloid peptides, formation of neufibrillary tangles and neurodegeneration (Akiyama et al. 2000). The major players involved in these inflammatory processes are thought to be the activation of microglia and astrocytes, and possibly to a lesser extent the neurons, leading to the production of many inflammatory mediators in the brain such as cytokines, growth factors, prostaglandins, leukotrienes, thromboxanes, complement factors or cell adhesion molecules, among others (Tuppo and Arias 2005). However, considerable evidence supports that inflammation in AD is not only localized in the central nervous system, but there are also significant disturbances in the periphery (Butchart and Holmes 2012). Thus, the immune system and inflammatory processes have become a primary topic in AD research in the last years for the discovery of potential markers of diagnosis, the elucidation of pathological mechanisms underlying to disease and the development of novel therapeutic approaches.
Numerous transgenic mouse models that mimic the main features of AD have been developed to better understand the pathogenic mechanisms of neurodegeneration and to test potential therapies (Hall and Roberson 2012). Nevertheless, these mice usually do not present significant neuronal loss, so it has been theorized that the addition of the inflammatory component of AD could generate best models to investigate the disease (Birch et al. 2014). Glial modulation in these mouse models of AD via stimulation of a variety of receptors has been considered for the elucidation of the role of microglia activation in the development of disease, including genetic manipulation of scavenger receptors (Frenkel et al. 2013), Toll-like receptors (Jin et al. 2008) or chemokine receptors (Naert and Rivest 2011). Moreover, different studies have demonstrated that over-expression of pro-inflammatory mediators in transgenic mice of AD may potentiate pathology by increasing Aβ generation, aggregation and by affecting its clearance, in the same way that their deletion causes a significant improvement. Thereby, common inflammatory targets investigated in these transgenic models are interleukins (Vom Berg et al. 2012), tumor necrosis factor (He et al. 2007) or nitric oxide synthase (Colton et al. 2006). Among these mediators, there is growing evidence that interleukin-4 (IL-4) plays a prominent role in pathology of AD. This interleukin is an anti-inflammatory cytokine produced by mature T type 2 helper cells (Th2), mast cells or basophils, involved in T cell proliferation and in higher functions of the normal brain, such as memory and learning (Gadani et al. 2012). Previous studies demonstrated an association between single nucleotide polymorphisms of the IL-4 gene and risk for AD development (Ribizzi et al. 2010), as well as reduced IL-4 production in peripheral blood mononuclear cells from AD patients (Reale et al. 2008). Furthermore, it is proved that induction of IL-4 production reduces Aβ deposition and attenuates AD pathogenesis in different transgenic mice (Kiyota et al. 2010; Kawahara et al. 2012). In fact, several studies proposed that amelioration of AD pathology in patients treated with acetylcholinesterase inhibitors is due to increased IL-4 expression (Reale et al. 2004; Gambi et al. 2004), supporting a pivotal immunomodulatory effect of IL-4 in AD.
Considering this apparent close relation between interleukin-4 and AD, the objective of this study was the elucidation of pathological mechanisms underlying to AD in response to IL-4 depletion. For this purpose, we performed a serum metabolomic investigation into a transgenic line obtained by crossing the APP/PS1 mice with IL-4 knockout mice. The IL-4 deficient mouse is a standard model of impaired immune function, which presents a disturbed production of Th2 cytokines and immunoglobulins E and G1 (Kühn et al. 1991; Kopf et al. 1993), so that we hypothesized that the triple transgenic mice APP/PS1/IL4-KO must develop enhanced AD pathology. Serum samples from wild type animals and transgenic models (APP/PS1/IL4 and APP/PS1) were subjected to metabolomic analysis by direct infusion mass spectrometry (DIMS), and then multivariate statistics was employed to find metabolites responsible for discrimination. Furthermore, pathway analysis allowed to identify metabolic perturbations associated with these pathological processes.
2 Materials and methods
2.1 Animal handling
Two transgenic mouse lines were compared in this study (C57BL/6 background). The double transgenic mice APP/PS1 were generated as previously described by Jankowsky et al. overexpressing the Swedish mutation of APP, together with PS1 deleted in exon 9 (Jankowsky et al. 2004). Furthermore, these APP/PS1 mice were crossed with IL-4 knockout mice (Kühn et al. 1991) to create homozygotic APP/PS1/IL4-KO mice. On the other hand, age-matched wild-type mice of the same genetic background (C57BL/6) were used as controls. In this study, male and female animals at 6 months of age were used for experiments (N = 7, male/female 5/2, for each group). Animals were acclimated for 3 days after reception in rooms with a 12-h light/dark cycle at 20-25 °C, with water and food available ad libitum. Then, mice were anesthetized by isoflurane inhalation and blood was extracted by cardiac puncture. Blood samples were immediately cooled and protected from light for 30 min to allow clot retraction, and then centrifuged at 3,500 rpm for 10 min at 4 °C. Serum was aliquoted in Eppendorf tubes and frozen at −80 °C until analysis. Animals were handled according to the directive 2010/63/EU stipulated by the European Community, and the study was approved by the Ethical Committee of University of Huelva.
2.2 Metabolomic analysis
Metabolomic analysis was performed by extracting serum samples in a two-stage sequential procedure, followed by direct infusion analysis with high resolution tandem mass spectrometry, using the electrospray (ESI) source in both positive and negative ionization modes, as described elsewhere (González-Domínguez et al. 2014a). For protein precipitation and the extraction of polar metabolites, 100 µL of serum were mixed with 400 µL of methanol/ethanol (1:1 v/v), vortexed for 5 min and centrifuged at 4,000 rpm for 10 min at 4 °C. The supernatant was transferred to another tube, and the precipitate was kept for further treatment. Then, supernatant was dried under nitrogen stream and the resulting residue was reconstituted with 100 μl of methanol/water (80:20 v/v) containing 0.1 % formic acid (aqueous extract). On the other hand, the precipitate isolated in the first step was again extracted in order to recover lipophilic compounds adsorbed into the protein pellet. For this purpose, the precipitate was mixed with 400 µL of chloroform:methanol (1:1 v/v) and then stirred during 5 min, followed by centrifugation at 10,000 rpm for 10 min at 4 °C. Finally, the resulting supernatant was taken to dryness under nitrogen stream and reconstituted with 100 μl of 60:40 dichloromethane:methanol containing 10 mM ammonium formate and 0.1 % formic acid (organic extract). Furthermore, quality control (QC) samples were prepared by pooling equal volumes of each sample, which allows monitoring the stability and performance of the system along the analysis period (Sangster et al. 2006).
Mass spectrometry experiments were performed in a quadrupole-time-of-flight mass spectrometry system (QTOF-MS), model QSTAR XL Hybrid system (Applied Biosystems, Foster City, CA, USA), using the electrospray (ESI) source. Samples were directly introduced into the mass spectrometer at 5 µL min−1 flow rate using an integrated apparatus pump and a 1,000µL volume Hamilton syringe. Data were obtained in both positive and negative ionization modes, acquiring full scan spectra during 0.2 min in the m/z range 50-1,100, with 1.005 s of scan time. In positive mode, the ion spray voltage (IS) was set at 3,300 V, and high-purity nitrogen was used as curtain and nebulizer gas at flow rates about 1.13 L min−1 and 1.56 L min−1, respectively. The source temperature was fixed at 60 °C, with a declustering potential (DP) of 60 V and a focusing potential (FP) of 250 V. For ESI(-)/MS only a few parameters were modified respect to the ESI(+)/MS method, with an ion spray voltage at -4,000 V, a declustering potential (DP) of -100 V and a focusing potential (FP) of -250 V. To acquire MS/MS spectra, nitrogen was used as collision gas.
2.3 Data analysis
Metabolomic fingerprints were submitted to peak detection using the Markerview™ software (Applied Biosystems) in order to filter the mass spectrometry results, and to carry out the reduction of raw data into a two-dimensional data matrix containing spectral peaks and their intensities. For this, all peaks above the noise level (10 counts, determined empirically from experimental spectra) were selected and binned in intervals of 0.1 Da, and then data were normalized according to the total area sum in order to correct inter-sample variability due to instrumental drifts. Subsequently, data were subjected to multivariate analysis by principal component analysis (PCA) and partial least squares discriminant analysis (PLS-DA) in order to compare metabolomic profiles obtained, using the SIMCA-P™ software (version 11.5, UMetrics AB, Umeå, Sweden). Before performing statistical analysis, data are usually scaled and transformed in order to minimize the technical variability between individual samples to extract the relevant biological information from these data sets (van den Berg et al. 2006). For this purpose, we performed Pareto scaling for reducing the relative importance of larger values, and logarithmic transformation, in order to approximate a normal distribution. Quality of the statistical models was assessed by the R2 and Q2 values, which provide information about the class separation and predictive power of the model, respectively. These parameters are ranged between 0 and 1, and they indicate the variance explained by the model for all the data analyzed (R2) and this variance in a test set by cross-validation (Q2). Potential biomarkers were selected by two-class comparisons: APP/PS1/IL4 vs. WT, APP/PS1 vs. WT, APP/PS1/IL4 vs. APP/PS1. To this end, loadings plots from PLS-DA models were inspected to select altered metabolites according to the Variable Importance in the Projection, or VIP (a weighted sum of squares of the PLS weight, which indicates the importance of the variable in the model), considering only variables with VIP values higher than 1.5, indicative of significant differences among groups. In addition, groups comparison was also conducted by one-way analysis of variance with Bonferroni correction for multiple testing, using the STATISTICA 8.0 software (StatSoft, Tulsa, USA). Only p values below 0.05 were regarded as statistically significant.
2.4 Identification of discriminant metabolites
Discriminant metabolites were first putatively identified by matching the experimental accurate mass and tandem mass spectra (MS/MS) with those shown in metabolomic databases (HMDB and METLIN). Then, this identification was confirmed with commercial standards when available, according to the guidelines from the Metabolomics Standards Initiative (MSI). For this, standards were analyzed using identical experimental conditions to those described for the primary metabolomic analysis, and then fragmentation patterns and accurate masses obtained were compared to those observed in real serum samples. Standards of metabolites for this identification of potential biomarkers were obtained from Sigma (urea, histamine, threonine, aspartic acid, urocanic acid, dopamine, citrulline, tyrosine).
2.5 Metabolic pathway analysis
Metabolic pathway analysis was performed to identify and visualize the affected pathways in transgenic mice on the basis of potential biomarkers detected. For this purpose, the MetPA web tool was employed (http://metpa.metabolomics.ca), which conducts pathway analysis through pathway enrichment analysis and pathway topological analysis (Xia and Wishart 2010). In this work, we select the Mus musculus library and use the default ‘Hypergeometric Test’ and ‘Relative-Betweenness Centrality’ algorithms for pathway enrichment analysis and pathway topological analysis, respectively.
3 Results
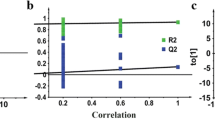
Metabolic fingerprints obtained by direct infusion mass spectrometry analysis of serum samples from APP/PS1/IL4, APP/PS1 and wild type mice were submitted to multivariate statistics in order to evaluate the potential of this high-throughput metabolomic approach for the investigation of mechanisms associated with the development of AD-type disorders in response to deficient interleukin-4. After peak detection, c.a. 5,000 molecular features were observed in DIMS profiles for each ionization mode, and all of them were used for statistical modelling. An initial principal components analysis (PCA) was applied to check trends and outliers (Fig. 1A), and then partial least squares discriminant analysis (PLS-DA) was performed to separate the study groups (Fig. 1B). Transgenic mice (APP/PS1/IL4 and APP/PS1) were clearly separated from control animals in the first latent variable, while a distinct classification of transgenic mice with/without IL-4 depletion was observed along the second latent variable (Fig. 1B). Statistical parameters also confirmed the goodness of this model for sample classification, with a variance explained (R2) of 99.8 % and variance predicted (Q2) of 72.5 %. Furthermore, the robustness of the methodology was assessed in terms of the clustering of quality control samples in the scores plots. For this purpose, quality control samples were prepared by pooling equal volumes from each sample and analyzed throughout the sequence run. As can be observed, QCs were tightly grouped in the PCA plot (Fig. 1A), and they were correctly predicted in the center of the scores plots for PLS-DA models (Fig. 1B), indicative of stability during the analyses (Sangster et al. 2006). Then, three additional PLS-DA models were generated according two-groups comparisons in order to facilitate the interpretation of results: APP/PS1/IL4 vs. WT (Fig. 1C, R2 = 0.998, Q2 = 0.794), APP/PS1 vs. WT (Fig. 1D, R2 = 1, Q2 = 0.729), APP/PS1/IL4 vs. APP/PS1 (Fig. 1E, R2 = 0.998, Q2 = 0.517). Discriminant metabolites influencing the differentiation between the three study groups were identified by inspecting loadings plots from these PLS-DA models (two-class comparisons) as well as by ANOVA. These classifiers are listed in Table 1, together with their ID numbers in HMDB database, the experimental mass, the ionization mode used for detection (P: positive mode; N: negative mode), the extract where they were found (A: aqueous extract; O: organic extract), and the fold change and p-value for each comparison. These findings showed that a number of metabolites are significantly perturbed in both APP/PS1/IL4 and APP/PS1 mice, including different amino acids (threonine, aspartic acid, tyrosine), eicosanoids (HEPE, prostaglandins, leukotriene B4) and other compounds (urea, citrulline, histamine, 1-methylhistamine, urocanic acid, dopamine), while only dopamine allowed to differentiate between the two transgenic lines. However, it is noteworthy that some of these metabolites suffered a gradual change between the three groups considered in this study (from WT to APP/PS1, and finally APP/PS1/IL4), as represented in the bar plots of Fig. 2. Therefore, it can be concluded that metabolic alterations detected in this work could be behind pathological mechanisms associated with the appearance of AD disorders in the APP/PS1 mice and their aggravation in response to IL-4 depletion. Finally, these altered metabolites were subjected to metabolic pathway analysis in order to elucidate the biochemical networks potentially impaired in these transgenic mice. An overview of this analysis is shown in Fig. 3, where each node represents a metabolic pathway and its size indicates the impact of this pathway in response to disease. Thus, the most important disturbances could be related to dyshomeostasis of different amino acids, such as altered biosynthesis of phenylalanine, tyrosine and tryptophan (a), and abnormal metabolism of histidine (b), tyrosine (c), alanine, aspartate and glutamate (d), arginine and proline (f), as well as perturbed arachidonic acid metabolism (e).
Scores plots of multivariate statistical models for DIMS data. PCA (a) and PLS-DA (b) models discriminating wild type controls (WT), APP/PS1 and APP/PS1/IL4 mice, with tight clustering of quality control samples (QC). Two-class comparisons: APP/PS1/IL4 vs. WT (c), APP/PS1 vs. WT (d) and APP/PS1/IL4 vs. APP/PS1 (e). QC: green stars (circled); WT: black squares; APP/PS1: red dots; APP/PS1/IL4: blue diamonds (Color figure online)
Pathway analysis overview showing altered metabolic pathways in serum from transgenic mice APP/PS1/IL4 and APP/PS1. (a) phenylalanine, tyrosine and tryptophan biosynthesis; (b) histidine metabolism; (c) tyrosine metabolism; (d) alanine, aspartate and glutamate metabolism; (e) arachidonic acid metabolism; (f) arginine and proline metabolism
4 Discussion
Numerous efforts have been made in the last years to identify metabolic failures associated with pathological mechanisms underlying to AD. To this end, different authors have previously addressed the metabolomic investigation of several mouse models of AD using both brain tissue and blood samples, such as the APP/PS1ΔE9 (González-Domínguez et al. 2014b, 2015a), TASTPM (Hu et al. 2012; Forster et al. 2012), CRND8 (Salek et al. 2010; Lin et al. 2013; Lin et al. 2014), APP/PS1M146L (Woo et al. 2010; Trushina et al. 2012), APPTg2576 (Dedeoglu et al. 2004; Lalande et al. 2014) or SAMP8 (Jiang et al. 2008; Wang et al. 2014), among others. Thereby, multiple associations have been described between AD and metabolic perturbations such as oxidative stress, mitochondrial dysfunction, abnormal lipid metabolism or inflammatory processes. The main objective of this work was to investigate a new triple transgenic line of AD characterized by deficient production of the anti-inflammatory interleukin 4 in order to determine the specific contribution of inflammation to AD development, and to elucidate the metabolic mechanisms underlying these pathological processes. For this purpose, we employed a screening tool previously optimized based on direct infusion mass spectrometry, whose potential has been demonstrated in terms of wide metabolome coverage and fast analysis (González-Domínguez et al. 2014a). The application of this metabolomic approach revealed significant alterations in levels of 13 metabolites: 1-methylhistamine, prostaglandins (series 2 and 3), HEPE, and leukotriene B4 were increased in transgenic mice, while urea, histamine, threonine, aspartate, urocanic acid, dopamine, citrulline and tyrosine were decreased in these samples. Furthermore, these perturbations could be linked to different metabolic pathways (Fig. 3), allowing the elucidation of biochemical processes underlying the pathology.
Reduced levels of histamine and urocanic acid, as well as increased 1-methylhistamine (Table 1) suggest a perturbed metabolism of histidine in these transgenic mice, as revealed metabolic pathway analysis. In this sense, previous works reported decreased content of this amino acid in brain (Mazurkiewicz-Kwilecki and Nsonwah 1989), cerebrospinal fluid (Ibañez et al. 2012) and serum (González-Domínguez et al. 2014c, d, 2015b) from AD patients, probably due to impaired biosynthesis of histamine. This compound is involved in the inflammatory response against pathogens by increasing the production of pro-inflammatory cytokines, but in addition may play an important role in neurotransmission failures in AD (Naddafi and Mirshafiey 2013). Thereby, AD has been previously associated with a loss of histaminergic neurons in the tuberomamillary nucleus (Nakamura et al. 1993), leading to a decline of histamine levels in different brain areas (Mazurkiewicz-Kwilecki and Nsonwah 1989; Panula et al. 1998), in agreement with our findings in serum samples. However, this work describes for the first time that reduced histamine is accompanied by altered serum levels of related metabolites (i.e. 1-methylhistamine and urocanic acid), supporting profound failures in the histaminergic system. Moreover, these changes were slightly accentuated in the IL-4 knockout mice compared with the APP/PS1 model (Fig. 2), indicating an aggravation of pathology. Metabolism of interleukin 4 and histamine are interconnected through immunoglobulin E, given that IL-4 induces the production of IgE by B-cells, which in addition is involved in the release of histamine during the immunologic response against allergens (Gould et al. 2003). Therefore, the down-regulation of histamine synthesis in the APP/PS1/IL4 mice could be due to reduced liberation of IgE in response to IL-4 depletion (Kühn et al. 1991).
Metabolomic fingerprints also showed decreased levels of different amino acids and related compounds in transgenic mice (threonine, aspartate, tyrosine, dopamine), highlighting the importance of a proper metabolism of amino acids for the immune system. Thereby, it is recognized that threonine and aspartate are important immunostimulants that promote growth of thymus and contribute to the modulation of immune function by controlling proliferation and activation of lymphocytes (Li et al. 2007). Moreover, dopamine and other tyrosine-derived compounds also may influence the immune system, by reducing the synthesis of proinflammatory cytokines and inducing the production of anti-inflammatory mediators, as well as by regulation of lymphocyte proliferation, platelet aggregation and the phagocytic activity of neutrophils (Basu and Dasgupta 2000).
Abnormal content of metabolites involved in the urea cycle has been previously related to impaired ammonia detoxification in AD (González-Domínguez 2014c, 2015c), in accordance with reduced levels observed in this study for urea and citrulline in serum from APP/PS1 and APP/PS1/IL4 mice. The accumulation of ammonia in AD (hyperammonemia) and consequent dyshomeostasis of urea-related metabolites has been associated with reduced glutamine synthetase activity (Seiler 2002) and enzymatic deficiencies in the urea cycle (Hansmannel et al. 2010). However, the extra decrease found in the IL-4 knockout mice compared with the APP/PS1 mice (although not statistically significant, Fig. 2) suggest an implication of interleukin 4 in this process. In this context, it is noteworthy that IL-4 promotes alternative activation of macrophages into M2 cells, leading to a differential utilization of arginine due to the release of proline and arginase, the enzyme that catalyzes the conversion of arginine into ornithine and urea (Gordon 2003). Thus, we hypothesized that IL-4 depletion in the APP/PS1 model may aggravate the already perturbed urea cycle.
Finally, a considerable accumulation of several eicosanoids was also found in serum from these transgenic models, including prostaglandins (PG), hydroxy-eicosapentaenoic acid (HEPE) and leukotriene B4 (LTB4), with the highest fold changes with regard to wild type controls (Table 1). The increase of these pro-inflammatory mediators in brain and biofluids has been repeatedly described in both human AD (Bazan et al. 2002; Trushina et al. 2013) and transgenic models (Piro et al. 2012; Lin et al. 2013; Lin et al. 2014). This inflammatory process has been traditionally associated with the over-activation of phospholipases, principally PLA2, leading to the release of polyunsaturated fatty acids from the hydrolysis of phospholipids (Farooqui et al. 2004), and subsequent oxidation by cyclooxygenases and 5-lipoxygenase (Manev et al. 2011), generating a complex meshwork of lipid mediators closely associated with neuronal pathways involved in AD neurobiology (Frisardi et al. 2011). However no substantial differences were observed between the APP/PS1 and APP/PS1/IL4 mice (Fig. 2), despite the demonstrated anti-inflammatory effects of interleukin 4 (Hart et al. 1989), suggesting that IL-4 does not play a relevant role in the production of eicosanoids in pathogenesis of AD.
5 Concluding remarks
In this study, we demonstrated that depletion of interleukin 4 may potentiate the pathology in the APP/PS1 mouse model of AD. Metabolomic fingerprinting of serum samples from transgenic mice (APP/PS1 and APP/PS1/IL4-KO) and wild type controls using direct infusion mass spectrometry revealed significant alterations in levels of 13 metabolites (urea, histamine, threonine, 1-methylhistamine, aspartic acid, urocanic acid, dopamine, citrulline, tyrosine, HEPE, prostaglandins and leukotriene B4), which could be related to abnormalities in different metabolic pathways. Thus, the most important failures might be associated with impaired biosynthesis of histamine, altered metabolism of amino acids, deregulated urea cycle and increased production of pro-inflammatory eicosanoids. Therefore, these findings highlight the potential of this triple transgenic model (APP/PS1/IL4) for the study of pathological mechanisms associated with inflammation in AD.
References
Akiyama, H., Barger, S., Barnum, S., Bradt, B., Bauer, J., Cole, G. M., et al. (2000). Inflammation and Alzheimer’s disease. Neurobiology of Aging, 21, 383–421.
Basu, S., & Dasgupta, P. S. (2000). Dopamine, a neurotransmitter, influences the immune system. Journal of Neuroimmunology, 102, 113–124.
Bazan, N. G., Colangelo, V., & Lukiw, W. J. (2002). Prostaglandins and other lipid mediators in Alzheimer’s disease. Prostaglandins & Other Lipid Mediators, 68–69, 197–210.
Birch, A. M., Katsouri, L., & Sastre, M. (2014). Modulation of inflammation in transgenic models of Alzheimer’s disease. J Neuroinflammation, 11, 25.
Butchart, J., & Holmes, C. (2012). Systemic and central immunity in Alzheimer’s disease: therapeutic implications. CNS Neuroscience & Therapeutics, 18, 64–76.
Colton, C. A., Vitek, M. P., Wink, D. A., Xu, Q., Cantillana, V., Previti, M. L., et al. (2006). NO synthase 2 (NOS2) deletion promotes multiple pathologies in a mouse model of Alzheimer’s disease. Proceedings of the National Academy of Sciences of the United States of America, 103, 12867–12872.
Dedeoglu, A., Choi, J. K., Cormier, K., Kowall, N. W., & Jenkins, B. G. (2004). Magnetic resonance spectroscopic analysis of Alzheimer’s disease mouse brain that express mutant human APP shows altered neurochemical profile. Brain Research, 1012, 60–65.
Farooqui, A. A., Ong, W. Y., & Horrocks, L. A. (2004). Biochemical aspects of neurodegeneration in human brain: Involvement of neural membrane phospholipids and phospholipases A2. Neurochemical Research, 29, 1961–1977.
Forster, D. M., James, M. F., & Williams, S. R. (2012). Effects of Alzheimer’s disease transgenes on neurochemical expression in the mouse brain determined by 1H MRS in vitro. NMR in Biomedicine, 25, 52–58.
Frenkel, D., Wilkinson, K., Zhao, L., Hickman, S. E., Means, T. K., Puckett, L., et al. (2013). Scara1 deficiency impairs clearance of soluble amyloid-beta by mononuclear phagocytes and accelerates Alzheimer’s-like disease progression. Nat Comm, 4, 2030.
Frisardi, V., Panza, F., Seripa, D., Farooqui, T., & Farooqui, A. A. (2011). Glycerophospholipids and glycerophospholipid-derived lipid mediators: A complex meshwork in Alzheimer’s disease pathology. Progress in Lipid Research, 50, 313–330.
Gadani, S. P., Cronk, J. C., Norris, G. T., & Kipnis, J. (2012). Interleukin-4: A cytokine to remember. Immunol, 189, 4213–4219.
Gambi, F., Reale, M., Iarlori, C., Salone, A., Toma, L., Paladini, C., et al. (2004). Alzheimer patients treated with an AchE inhibitor show higher IL-4and lower IL-1 beta levels and expression in peripheral blood mononuclearcells. J Clin Psychopharm, 24, 314–321.
González-Domínguez, R., García, A., García-Barrera, T., Barbas, C., & Gómez-Ariza, J. L. (2014d). Metabolomic profiling of serum in the progression of Alzheimer’s disease by capillary electrophoresis–mass spectrometry. Electrophoresis, 35, 3321–3330.
González-Domínguez, R., García-Barrera, T., & Gómez-Ariza, J. L. (2014a). Using direct infusion mass spectrometry for serum metabolomics in Alzheimer’s disease. Analytical and Bioanalytical Chemistry, 406, 7137–7148.
González-Domínguez, R., García-Barrera, T., & Gómez-Ariza, J. L. (2014c). Metabolomic study of lipids in serum for biomarker discovery in Alzheimer’s disease using direct infusion mass spectrometry. Journal of Pharmaceutical and Biomedical Analysis, 98, 321–326.
González-Domínguez, R., García-Barrera, T., & Gómez-Ariza, J. L. (2015b). Metabolite profiling for the identification of altered metabolic pathways in Alzheimer’s disease. Journal of Pharmaceutical and Biomedical Analysis, 107, 75–81.
González-Domínguez, R., García-Barrera, T., & Gómez-Ariza, J. L. (2015c). Application of a novel metabolomic approach based on atmospheric pressure photoionization mass spectrometry using flow injection analysis for the study of Alzheimer’s disease. Talanta, 131, 480–489.
González-Domínguez, R., García-Barrera, T., Vitorica, J., & Gómez-Ariza, J. L. (2014b). Region-specific metabolic alterations in the brain of the APP/PS1 transgenic mice of Alzheimer’s disease. Biochimica et Biophysica Acta, 1842, 2395–2402.
González-Domínguez, R., García-Barrera, T., Vitorica, J., & Gómez-Ariza, J. L. (2015a). Metabolomic screening of regional brain alterations in the APP/PS1 transgenic model of Alzheimer’s disease by direct infusion mass spectrometry. Journal of Pharmaceutical and Biomedical Analysis, 102, 425–435.
Gordon, S. (2003). Alternative activation of macrophages. Nature Reviews Immunology, 3, 23–35.
Gould, H. J., Sutton, B. J., Beavil, A. J., Beavil, R. L., McCloskey, N., Coker, H. A., et al. (2003). The biology of IgE and the basis of allergic disease. Annual Review of Immunology, 21, 579–628.
Hall, A. M., & Roberson, E. D. (2012). Mouse models of Alzheimer’s disease. Brain Research Bulletin, 88, 3–12.
Hansmannel, F., Sillaire, A., Kamboh, M. I., Lendon, C., Pasquier, F., Hannequin, D., et al. (2010). Is the urea cycle involved in Alzheimer’s disease? Journal of Alzheimer’s Disease, 21, 1013–1021.
Hart, P. H., Vitti, G. F., Burgess, D. R., Whitty, G. A., Piccoli, D. S., & Hamilton, J. A. (1989). Potential antiinflammatory effects of interleukin 4: suppression of human monocyte tumor necrosis factor alpha, interleukin 1, and prostaglandin E2. Proc Natl Acad Sci USA, 86, 380–33807.
He, P., Zhong, Z., Lindholm, K., Berning, L., Lee, W., Lemere, C., et al. (2007). Deletion of tumor necrosis factor death receptor inhibits amyloid beta generation and prevents learning and memory deficits in Alzheimer’s mice. Journal of Cell Biology, 178, 829–841.
Hu, Z. P., Browne, E. R., Liu, T., Angel, T. E., Ho, P. C., & Chan, E. C. Y. (2012). Metabonomic profiling of TASTPM transgenic alzheimer’s disease mouse model. Journal of Proteome Research, 11, 5903–5913.
Ibáñez, C., Simó, C., Martín-Álvarez, P. J., Kivipelto, M., Winblad, B., Cedazo-Mínguez, A., et al. (2012). Toward a predictive model of Alzheimer’s disease progression using capillary electrophoresis–mass spectrometry metabolomics. Analytical Chemistry, 84, 8532–8540.
Jankowsky, J. L., Fadale, D. J., Anderson, J., Xu, G. M., Gonzales, V., Jenkins, N. A., et al. (2004). Mutant presenilins specifically elevate the levels of the 42 residue beta-amyloid peptide in vivo: evidence for augmentation of a 42-specific g secretase. Human Molecular Genetics, 13, 159–170.
Jiang, N., Yan, X., Zhou, W., Zhang, Q., Chen, H., Zhang, Y., et al. (2008). NMR-based metabonomic investigations into the metabolic profile of the senescence-accelerated mouse. Journal of Proteome Research, 7, 3678–3686.
Jin, J. J., Kim, H. D., Maxwell, J. A., Li, L., & Fukuchi, K. (2008). Toll-like receptor 4-dependent upregulation of cytokines in a transgenic mouse model of Alzheimer’s disease. J Neuroinflammation, 5, 23.
Kawahara, K., Suenobu, M., Yoshida, A., Koga, K., Hyodo, A., Ohtsuka, H., et al. (2012). Intracerebral microinjec-tion of interleukin-4/interleukin-13 reduces beta-amyloid accumulation in theipsilateral side and improves cognitive deficits in young amyloid precursorprotein 23 mice. Neuroscience, 207, 243–260.
Kiyota, T., Okuyama, S., Swan, R. J., Jacobsen, M. T., Gendelman, H. E., & Ikezu, T. (2010). CNS.expression of anti-inflammatory cytokine interleukin-4 attenuates Alzheimer’s disease-like pathogenesis in APP+PS1 bigenic mice. FASEB J, 24, 3093–3102.
Kopf, M., Le Gros, G., Bachmann, M., Lamers, M. C., Bluethmann, H., & Köhler, G. (1993). Disruption of the murine IL-4 gene blocks Th2 cytokine responses. Nature, 362, 245–248.
Kühn, R., Rajewsky, K., & Müller, W. (1991). Generation and analysis of interleukin-4 deficient mice. Science, 254, 707–710.
Lalande, J., Halley, H., Balayssac, S., Gilard, V., Déjean, S., Martino, R., et al. (2014). 1H NMR metabolomic signatures in five brain regions of the AβPPswe Tg2576 mouse model of Alzheimer’s disease at four ages. Journal of Alzheimer’s Disease, 39, 121–143.
Li, P., Yin, Y. L., Li, D., Kim, S. W., & Wu, G. (2007). Amino acids and immune function. British Journal of Nutrition, 98, 237–252.
Lin, S., Kanawati, B., Liu, L., Witting, M., Li, M., Huang, J., et al. (2014). Ultra high resolution mass spectrometry-based metabolic characterization reveals cerebellum as a disturbed region in two animal models. Talanta, 118, 45–53.
Lin, S., Liu, H., Kanawati, B., Liu, L., Dong, J., Li, M., et al. (2013). Hippocampal metabolomics using ultrahigh-resolution mass spectrometry reveals neuroinflammation from Alzheimer’s disease in CRND8 mice. Analytical and Bioanalytical Chemistry, 405, 5105–5117.
Manev, H., Chen, H., Dzitoyeva, S., & Manev, R. (2011). Cyclooxygenases and 5-lipoxygenase in Alzheimers disease. Progress in Neuropsychopharmacology and Biological Psychiatry, 35, 315–319.
Mazurkiewicz-Kwilecki, I. M., & Nsonwah, S. (1989). Changes in the regional brain histamine and histidine levels in postmortem brains of Alzheimer patients. Canadian Journal of Physiology and Pharmacology, 67, 75–78.
Naddafi, F., & Mirshafiey, A. (2013). The neglected role of histamine in Alzheimer’s disease. American Journal of Alzheimer’s Disease and Other Dementias, 28, 327–336.
Naert, G., & Rivest, S. (2011). CC chemokine receptor 2 deficiency aggravates cognitive impairments and amyloid pathology in a transgenic mouse model of Alzheimer’s disease. Journal of Neuroscience, 31, 6208–6220.
Nakamura, S., Takemura, M., Ohnishi, K., Suenaga, T., Nishimura, M., Akiguchi, I., et al. (1993). Loss of large neurons and occurrence of neurofibrillary tangles in the tuberomammillary nucleus of patients with Alzheimer’s disease. Neuroscience Letters, 151, 196–199.
Panula, P., Rinne, J., Kuokkanen, K., Eriksson, K. S., Sallmen, T., Kalimo, et al. (1998). Neuronal histamine deficit in Alzheimer’s disease. Neuroscience, 82, 993–997.
Piro, J. R., Benjamin, D. I., Duerr, J. M., Pi, Y. Q., Gonzales, C., Wood, K. M., et al. (2012). A dysregulated endocannabinoid-eicosanoid network supports pathogenesis in a mouse model of Alzheimer’s disease. Cell Rep, 1, 617–623.
Reale, M., Iarlori, C., Feliciani, C., & Gambi, D. (2008). Peripheral chemokine receptors, their ligands, cytokines and Alzheimer’s disease. Journal of Alzheimer’s Disease, 14, 147–159.
Reale, M., Iarlori, C., Gambi, F., Feliciani, C., Salone, A., Toma, L., et al. (2004). Treatment with an acetylcholinesteraseinhibitor in Alzheimer patients modulates the expression and production of the pro-inflammatory and anti-inflammatory cytokines. Journal of Neuroimmunology, 148, 162–171.
Ribizzi, G., Fiordoro, S., Barocci, S., Ferrari, E., & Megna, M. (2010). Cytokine polymorphisms and Alzheimer disease: possible associations. Neurological Sciences, 31, 321–325.
Salek, R. M., Xia, J., Innes, A., Sweatman, B. C., Adalbert, R., Randle, S., et al. (2010). A metabolomic study of the CRND8 transgenic mouse model of Alzheimer’s disease. Neurochemistry International, 56, 937–943.
Sangster, T., Major, H., Plumb, R., Wilson, A. J., & Wilson, I. D. (2006). A pragmatic and readily implemented quality control strategy for HPLC-MS and GC-MS-based metabonomic analysis. Analyst, 131, 1075–1078.
Seiler, N. (2002). Ammonia and Alzheimer’s disease. Neurochemistry International, 41, 189–207.
Trushina, E., Dutta, T., Persson, X. M. T., Mielke, M. M., & Petersen, R. C. (2013). Identification of altered metabolic pathways in plasma and CSF in mild cognitive impairment and Alzheimer’s disease using metabolomics. PLoS ONE, 8, e63644.
Trushina, E., Nemutlu, E., Zhang, S., Christensen, T., Camp, J., Mesa, J., et al. (2012). Defects in mitochondrial dynamics and metabolomic signatures of evolving energetic stress in mouse models of familial Alzheimer’s disease. PLoS ONE, 7, e32737.
Tuppo, E. E., & Arias, H. R. (2005). The role of inflammation in Alzheimer’s disease. International Journal of Biochemistry & Cell Biology, 37, 289–305.
van den Berg, R. A., Hoefsloot, H. C. J., Westerhuis, J. A., Smilde, A. K., & van der Werf, M. J. (2006). Centering, scaling, and transformations: improving the biological information content of metabolomics data. BMC Genomics, 7, 142.
Vom Berg, J., Prokop, S., Miller, K. R., Obst, J., Kalin, R. E., Lopategui-Cabezas, I., et al. (2012). Inhibition of IL-12/IL-23 signaling reduces Alzheimer’s disease-like pathology and cognitive decline. Nature Medicine, 18, 1812–1819.
Wang, H., Lian, K., Han, B., Wang, Y., Kuo, S. H., Geng, Y., et al. (2014). Age-related alterations in the metabolic profile in the hippocampus of the senescence-accelerated mouse prone 8: a spontaneous Alzheimer’s disease mouse model. Journal of Alzheimer’s Disease, 39, 841–848.
Woo, D. C., Lee, S. H., Lee, D. W., Kim, S. Y., Kim, G. Y., Rhim, H. S., et al. (2010). Regional metabolic alteration of Alzheimer’s disease in mouse brain expressing mutant human APP-PS1 by 1H HR-MAS. Behavioural Brain Research, 211, 125–131.
Xia, J., & Wishart, D. S. (2010). MetPA: a web-based metabolomics tool for pathway analysis and visualization. Bioinformatics, 26, 2342–2344.
Acknowledgments
This work was supported by the Projects CTM2012-38720-C03-01 from the Ministerio de Ciencia e Innovación, and projects P012-FQM-0442 and P009-FQM-4659 from the Consejería de Innovación, Ciencia y Empresa (Junta de Andalucía). Raúl González Domínguez thanks the Ministerio de Educación for a predoctoral scholarship.
Conflict of interest
The authors declare no conflict of interest.
Compliance with Ethical Requirements
Animals were handled according to the directive 2010/63/EU stipulated by the European Community, and the study was approved by the Ethical Committee of University of Huelva.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
González-Domínguez, R., García-Barrera, T., Vitorica, J. et al. Metabolomic research on the role of interleukin-4 in Alzheimer’s disease. Metabolomics 11, 1175–1183 (2015). https://doi.org/10.1007/s11306-015-0773-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11306-015-0773-z