Abstract
In the wake of genomics, metabolomics characterizes the small molecular metabolites revealing the phenotypes induced by gene mutants. To address the metabolic signatures in the hippocampus of the amyloid-beta (Aβ) peptides produced in transgenic (Tg) CRND8 mice, high-field ion cyclotron resonance–Fourier transform mass spectrometry supported by LC-LTQ-Orbitrap was introduced to profile the extracted metabolites. More than 10,000 ions were detected in the mass profile for each sample. Subsequently, peak alignment and the 80 % rule followed by feature selection based on T score computation were performed. The putative identification was also conducted using the highly accurate masses with isotopic distribution by interfacing the MassTRIX database as well as MS/MS fragmentation generated in the LTQ-Orbitrap after chromatographic separation. Consequently, 58 differentiating masses were tentatively identified while up to 44 differentiating elemental compositions could not be biologically annotated in the databases. Nonetheless, of the putatively annotated masses, eicosanoids in arachidonic acid metabolism, fatty acid beta-oxidation disorders as well as disturbed glucose metabolism were highlighted as metabolic traits of Aβ toxicity in Tg CRND8 mice. Furthermore, a web-based bioinformatic tool was used for simulation of the metabolic pathways. As a result of the obtained metabolic signatures, the arachidonic acid metabolism dominates the metabolic perturbation in hippocampal tissues of Tg CRND8 mice compared to non-Tg littermates, indicating that Aβ toxicity functions neuroinflammation in hippocampal tissue and new theranostic opportunities might be offered by characterization of altered arachidonic acid metabolism for Alzheimer’s disease.

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The small molecular metabolites (Da < 1,500) have received our attention in recent years, because they represent the end point in the “omics” cascade. As substrates of the enzymes in metabolic pathways, these metabolites (namely metabolomes) could delineate the metabolic phenotypes and reveal the underlying mechanisms in tissues or cells [1]. Herein, metabolomics was regarded as a powerful technology to manifest the dynamic and amplified readout induced by genetic modification and/or environmental influences [2]. The current revolution in metabolomics and systems biology is driven by new analytical tools that are both fast and sensitive. Among these tools, mass spectrometry has become the method of choice for rapidly identifying metabolites and determining their concentrations. Ion cyclotron resonance–Fourier transform mass spectrometry (ICR-FT/MS) could yield the fine resolved ions for small molecule analysis, especially when combined to electrospray ionization source [3]. High-field ICR-FT/MS can enable a global analysis of the compounds in a complex matrix. It is well known that unambiguous identification in non-targeted analysis is of particular difficulty. High mass accuracy with fine isotopic distribution allows the rapid identification of the metabolic pathways, although the isomers could not be differentiated by direct infusion. Notwithstanding, the feasibility and utility of the FTMS approach for a non-targeted metabolic profiling, greatly speed up the holistic understanding of gene function in any biological system [4]. So far, ICR-FT/MS has widely been used in metabolomic studies, such as, i.e., Crohn’s disease and insulin sensitivity in diabetes [5–7].
Alzheimer’s disease (AD), the most common cause of dementia, has a complex etiology involving both genetic and environmental determinants. The Tg CRND8 line carries the Swedish double mutation and the Indiana (V717F) mutation on a human APP695 isoform under the control of the hamster PrP promoter, and on a C3H/B6 background [8]. Gamma-secretase site APP mutations lead to a drastic reduction of Aβ40 or total Aβ, and Swedish APP mutation ensures a high Aβ secretion level, for instance, levels of Aβ42 in 6-month-old Tg CRND8 mice are close to those seen in PDAPP mice at 16 months of age [9, 10]. Amyloid deposition started at ∼6 weeks of age in Tg CRND8 mice, appearing first in the subiculum and frontal cortex, and later also involving corpus callosum, hippocampus, etc. Dense plaques associated with dystrophic neuritis, synaptic degeneration and inflammatory response were also observed. The Swedish mutation causes early-onset Alzheimer’s disease and the decline of cognitive function in carriers of the Swedish APP mutation was also investigated using another mouse model T2576. Therefore, the characterization of borderline syndrome between normal cognitive function and the Swedish APP mutation-induced cognitive decline should be explored for testing therapeutic agents in AD mouse models [11–13]. The metabolic mechanisms would be complex and involve a large number of metabolites. For instance, the arachidonate oxidation products, such as leukotriene B4 (LTB4), 15-hydroxyeicosatetraenoic acid (15-HETE), associated with inflammation might be hit as the targets in metabolomic analysis. In our previous work, hippocampus has been treated into metabolomic analysis to delineate the metabolite profiling for the speculation of the metabolic mechanisms caused by high-fructose diet or 2,3,7,8-tetrachlorodibenzo-p-dioxin exposure in rats [14, 15]. The biochemical changes in hippocampus are mirrored by mass spectrometric analysis of the metabolites. In this study, we presented ICR-FT/MS to depict the specific chemical space, and offer the mirror of the neuropathological abnormalities in hippocampus due to gene mutant. Multivariate data analysis enables the evaluation of the grouping and selection of differential biomarkers. Moreover, the sophisticated chemometrics and bioinformatics tools highlight the data visualization and pathway analysis.
Materials and methods
Chemicals and reagents
LTB4, 15-HETE, and 12-HETE were purchased from Cayman Chemical (Ann Arbor, MI, USA). Acetonitrile and methanol in HPLC grade were obtained from Tedia (Fairfield, OH, USA). Water was purified by using a Milli-Q purification system (Millipore, Billerica, USA).
Sample preparation
Twenty-week male Tg CRND8 and age-matched male non-Tg littermates (n = 6 for each group) were decapitated for dissecting the hippocampus tissues. For transgenic discrimination, animals were confirmed and divided by polymerase chain reaction for assaying the brain tissues. The tissues were stored at −80 °C prior to sample preparation.
Twenty milligrams of each tissue was stored at −20 °C. Protein precipitation was performed by adding 1 mL solvent mixture (methanol/water 4:1, v/v) to 2-mL Eppendorf tubes. Samples were vortexed for 30 s and submerged for 2 min in liquid nitrogen and then thawed for 3–4 min and sonicated for 5 min. This process was repeated three times and samples were stored at −20 °C for 1 h. The supernatant was removed by centrifugation at 12,000 rpm for 10 min. The supernatant was transferred to a clean tube and dried in SpeedVac. The residue was re-dissolved in 500 μL of cold acetonitrile/water (50:50, v/v) containing 0.1 % formic acid. The supernatant was collected after centrifugation at 12,000 rpm for 10 min and the solution was stored at −20 °C prior to analysis. The solution was diluted 100 times using 70 % methanol for ICR-FT/MS measurement.
Mass spectrometric conditions
High-resolution mass spectra for molecular formula assignment were acquired on an ICR-FT/MS (Solarix, Bruker Daltonics, Bremen, Germany) equipped with a 12-T superconducting magnet and an Apollo II electrospray ion (ESI) source. The electric potential difference between the ESI needle and the counter electrode cone was 3,500 V. The electric acceleration potential difference between the counter electrode cone and an additional installed metal cone was 500 V. Each sample was introduced into the ionization source at a flow rate of 2 μL min−1 by a microliter pump with a nebulizer gas pressure of 20 psi and a drying gas pressure of 15 psi (heated to 200 °C). All samples were measured three times in negative ionization electrospray mode. To reduce systematic error associated with instrumental drift, samples were run in an order that alternated between Tg and non-Tg.
The instrument was calibrated based on cluster ions of 5 mg/L arginine in methanol. Calibration errors in the relevant mass range were always below 0.05 ppm and the standard deviation was always less than 10 % by consecutive ten injections in negative ion mode. The spectra were acquired with a time domain transient of two Mega words with a mass-to-charge ratio (m/z) range between 150 and 1,200 amu. This time domain transient length used for these measurements gave a mass resolving power of more than 190,000 at m/z = 400. Three hundred scans were summed up in each acquisition. The ion accumulation time in the ion source was set to 100 ms.
The MS/MS analysis was performed in LC/MS mode on an Accela liquid chromatographic system (Thermo Fisher Scientific Inc.) coupled to an LTQ-Orbitrap XL MS (Thermo Fisher Scientific Inc.) and fitted with an electrospray source operated in the negative ionization mode. The chromatographic conditions were described previously [16]. Briefly, the mobile phase was consisted of A (0.1 % formic acid in water) and B (0.1 % formic acid in acetonitrile). The gradient program started from 30 % B, increased to 40 % B in 5 min and held for 5 min, then increased to 95 % B in 8 min and held for 4 min followed by re-equilibration in 8 min. The samples were separated on Zorbax RRHD C18 column (2.1 × 100 mm, 1.8 μm). The flow rate was set up to 300 μL min−1. Heater temperature, sheath gas flow rate, aux gas flow rate, spray voltage, capillary temperature, capillary voltage, tube lens and collision-induced dissociation were set up to 350 °C, 40 arb, 5 arb, 2.5 kV, 300 °C, −9.0 V,–10.0 V and 35.0 eV, respectively. All data were required with scanning mass range from m/z 200 to 1,000.
Data analysis
The mass spectra were exported to peak lists at a signal to noise ratio (S/N) of 3. The peak alignment was performed by using the home made software matrix generator at a window width set at 1 ppm. 80 % rule was used for data pre-processing. 80 % rule was conducted for a variable that had a non-zero measurement value in at least 80 % of the variables within one of the two subsets; otherwise the variable was removed [17]. After performing this data filtration, T score was performed for feature selection in MATLAB R2009b programming environment (MathWorks, Inc. USA). Principal components analysis (PCA) and partial least squares-discriminate analysis (PLS-DA) were performed in SIMCA-P 12.0.1 (Umetrics, Sweden). In addition, the differential ions were implemented into MassTRIX interface (http://www.masstrix.org) to search for differentiating metabolites [18]. The criteria in database searching were as follows: negative ionization (correct for H+ loss) was selected as scan mode; maximum error was up to 1.0 ppm. KEGG with expanded lipids (HMDB, LipidMaps) (2006–2009) and Mus musculus (mouse) were chosen as database and organism, respectively. Cluster heat map was generated using hierarchical clustering explorer (http://www.cs.umd.edu/hcil/hce/). Because metabolic pathways were proposed for a better understanding, a web-tool MetPA (http://metpa.metabolomics.ca) was used for the pathway analysis and visualization.
Results and discussion
Data analysis for the chemical profiles
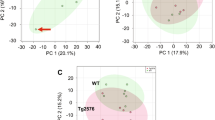
High-field (12-T) ICR-FT/MS was used to measure the metabolites in hippocampal tissues of Tg CRND8 mice as well as non-Tg littermates. The high end instrument yielded the high mass accuracy after calibration. More than 15,000 ions in the mass profiling were obtained for each sample. Subsequently, we utilized the matrix generator (homemade program, Germany) for peak alignment within 1 ppm of the mass error resulting in more than 100,000 peaks in the limited table. In this regard, a lot of meaningless zeroes filled the gaps in the table. Hence, 80 % rule was carried out to filter the raw data. For example, 138,360 variables detected in negative ionization mode of ICR-FT/MS measurement were reduced to 2,948 ions after peak alignment and 80 % rule filtering (Fig. 1A). Among the 2,948 ions, only 5 ions are unique in non-Tg sample sets while 38 ions are unique in Tg sample sets. The mass profiles were imported into SIMCA-P software. In the PCA scores plot using the first two principal components (R 2 X(cum) = 69.2 %, Fig. 1B), the grouping did not exhibit sufficient separation. Therefore, a supervised PLS-DA was established by maximizing the difference of mass profiles of the two groups. The resulting two-component PLS-DA classification plot (Fig. 1C, R 2 X(cum) = 68.3 %, R 2 Y(cum) = 76.8 %, Q 2 Y(cum)=59.1 %) presents a clear separation between Tg and non-Tg sample sets. It is well known that R 2 and Q 2 were used to evaluate the robustness of a pattern recognition model within SIMCA. R 2 is the fraction of variance explained by a component, and cross-validation was performed using the “validate” option, provided in SIMCA program. Furthermore, R 2 gives Q 2 which reveals the fraction of the total variation predicted by a component. According to the typical model, our results meet the requirement that a robust model has R 2 > 50 % and Q 2 > 40 %. Moreover, feature selection was conducted based on T score computation for the potential biomarkers using the algorithms reported previously [19]. T score allows us to highlight the differential ions whose intensities vary between two groups. In this study, T score ≥ 3.0 was considered as the statistical significance for feature selection.
A 138360 variables were produced after peak alignment, and performed 80 % rule to reduce to 2,948 ions; Venn diagram displays 5 ions unique in non-Tg sample sets while approximately sevenfold unique in Tg sample sets. B PCA score plots by Pareto scaling (R 2 X(cum) = 69.2 %); C PLS-DA score plots by Pareto scaling (blue point non-Tg; red triangle Tg) (R 2 X(cum) = 68.3 %, R 2 Y(cum) = 76.8 %, Q 2 Y(cum) = 59.1 %)
Ultrahigh resolution mass spectrometry is a promising tool for unraveling the compositional space. The mass profiles were converted into elementary compositions and visualized and displayed in Van Krevelen diagrams [20]. After calculating the elemental compositions from the discriminant mass list of the PLS-DA, the presence of typical CHO, CHOS, CHON, and CHONS could be displayed in Van Krevelen diagrams (e.g., plots of H/C versus O/C for each peak). Hundreds of masses identified in the mass range 150 to 1200 amu, shown in Figure S1 (see Electronic Supplementary Material), provide a clear portrait of the molecular partitioning assigned to unique absolute formulas. These formulas were generated to meet our criteria with 0.2 ppm mass deviation tolerance and confirmed with 13C-isotope. By comparing Van Krevelen diagram in Tg to that in non-Tg mass data, the number of CHO compositions of Tg samples (see Electronic Supplementary Material, Figure S1 (A1 and A2)) is higher ∼42 % than that of non-Tg (see Electronic Supplementary Material, Figure S1 (B1 and B2)), indicating that more oxygenated compounds and less nitrogen containing compounds were observed in Tg samples. Moreover, the ratio of Tg to non-Tg samples was also calculated and the ratios were represented as bubble size in the Van Krevelen diagrams (see Electronic Supplementary Material, Figure S1(C)). Mass profiles revealed the difference of elemental compositions between these two tested groups and suggested the oxidative stress in gene mutant mouse model. For those masses with significant changes between Tg and non-Tg groups, the putative annotation is discussed in the next section.
Mass spectrometric analysis
The representative ICR-FT/MS mass spectrum is shown in Fig. 2A. With respect to metabolite identification, high mass accuracy was used for the annotation of elemental compositions and searching against in MassTRIX supported databases for putative identification. For example, the ion at m/z 355.24903 was assigned to C20H36O5 and tentatively identified to be prostaglandin F1α. Because of high resolution data, two neighboring ions at m/z 355.26436 and 355.28543 were assigned to C24H36O2 and C21H40O4, respectively. The mass errors are less than 0.5 ppm and these two ions were tentatively identified to be tetracosahexaenoic acid and monoacylglyceride (MG (18:1)), respectively (Fig. 2B). This example suggested that the ultrahigh resolution mass spectrometry could provide the fine spectrum in a well-calibrated method, although these two neighboring ions are not significant changes between two tested groups and did not be listed in the tables. Another ion at m/z 353.23344 was assigned to C20H34O5 and its isotopic distribution was also confirmed by the simulation (Fig. 2C and D). As mentioned above, the differential analysis of ICR-FT/MS spectra was conducted by the high mass accuracy and clear overlapping peaks. With respect to feature selection, the selected ions were transported into MassTRIX database for the identification. As a consequence, 58 differential ions of our interest were tentatively identified by high mass accuracy (see Electronic Supplementary Material Table S1). For instance, hippurate and salicylurate as the two leading metabolites sorted by T score belong to acyl glycines. In addition, l-gulonolactone, also known as reduced ascorbic acid, was measured as a unique compound in hippocampus of Tg mice when compared to non-Tg littermates. Alpha-N-phenylacetyl-l-glutamine was present in Tg mice uniquely, formed by the conjugation of phenylacetate and glutamine. Out of 58 annotated masses with significant changes, 11 elemental compositions covering 70 differentiating metabolites of eicosanoids were of important interest. For example, the ion at m/z 317.21221 in negative ion mode was assigned to 6 metabolite isomers: Leukotriene A4 (C00909), 4,5-Leukotriene A4 (C02645), 15-Oxo-5,8,11-cis-13-trans-eicosatetraenoate (C04577), 5-Oxoeicosatetraenoic acid (C14732), 12-OxoETE (C14807) and 17R,18S-Epoxy-5Z,8Z,11Z,14Z-icosatetraenoic acid (C13843). These isomers have very similar chemical structures and belong to eicosanoids. The isomers could not be discriminated under the current conditions, although direct infusion in ICR-FT/MS is a rapid and sensitive approach to measure a large number of small molecules in an unbiased manner. The mass spectrometric data of 11 elemental compositions were listed in Table 1. The mass accuracy (<0.5 ppm) and concentration changes were easily evaluated. The fold-change is around two- to threefold and P values are less than 0.001. The confirmation of these metabolites is displayed as a list of overlaid mass spectra, which is ranked using the descending T scores (Fig. 3). Good calibration has been obtained for the high mass accuracy to limit the potential candidates as biomarkers. Overall, the T score algorithm can detect discriminating peaks when most of the peaks in a group are clearly higher or lower than the peaks in the other one. It should be noted that isotopic distribution including isotopic abundance in high resolution mass spectrometry would also facilitate to improve the confidence of metabolite identification [21].
Mass spectra for detection and identification. A The representative total ion chromatogram of ICR-FT/MS measurement by direct infusion; B the high mass accuracy used for the interpretation of elemental composition, the ion at m/z 355.24903 (upper) with simulation of the ion at m/z 355.24900; C the ion at m/z 353.23344 with simulation of the ion at m/z 353.23335; D the isotopic ion at m/z 354.23672 with simulation of the isotopic ion at m/z 354.23670
However, exact mass alone does not enable to differentiate isomers and thus the MS/MS analysis was also conducted in UPLC/LTQ-Orbitrap FTMS for confirmation in Fig. 4. The two isomers of 15-HETE and 12-HETE were separated in the chromatographic condition. The metabolite at retention time of 16.51 min was assigned to 15-HETE by the high mass accuracy with mass error of 1.625 ppm in LTQ-Orbitrap FTMS and the MS/MS fragment ions. The compound at retention time 16.95 min in the sample set was identified to be 12-HETE by the high mass accuracy although it could not yield daughter ions in MS/MS spectrum due to the low intensity (Fig. 4A to E). Another compound in hippocampus sample was also confirmed by the MS/MS spectrum of the authentic standard to be LTB4 (Fig. 4F and G). For those un-annotated masses with significant changes between both groups, we adopted the strict criteria: mass error ≤ 0.2 ppm and coinciding with next rules for the elemental compositions. Firstly, double-bond equivalents (DBEs) were calculated from elemental compositions using the next equation, on the basis of the elemental formula CcHhNnOoSsPp: DBE = c − h / 2 + n / 2 + p / 2 + 1. The number of valence electrons considered for phosphorus is 3 [22]. Secondly, the upper limit for the DBE to carbon is 90 % of the carbon number and the DBE lower limit is zero, which is termed “90 % rule” [23]. Thirdly, other two rules are that the number of n over c should be equal to or less than 1, and (o + n) / c should be less than 3 to make sure the sum formulas have chemical meaning. According to these rules, several masses have two molecular formulas. For instance, the ion at m/z 541.27652 was assigned to C26H42N2O10 or C24H30N16. By searching against ChemSpider database (http://www.chemspider.com/), there are two hits for C26H42N2O10 and zero hits for C24H30N16, respectively. In this regard, only C26H42N2O10 was selected as the elemental composition for m/z 541.27652 in negative ion mode. Similarly, all the un-annotated masses were validated in ChemSpider database, although we did not list the possible assignments because of the un-annotated biological meaning. As a result, 44 differentiating un-annotated masses were listed in Table S1 (Electronic Supplementary Material) for the discrimination of the two tested groups. Interestingly, the unknown masses also reflected the metabolic burden in hippocampal tissues of Tg mice compared to non-Tg littermates. Although the unknown masses could not be annotated for their biological meanings, the chemical profiles generated from ultrahigh resolution mass spectrometry could cluster the different groups by the high throughput screening way as mentioned above.
A LC/MS/MS chromatogram of 15-HETE and 12-HETE. B The mass spectrum of the ion at m/z 319.22729 with mass error of 1.625 ppm. C The MS/MS spectrum of 15-HETE at retention time of 16.51 min generated from the parent ion at m/z 319. D The MS/MS spectrum of the compound at retention time of 16.51 min in hippocampus sample generated from the parent ion at m/z 319. E The MS/MS spectrum of 12-HETE at retention time of 16.95 min generated from the parent ion at m/z 319. F The MS/MS spectrum of LTB4 generated from the parent ion at m/z 335. G The MS/MS spectrum of the compound in the hippocampus sample generated from the parent ion at m/z 335
Metabolic function analysis
The strategy that identification was approached by using high mass accuracy with elemental compositions and MS/MS was adopted in the present study. To annotate their metabolic pathways, we used MetPA, a web-based metabolomics tool for pathway visualization. Fisher’s extract test as over representation analysis and relative—between centrality as pathway topology analysis method were chosen for construction of the model. Consequently, the metabolic pathways were visualized by their impacts (Fig. 5A) and summarized in Table 2. Furthermore, the heat map shown in Fig. 5B to visualize the metabolic burden in Tg mice compared to non-Tg littermates. Strikingly, 11 ions pinpoint the 70 intermediate metabolites because of the existing isomers. Out of 70 prostaglandin analogs, 61 intermediate metabolites were tentatively identified based on KEGG database, indicating arachidonic acid metabolism as the most important target in hippocampus tissues of Tg mice (Fig. 5C). Two- to threefold elevated levels of the metabolites in arachidonic acid metabolism could be recognized to be associated with inflammatory response, postulating that neuroinflammation occurred within 20 weeks of Tg mice in comparison with their non-Tg littermates. Inflammatory aspects of amyloid accumulation have long been recognized, including reports of elevated phospholipase A2 in brain tissue of Alzheimer’s patients [24]. Similarly, it was demonstrated that serum amyloid increases cyclooxygenase metabolites of activated human monocytes [25]. This notion was also supported by recent evidence that the altered prostaglandin system was observed in presenilin transgenic mouse model of Alzheimer’s disease [26]. Moreover, in line with the alterations of essential fatty acids associated with AD development in previous reports, inflammation is believed to be a critical factor for contributing to the Aβ-induced pathogenesis [27, 28]. Besides the intermediate metabolites in arachidonic acid metabolism, notably, the metabolites with first two top T scores were hippurate (T score: 12.39) and salicylurate (T score: 7.49). The accumulation of hippurate and salicylurate as the acyl glycines were determined to be the potential biomarkers. The excess excretion of certain acyl glycines is a hint for mitochondrial fatty acid beta-oxidation disorders [29, 30] which is consistent with the interpretation of oxidative stress from CHO portraits of Van Krevelen diagrams. The metabolic pathways were proposed from benzoate to hippurate through biochemical reaction with glycine. Another pathway is the formation of salicylurate from the precursor salicylate which may be also catalyzed with glycine (Fig. 6). The accumulation of hippurate and salicylurate suggested the alteration of phenylalanine metabolism in hippocampus of Tg mice, although the contribution of phenylalanine metabolism to underlying pathogenesis of the disease should be explored further. Moreover, in agreement with oxidative stress in hippocampus of Tg mice, l-gulonolactone is present in hippocampus was induced by gene mutant in Tg mice [31]. l-gulonolactone (also known as reduced ascorbic acid), which was recognized to be a marker in oxidative stress, is the substrate of the enzyme l-gulono-1,4-lactone oxidoreductase (EC 1.1.3.8). Additionally, d-glucose 6-phosphate (or glucose 1-phosphate, fructose 6-phosphate) in glycolysis and 6-phosphogluconic acid in pentose phosphate pathway were highlighted to point out that the glucose metabolism was also disturbed by gene mutant. Previously, an increase in concentration of fatty acids in hippocampus of Tg CRND8 mice has been observed in the NMR analysis [32]. However, NMR could not provide such detailed structural information of fatty acids as high mass accuracy. It has also been demonstrated that as harboring the human amyloid precursor protein gene with the Indiana and Swedish mutations, Tg CRND8 mouse model allows for a rapid, time-dependent study of the relationship between the fibrillogenic process and the inflammatory response during the brain amyloidogenic process [33]. In line with Tg CRND8 mouse model, the inflammatory onset in the mouse model of AD was also found in transgenic APPswe/PS1dE9 mice and McGill-Thy1-APP mice [34, 35]. The neuron-specific cyclooxygenase (COX-2) enzyme was found to be upregulated in the latter mouse model, indicating that neuroinflammation is the characteristic phenotype caused by the genetic modification in AD mouse models. Indeed, a direct correlation has been established between the Abeta-induced neurodegeneration and inflammatory factors (e.g., cytokine) release [36]. From the clinical aspect, growing evidence was demonstrated that neurotoxicity is mediated by inflammatory processes in AD patients [37, 38]. Collectively, in good agreement with our finding that the upregulation of arachidonic acid metabolism reveals the neuroinflammation in hippocampus, the novel anti-AD drugs might be developed by targeting arachidonic acid metabolism.
A Proposed pathway library construction and visualization for the metabolites of interest in hippocampus tissues of Tg CRND8 mice using MetPA web tool (1 arachidonic acid metabolism; 2 Fructose and mannose metabolism; 3 amino sugar and nucleotide sugar metabolism; 4 starch and sucrose metabolism; 5 ascorbate and aldarate metabolism; 6 pentose phosphate pathway; 7 taurine and hypotaurine metabolism; 8 galactose metabolism; 9 pentose and glucuronate interconversions). B Heat map was visualized for the 58 differential ions with biological annotation to reveal the metabolic burden in hippocampus of transgenic mice (red increase in concentration; green decrease in concentration). C Arachidonic acid metabolism as the most important pathway was highlighted
The most important metabolites hippurate and salicylurate were visualized in box plots. The interpretation of the glycine involving metabolism for hippurate and salicylurate production was also illustrated for the proposed metabolic pathways. There are two pathways to form hippurate and salicylurate from benzoate, respectively. Both pathways are catalyzed with glycine
Conclusions
Rather than evaluating single molecule at a time, a metabolomic analysis provides a broad-based portrait of the changes in hippocampal metabolomes induced by gene mutant. Direct infusion electrospray ICR-FT/MS combined with LC/MS analysis offers high mass accuracy and high resolution for analyzing complex metabolite mixtures with increasing metabolome coverage. The grouping between Tg and non-Tg sample sets was evaluated by unsupervised as well as supervised machine learning. With regard to data mining, feature selection was conducted by calculating T scores. Moreover, high-resolution mass spectra display the distinct ions with their isotopic distributions, facilitating identification of the metabolites of interest. These statistically significant metabolites were identified by interfacing with databases for the interpretation of the metabolic traits. Strikingly, the metabolites associated with inflammation dominate the metabolic changes, revealing neuroinflammation in transgenic hippocampus tissues due to the gene mutant. The concentrations of the metabolites of interest were enhanced up to two- to threefolds, indicating that the neurotoxicity was associated with metabolic burden. Therefore, such rapid approach enables to hit the therapeutic target and elucidate the metabolic clues for a better understanding of the pathogenesis in the transgenic mouse model.
References
Kaddurah-Daouk R, Kristal BS, Weinshilboum RM (2008) Metabolomics: a global biochemical approach to drug response and disease. Annu Rev Pharmacol Toxicol 48:653–683
Denkert C, Budczies J, Kind T, Weichert W, Tablack P, Sehouli J, Niesporek S, Konsgen D, Dietel M, Fiehn O (2006) Mass spectrometry-based metabolic profiling reveals different metabolite patterns in invasive ovarian carcinomas and ovarian borderline tumors. Cancer Res 66(22):10795–10804
Southam AD, Payne TG, Cooper HJ, Arvanitis TN, Viant MR (2007) Dynamic range and mass accuracy of wide-scan direct infusion nanoelectrospray fourier transform ion cyclotron resonance mass spectrometry-based metabolomics increased by the spectral stitching method. Anal Chem 79(12):4595–4602
Aharoni A, Ric de Vos CH, Verhoeven HA, Maliepaard CA, Kruppa G, Bino R, Goodenowe DB (2002) Nontargeted metabolome analysis by use of fourier transform ion cyclotron mass spectrometry. OMICS 6(3):217–234
Ohta D, Kanaya S, Suzuki H (2010) Application of Fourier-transform ion cyclotron resonance mass spectrometry to metabolic profiling and metabolite identification. Curr Opin Biotechnol 21(1):35–44
Jansson J, Willing B, Lucio M, Fekete A, Dicksved J, Halfvarson J, Tysk C, Schmitt-Kopplin P (2009) Metabolomics reveals metabolic biomarkers of Crohn’s disease. PLoS One 4(7):e6386
Lucio M, Fekete A, Weigert C, Wagele B, Zhao XJ, Chen J, Fritsche A, Haring HU, Schleicher ED, Xu GW, Schmitt-Kopplin P, Lehmann R (2010) Insulin sensitivity is reflected by characteristic metabolic fingerprints—a fourier transform mass spectrometric Non-targeted metabolomics approach. PLoS One 5(10):e13317
Chishti MA, Yang DS, Janus C, Phinney AL, Horne P, Pearson J, Strome R, Zuker N, Loukides J, French J, Turner S, Lozza G, Grilli M, Kunicki S, Morissette C, Paquette J, Gervais F, Bergeron C, Fraser PE, Carlson GA, George-Hyslop PS, Westaway D (2001) Early-onset amyloid deposition and cognitive deficits in transgenic mice expressing a double mutant form of amyloid precursor protein 695. J Biol Chem 276(24):21562–21570
Kumar-Singh S, De Jonghe C, Cruts M, Kleinert R, Wang R, Mercken M, De Strooper B, Vanderstichele H, Lofgren A, Vanderhoeven I, Backhovens H, Vanmechelen E, Kroisel PM, Van Broeckhoven C (2000) Nonfibrillar diffuse amyloid deposition due to a gamma(42)-secretase site mutation points to an essential role for N-truncated A beta(42) in Alzheimer’s disease. Hum Mol Genet 9(18):2589–2598
De Jonghe C, Esselens C, Kumar-Singh S, Craessaerts K, Serneels S, Checler F, Annaert W, Van Broeckhoven C, De Strooper B (2001) Pathogenic APP mutations near the gamma-secretase cleavage site differentially affect Abeta secretion and APP C-terminal fragment stability. Hum Mol Genet 10(16):1665–1671
Haass C, Lemere CA, Capell A, Citron M, Seubert P, Schenk D, Lannfelt L, Selkoe DJ (1995) The Swedish mutation causes early-onset Alzheimer’s disease by β-secretase cleavage within the secretory pathway. Nat Med 1:1291–1296
Rustay NR, Cronin EA, Curzon P, Markosyan S, Bitner RS, Ellis TA, Waring JF, Decker MW, Rueter LE, Browman KE (2010) Mice expressing the Swedish APP mutation on a 129 genetic background demonstrate consistent behavioral deficits and pathological markers of Alzheimer’s disease. Brain Res 1311:136–147
Rabe S, Reichwald J, Ammaturo D, de Strooper B, Saftig P, Neumann U, Staufenbiel M (2011) The Swedish APP mutation alters the effect of genetically reduced BACE1 expression on the APP processing. J Neurochem 119(1):231–239
Lin S, Yang Z, Liu H, Tang L, Cai Z (2011) Beyond glucose: metabolic shifts in responses to the effects of the oral glucose tolerance test and the high-fructose diet in rats. Mol Biosyst 7(5):1537–1548
Lin S, Yang Z, Zhang X, Bian Z, Cai Z (2011) Hippocampal metabolomics reveals 2,3,7,8-tetrachlorodibenzo-p-dioxin toxicity associated with ageing in Sprague–Dawley rats. Talanta 85(2):1007–1012
He C, Wu Y, Lai Y, Cai Z, Liu Y, Lai L (2012) Dynamic eicosanoid responses upon different inhibitor and combination treatments on the arachidonic acid metabolic network. Mol Biosyst 8(5):1585–1594
Bijlsma S, Bobeldijk I, Verheij ER, Ramaker R, Kochhar S, Macdonald IA, van Ommen B, Smilde AK (2006) Large-scale human metabolomics studies: a strategy for data (pre-) processing and validation. Anal Chem 78(2):567–574
Suhre K, Schmitt-Kopplin P (2008) MassTRIX: mass translator into pathways. Nucleic Acids Res 36(Web Server issue):W481–W484
Wang L, Chu F, Xie W (2007) Accurate cancer classification using expressions of very few genes. IEEE/ACM Trans Comput Biol Bioinform 4(1):40–53
Liger-Belair G, Cilindre C, Gougeon RD, Lucio M, Gebefugi I, Jeandet P, Schmitt-Kopplin P (2009) Unraveling different chemical fingerprints between a champagne wine and its aerosols. Proc Natl Acad Sci U S A 106(39):16545–16549
Weber RJ, Southam AD, Sommer U, Viant MR (2011) Characterization of isotopic abundance measurements in high resolution FT-ICR and Orbitrap mass spectra for improved confidence of metabolite identification. Anal Chem 83(10):3737–3743
Bae E, Yeo IJ, Jeong B, Shin Y, Shin KH, Kim S (2011) Study of double bond equivalents and the numbers of carbon and oxygen atom distribution of dissolved organic matter with negative-mode FT-ICR MS. Anal Chem 83(11):4193–4199
Lobodin VV, Marshall AG, Hsu CS (2012) Compositional space boundaries for organic compounds. Anal Chem 84(7):3410–3416
Stephenson DT, Lemere CA, Selkoe DJ, Clemens JA (1996) Cytosolic phospholipase A(2) (cPLA(2)) immunoreactivity is elevated in Alzheimer’s disease brain. Neurobiol Dis 3(1):51–63
Malle E, Bollmann A, Steinmetz A, Gemsa D, Leis HJ, Sattler W (1997) Serum amyloid A (SAA) protein enhances formation of cyclooxygenase metabolites of activated human monocytes. FEBS Lett 419(2–3):215–219
Schulte EC, Slawik H, Schule R, Gunther T, Hull M (2009) Alterations in excitotoxicity and prostaglandin metabolism in a transgenic mouse model of Alzheimer’s disease. Neurochem Int 55(7):689–696
Sanchez-Mejia RO, Mucke L (2010) Phospholipase A2 and arachidonic acid in Alzheimer’s disease. Biochim Biophys Acta 1801(8):784–790
Amtul Z, Uhrig M, Wang L, Rozmahel RF, Beyreuther K (2012) Detrimental effects of arachidonic acid and its metabolites in cellular and mouse models of Alzheimer’s disease: structural insight. Neurobiol Aging 33(4):831, e21-31
Kimura M, Yamaguchi S (1999) Screening for fatty acid beta oxidation disorders. Acylglycine analysis by electron impact ionization gas chromatography-mass spectrometry. J Chromatogr B: Biomed Sci Appl 731(1):105–110
Fourcade S, Ruiz M, Camps C, Schluter A, Houten SM, Mooyer PA, Pampols T, Dacremont G, Wanders RJ, Giros M, Pujol A (2009) A key role for the peroxisomal ABCD2 transporter in fatty acid homeostasis. Am J Physiol Endocrinol Metab 296(1):E211–E221
Steinberg JG, Delliaux S, Jammes Y (2006) Reliability of different blood indices to explore the oxidative stress in response to maximal cycling and static exercises. Clin Physiol Funct Imag 26(2):106–112
Salek RM, Xia J, Innes A, Sweatman BC, Adalbert R, Randle S, McGowan E, Emson PC, Griffin JL (2010) A metabolomic study of the CRND8 transgenic mouse model of Alzheimer’s disease. Neurochem Int 56(8):937–947
Dudal S, Krzywkowski P, Paquette J, Morissette C, Lacombe D, Tremblay P, Gervais F (2004) Inflammation occurs early during the Abeta deposition process in TgCRND8 mice. Neurobiol Aging 25(7):861–871
Ruan L, Kang Z, Pei G, Le Y (2009) Amyloid deposition and inflammation in APPswe/PS1dE9 mouse model of Alzheimer’s disease. Curr Alzheimer Res 6(6):531–540
Ferretti MT, Bruno MA, Ducatenzeiler A, Klein WL, Cuello AC (2012) Intracellular Abeta-oligomers and early inflammation in a model of Alzheimer’s disease. Neurobiol Aging 33(7):1329–1342
Rojo LE, Fernandez JA, Maccioni AA, Jimenez JM, Maccioni RB (2008) Neuroinflammation: implications for the pathogenesis and molecular diagnosis of Alzheimer’s disease. Arch Med Res 39(1):1–16
Rosenberg PB (2005) Clinical aspects of inflammation in Alzheimer’s disease. Int Rev Psychiatry 17(6):503–514
Trepanier CH, Milgram NW (2010) Neuroinflammation in Alzheimer’s disease: are NSAIDs and selective COX-2 inhibitors the next line of therapy? J Alzheimers Dis 21(4):1089–1099
Acknowledgments
The authors would like to thank Dr. Sabina Bijlsma (TNO Quality of Life, The Netherlands) shared the MATLAB code for 80 % rule and Prof. Zhao Wang (School of Medicine, Tsinghua University, Beijing, China) offered the helpful discussion for the manuscript. Financial supports from General Research Grant (HKBU200310) from Hong Kong SAR and collaborative research fund (IRMC/12-13/1A) from Hong Kong Baptist University are acknowledged. Dr. Shuhai Lin was supported by German Academic Exchange Service (DAAD) scholarship (A/12/00412) for the research stay in Germany. The authors would also like to thank Mr. Yongle Li in Shenzhen Academy of Metrology & Quality Inspection of China for technical assistance of UPLC/LTQ-Orbitrap MS performance.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Published in the topical collection Metabolomics and Metabolite Profiling with guest editors Rainer Schuhmacher, Rudolf Krska, Roy Goodacre, and Wolfram Weckwerth.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 331 kb)
Rights and permissions
About this article
Cite this article
Lin, S., Liu, H., Kanawati, B. et al. Hippocampal metabolomics using ultrahigh-resolution mass spectrometry reveals neuroinflammation from Alzheimer’s disease in CRND8 mice. Anal Bioanal Chem 405, 5105–5117 (2013). https://doi.org/10.1007/s00216-013-6825-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-013-6825-1