Human hepatopathies are a diagnostic challenge, with many distinct diseases having similar clinical signs and laboratory findings. Naturally occurring canine hepatic disease provides an excellent model for human diseases and similar diagnostic dilemmas exist; differentiating canine congenital portosystemic vascular anomalies (PVA) from acquired hepatopathies is difficult and traditionally requires invasive diagnostic procedures. The emerging post-genomic science of metabolomics is concerned with detecting global changes of populations of endogenous low molecular weight metabolites in biological samples and offers the possibility of identifying surrogate profiles of disease. Metabolomics couples sensitive metabolite analysis with sophisticated pattern recognition techniques. In this study, a metabolomic strategy has been employed to assess metabolite changes in the plasma of dogs with congenital PVA and acquired hepatic disease. Plasma samples were collected from 25 dogs, comprising 9 dogs with congenital PVA, 6 with acquired hepatopathy and 10 with non-hepatic disorders. Low molecular weight metabolites were analyzed by liquid chromatography-mass spectrometry (LC-MS). Following identification of metabolites, multivariate data analysis was used to compare profiles amongst groups. The analysis demonstrated significant disturbances in the plasma bile acid and phospholipid profiles of dogs with portovascular anomalies. In contrast to traditional laboratory parameters, the metabolomic strategy was able to produce a clear segregation between all three study groups. In conclusion, this study demonstrates the potential of metabolomics as a diagnostic tool for naturally occurring hepatic disease. With further validation, this approach will improve diagnostic capabilities, provide an insight into pathogenetic mechanisms, and ultimately inform therapeutic decision making in clinical hepatology.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The liver is the most metabolically diverse organ of the body, and is involved in a number of critical metabolic processes. This organ is sensitive to many pathological insults, and the result of dysfunction leads to an array of clinical signs and symptoms. Clinicians can utilize such metabolic derangements in diagnosis. Conventional diagnostic approaches have typically focussed on a small number of biomarkers, which are characteristic of a particular disease state. Although, for some diseases with a straightforward cause and effect relationship, this approach can reliably group a population into affected and unaffected individuals, such links are difficult to establish for complex multifactorial diseases (German et al., 2003). In many cases, no single biomarker is sufficiently specific to provide a definitive diagnosis. Clinicians must, therefore, combine tests to diagnose the disease accurately, and use pattern recognition to differentiate conditions which lead to similar clinical signs. However, the ability to make a correct diagnosis is often limited by the number of variables that the clinician can assimilate in parallel.
Thousands of low molecular weight metabolites exist in the body fluids and tissues. Metabolite profiles can be altered by a variety of physiological and pathological processes, and therefore global changes in such profiles may signal the presence of a particular disease state (Nicholson et al., 2002; Griffin and Shockcor, 2004). Metabolomics is an emerging post-genomic science that uses analytical techniques such as nuclear magnetic resonance (NMR) spectroscopy and mass spectrometry to measure low molecular weight metabolites in biological samples (Griffin, 2003; Dunn et al., 2005; Wilson et al., 2005). Advanced statistical analysis is then employed to maximize the recovery of information and interpret the large data sets that are generated. Whilst still in its infancy, metabolomics is already beginning to make a significant impact in biological and medical research. Metabolomic strategies have been employed to characterize metabolic perturbations resulting from altered gene function in biological systems (Raamsdonk et al., 2000; Roessner et al., 2001; Allen et al., 2003) and as a means of assessing the toxicological effects of chemical compounds (Waters et al., 2002; Lindon et al., 2003; Nicholson et al., 2003). To date there have been few studies, which focus on naturally occurring disease states. However, metabolomic approaches are increasingly being employed by investigators as a means of characterizing metabolic alterations in body fluids and tissues of humans and animals in order to improve clinical diagnosis and explore the pathogenesis of diseases (Brindle et al., 2002; Lamers et al., 2003; Yang et al., 2004).
Congenital portovascular anomalies (PVA; also known as portosystemic shunts, PSS) have been recognized in many species, and are the most common congenital liver disease in dogs (Center, 1996). In all cases, an abnormal vascular communication exists between the portal and systemic venous circulations, which allows blood from splanchnic organs to bypass the liver and avoids normal hepatic metabolism. The reduced ability of the liver to detoxify and excrete waste results in dramatic changes in the levels of a wide range of metabolites. PVA are associated with hyperammonaemia and disturbances in the concentrations of aromatic and branched chain amino acids in the plasma of affected dogs have been reported (Schafer et al., 1985). The disease state is characterized by the development of neurological (dementia, staggering, circling, central blindness, tremors, seizures and coma) and systemic (stunting, poor body condition, polydipsia, polyuria, vomiting and diarrhoea) signs. Whilst congenital PVA are rare abnormalities of liver vasculature in humans, surgically induced transjugular intrahepatic portosystemic shunts (TIPS) are a valuable tool in the management of patients with cirrhosis and treatment of complications of portal hypertension (Watanabe, 2000; Rosado and Kamath, 2003). Interestingly, a common complication of TIPS is hepatic encephalopathy. Therefore, congenital PVA in dogs can provide an excellent animal model with which to study hepatic dysfunction.
Diagnosis of congenital PVA in dogs can be problematic because a number of acquired hepatic diseases can also arise in young dogs, and the presenting clinical signs can be identical. Congenital PVA are typically diagnosed by a combination of laboratory analyses, and diagnostic imaging. Although diagnosis can be achieved in most instances there are numerous pitfalls. Preliminary investigations include routine haematological and clinical chemistry analysis, and measurement of fasting and post-prandial bile acid concentrations. However, none of these tests are pathognomonic for congenital PVA and often can not differentiate from acquired PVA. Alternatively, the abnormal shunting vessel can be detected by abdominal ultrasonography, but the technique is technically challenging and many shunts are missed (Center, 1996). At present, the most informative diagnostic approach is intravenous portovenography. However, this technique also requires considerable expertise and given the requirement for general anaesthesia and celiotomy, it is invasive and not without risk. Therefore, additional non-invasive diagnostic modalities are urgently required. Our overall hypothesis is that metabolomics will provide a powerful, non-invasive, diagnostic tool for both human and veterinary hepatology. To test this hypothesis we have employed metabolomic strategies to assess the perturbations of the metabolite profiles in the plasma of dogs with congenital PVA and compared them with dogs with acquired liver disease.
Materials and methods
Study animals
The study involved 25 dogs referred to the Department of Veterinary Clinical Sciences, University of Liverpool. Of these dogs, 15 were referred for the investigation of suspected hepatic disease (table 1). After diagnostic investigations, these dogs were assigned to two groups; Group 1 (n=9) represented dogs with a final diagnosis of congenital portovascular anomaly (PVA); Group 2 (n=6) represented dogs with a final diagnosis of an acquired hepatopathy. The remaining dogs (n=10) were assigned to Group 3, and were a control population, referred for unrelated i.e. non-hepatic disorders over the same period of study as the clinical cases (table 1). The study was performed in adherence to the University of Liverpool Animal Ethics Guidelines and the owners of all animals participating in the study gave written consent.
Clinical investigations and diagnosis
The exact diagnostic investigations performed depended upon the presenting signs of the individual case. All dogs had routine haematological analysis, whilst the majority of dogs (5/9, 4/6 and 7/10 for Groups 1, 2 and 3, respectively) had routine urinalysis (dipstick, specific gravity by refractometer, sediment analysis). Fasting (9/9, 6/6 and 8/10; for Groups 1, 2 and 3, respectively) and post-prandial (9/9, 4/6 and 4/10; for groups 1, 2 and 3, respectively) bile acid concentrations, and abdominal ultrasonography (9/9, 4/6 and 7/10; for Groups 1, 2 and 3, respectively) were also performed in most cases. All routine laboratory investigations were performed by the diagnostic laboratories in the Department of Veterinary Pathology, University of Liverpool.
Definitive diagnosis of congenital PVA was made at celiotomy either by direct visualization of the anomalous blood vessel (n=4) or by direct visualization and portovenography (n=5). Seven dogs had extra-hepatic PVA, of which 5 were portocaval, one was porto-azygous and one was a communication between a gastric vein and the vena cava. The remaining two dogs had intrahepatic PVA, one of which was right divisional, and the other was left divisional. Definitive diagnosis of acquired hepatopathy was made by histopathological assessment of hepatic biopsy specimens in most cases, collected either at celiotomy (n=1), laparoscopy (n=3), or postmortem examination (n=1). In the remaining case, a presumptive diagnosis of glucocorticoid hepatopathy was made after documenting dramatic increases in liver enzymes (alkaline phosophatase and alanine aminotransferase), whilst the dog was undergoing high-dose glucocorticoid therapy (prednisolone at a dosage of 2 mg/kg). Elevated fasting serum bile acid concentrations were documented and fine needle aspiration cytology revealed findings consistent with vacuolar hepatopathy. Hepatic parameters had been normal prior to glucocorticoid use. The specific clinical investigations performed in the control dogs varied depending on reason for referral. However, there was no evidence of hepatic disease, and an alternative diagnosis was documented in every case.
Blood was collected from the dogs by jugular venepuncture for routine haematological and biochemical analyses. Any surplus heparinized plasma (approximately 1 mL) was used for the study, aliquoted into 20 μL units, and immediately stored at −20 °C for metabolite analyses. All the investigative procedures performed were for the direct benefit of the patient. After diagnosis, all animals were treated according to standard hospital protocols and received the most appropriate therapy.
Statistical analysis of clinical data
Statistical analysis was performed using Minitab for Windows release 14.1 (Minitab Inc., State College, PA, USA). Prior to statistical analysis, all continuous data were assessed for normal distribution. Given that none of the data for any outcome were normally distributed, data were logarithmically transformed and again assessed for normal distribution. The data for age and fasting bile acid concentrations were normalized by this transformation. When a square root transformation was used instead, data for post-prandial bile acid concentrations were normalized. Therefore, parametric tests were employed (after appropriate transformation).
The order of statistical analysis was as follows: baseline data (e.g. age, gender, neuter status) amongst groups first were compared with either a one-way analysis of variance (ANOVA) (age) or Chi squared tests (gender and neuter status). The data were then used in quantitative analyses among groups, using one-way ANOVA. Post hoc analysis was performed with Tukey’s pairwise comparisons. The level of significance for all statistical tests was set at p<0.05.
Extraction of low molecular weight metabolites
The plasma samples were thawed at room temperature and mixed thoroughly prior to analysis. The low molecular weight metabolites in plasma were extracted in organic solvents. A 50 μL aliquot of plasma was then extracted with 200 μL of acetonitrile. The mixture was left to stand at room temperature for 10 min and was then centrifuged at to remove the precipitated plasma proteins.
Liquid chromatography-mass spectrometry (LC-MS)
The low molecular weight metabolites were then analyzed by liquid chromatography-mass spectrometry (LC-MS) as previously described (Plumb et al., 2003; Lenz et al., 2004a, b). All mass spectrometric analyses were performed using a Micromass Q-TOF micro mass spectrometer (Waters Corporation, Manchester, UK) equipped with a Z-Spray electrospray source and coupled to a Waters Alliance 2795XC HPLC system.
Samples (10 μL) were injected on to a Waters Symmetry C18 reverse phase LC column (2.1× 100 mm, 3.5 μm) held at 40 °C. The column was eluted using a water/acetonitrile gradient at a flow rate of 600 μL/min. The mobile phases both contained 0.1% (volume/volume, v/v) formic acid. The gradient conditions were 0–20% acetonitrile over 4 min, followed by 20–95% acetonitrile over 4–8 min. The gradient was held for 1 min and then returned to 0% acetonitrile at 9.1 min. The flow rate was split to 120 μL/min into the mass spectrometer.
The mass spectrometer was operated in both the positive and negative ion mode. The positive and negative ion data was acquired separately in two runs. The capillary voltage was set to 3200 V in the positive mode and 2600 V in negative ion mode. The cone voltage was set to 30 V. The source temperature was set at 120 °C with a cone gas flow of 50 L/h, the desolvation heater at 250 °C and a desolvation gas flow of 500 L/h. Data were collected in centroid mode using a LockSpray interface with a scan time of 0.4 s and an inter-scan delay of 0.1 s. Leucine enkephalin was used as the lockmass standard at a concentration of 0.5 ng/μL in acetonitrile:water + 0.1% (v/v) formic acid in positive ion mode ([M+H]+=556.2771) and 1 ng/μL in acetonitrile:water + 0.1% (v/v) formic acid in negative ion mode ([M-H]−=554.2615). The LockSpray frequency was set as 5 s and the lockmass was averaged over 10 scans. Mass spectra were acquired over the mass to charge ratio range (m/z) 50–850. Metabolites were identified using liquid chromatography-tandem mass spectrometry (LC-MS/MS). Product ions were detected over a scanning range of m/z 50–850. The collision voltage was set between 30 and 70 V and argon was used as the collision gas at a pressure of 15 p.s.i. Identification of the metabolites was confirmed by comparison with authentic standards.
Data processing and statistical analysis of metabolite data
The raw LC-MS data was initially processed using the MarkerLynx (version 1.0) applications manager software (Lenz et al., 2004a, b). The software incorporates a peak deconvolution package which allows detection and retention time alignment of the peaks eluting in each date file across the whole dataset. MarkerLynx extracts components utilizing exact mass chromatograms and lists detected peaks as their mass and retention time along with their associated intensities.
Chromatographic peaks in the raw data files are detected by extracting nominal mass chromatograms and tracking the apex of the peaks in the chromatograms. The spectra from each of the detected peaks are saved as retention time and exact mass pairs along with associated intensities which can be saved as either normalized or absolute intensities. Once all the data has been extracted it is aligned within user defined mass and retention time windows. If necessary retention time correction can be applied using an internal standard. Once collected the data were then interpreted by principle component analysis (PCA) within MarkerLynx applications manager.
Partial least squares-discriminant analysis (PLS-DA) was performed by exporting the processed data to SIMCA-P+ (version 10.0) multivariate data analysis software package (Umetrics, Sweden). PLS-DA models were originally constructed using all the samples and the information supplied regarding their class membership. To validate the PLS-DA model an observation from each class was excluded in turn and a new model generated. The coefficients from each model were then taken and sorted in ascending order. The models were then compared to check that the order of the potential biomarkers were the same or similar between the different models. The models were also validated by predicting the excluded observations and checking their classifications against the models. If the probability of belonging to a class is >0.65 then this is considered positive. If the probability of belonging to a class is >0.35 to 0.65 then it is considered to be borderline and below that it is considered not to belong to that class.
Results and discussion
Clinical data
The clinical data from all cases is summarized in table 1. Preliminary statistical analyses did not reveal any significant differences in age, gender or neuter status between groups (p>0.14 for all). Further, a number of breeds were represented in all groups, and a particular breed bias was not obvious in any one group. However, given the small numbers of dogs in each group, the true effect of breed could not be determined. In the future, larger studies will be required to make certain that chance differences between such groups are not simply the effect of a chance association with a particular breed.
Prior to metabolomic analysis, we first assessed the ability of routine diagnostic investigations to differentiate the three clinical groups. Haematological and routine clinical chemistry results are reported in table 2. Although typical abnormalities were documented in Group 1 (including microcytic anaemia and low albumin concentrations), similar findings were evident in group 2 dogs. Therefore, none of the parameters measured could reliably distinguish cases with congenital PVA from dogs with acquired hepatic disease. Significant differences amongst groups were noted for both fasting and post-prandial bile acid concentrations (p<0.0001 for both). However, the group ranges for bile acid concentrations overlapped (table 1). Therefore, measurement of bile acid concentrations could not be used to differentiate accurately between dogs with PVA, dogs with acquired hepatic disease and controls. Similarly, multivariate data analysis of the conventional biochemical and haematological parameters was able to separate control and affected populations of dogs, but was unable to fully discriminate animals with congenital PVA from those with acquired hepatopathies (data not shown).
Negative ion mode LC-MS analysis
Plasma low molecular weight metabolites were characterized by LC-MS in both the positive and negative ion modes. The mass spectral data were then processed by multivariate analysis. Initial analyses employed an unsupervised statistical method, PCA. PCA is a pattern recognition method that reduces the dimensionality of data to a number of summary variables, principle components (PC). The PC are designed so that they encapsulate most of the variation present within the collected data. Data plotted using PC reveals inter-sample relationships via their spatial proximity. Trajectories can be identified in PC values between sample groups which reflect the perturbation of multiple metabolic pathways.
PCA can generate two main outputs, a scores plot and a loadings plot. A PCA scores plot displays any natural clustering or separation within a data set. The variability within each PC factor is attributed to the original measured individual and combined variables within the data set. This plot allows physiological or pathological similarities or differences to be explored. A PCA loadings plot identifies the relative contribution of individual ions to each PC and thus the most influential contributors to the natural sample class clustering.
Following PCA the data sets were processed using more powerful supervised statistical method, PLS-DA. PLS-DA like PCA is a projection method and is a classification method based on the regression extension of PCA. The regression against a dummy variable defining class identity yields latent variables explaining maximum separation between known classes of samples. The interpretation of the scores gives information about class separation while interpretation of loadings gives information about variables responsible for separating the predefined classes.
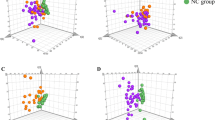
The principle components analysis (PCA) scores plot (PC1 versus PC2) of the negative ion data is shown in figure 1. Each point represents a single dog plasma sample. Control samples cluster in one area of the plot while samples exhibiting biochemical differences caused by disease states appear in regions away from the controls. The PCA was unable to discriminate fully between the different cohorts of plasma samples; there was considerable overlap of plasma samples from Group 2 (dogs with acquired hepatopathies) and Group 3 (“controls”), whilst the Group 1 dogs (congenital PVA) tended to cluster away from the other groups of dogs (figure 1). The results indicated that the major separation was achieved with PC1 and PC 2. No real improvement could be obtained in the separation by looking at PC3
PCA scores plot (PC1 versus PC2) with Pareto scaling of plasma metabolites generated by LC-MS in negative ion mode. Circles=group 1 (congenital PVA); triangles=group 2 (acquired hepatopathy); squares=group 3 (controls). Each point represents a single dog plasma sample. Samples exhibiting biochemical differences caused by the disease states are represented by points in regions away from the controls.
A trend plot was employed to interpret the data. In this plot the peak intensity for a particular analyte in the loadings plot is compared across the entire sample set, enabling those metabolites which are reduced or elevated in the disease state to be rapidly identified (table 3). PCA loading plots identified several metabolite ions whose levels appeared to be significantly disturbed (figure 2). The ions most responsible for the variance in the scores plot are indicated in the loading plot by their distance from the origin. The peaks are labelled according to their mass to charge ratio (m/z) and chromatographic retention time.
Two of the most significant metabolite ions, which increased in some of the dogs with congenital PVA were at m/z 498.2909 and 514.2858. Analysis by tandem mass spectrometry (MS/MS) generated ions at m/z 80, which corresponds to a sulfite (SO\(_3^{-}\)) moiety (Mills et al., 1998). These metabolites have been identified as [M-H]− ions of the conjugated bile acids, taurochenodeoxycholic acid (C26H45NO6S, [M-H]− theoretical =498.2890, error=1.9 mDa, 3.8 ppm) and taurocholic acid (C26H45NO7S, [M-H]− theoretical=514.2839, error=1.9 mDa, 3.7 ppm) respectively.
Other ions at m/z 540.3302, 564.3301 and 568.3621 were decreased in intensity. These are believed to correspond to the formate adducts [M+45]− of 16:0-lysophosphatidylcholine (C24H52NO7P, [M+HCOO]− theoretical=540.3301, error=0.1 mDa, 0.2 ppm) 18:2-lysophosphatidylcholine (C26H50NO7P, [M+HCOO]− theoretical=564.3301, error=0.6 mDa, 1.1 ppm) and 18:0-lysophosphatidylcholine (C26H54NO7P [M+HCOO]− theoretical=568.3614, error=0.7 mDa, 1.2 ppm) respectively (also see positive ion mode data). LC-MS/MS analysis gave rise to ions relating to the loss of the formate and [M-H]− ions of fatty acids from the lysophospholipids. The assignment of these ions is consistent with previous reports (Fang et al., 2003).
Having established the existence of clustering behaviour between the cohorts with PCA, more powerful, supervised statistical methods were then used to maximize the separation between the groups of plasma samples. To determine whether it was possible to distinguish congenital PVA from acquired hepatic disease and control groups, the LC-MS data was processed using partial least squares-discriminant analysis (PLS-DA). The PLS-DA scores plot is shown in figure 3.
PLS-DA revealed greater discrimination between the plasma samples from the control and congenital liver disease groups (figure 4). PLS-DA models were generated using the data from only 2 groups at a time e.g. controls and congenital PVA (figure 4). The models were then reconstructed excluding one sample from each group and the group membership of the excluded samples was then predicted. This process was repeated for each group with every sample being excluded and predicted. This resulted in 18 of the 25 samples being correctly predicted (probability p>0.10) with 6 being borderline to being borderline/correctly predicted (0.10>p>0.05) and the remaining sample number 10 being wrongly classified as a congenital PVA. Although encouraging these findings provide only a preliminary separation because of the small number of animals in the study. A more rigorous approach would be to use a training and validation data, and an independent test set to prove whether their PLS-DA model is valid or not.
Positive ion mode LC-MS analysis
The LC-MS data in positive ion mode using PCA was also unable to distinguish fully the different groups of disease and control dogs, although the congenital PVA dogs (Group 1), tended to cluster away from the acquired hepatopathies (Group 2) and controls (Group 3) as observed in negative ion mode (data not shown).
The supervised technique PLS-DA was again used to maximize the separation between the classes. As only limited numbers of samples were available, the robustness of the models were again tested by generating models with one sample from each class excluded and the model used to predict the class membership of the excluded samples. Figure 5 shows the PLS-DA scores plot (PC1 versus PC3) of group 1 (congenital PVA) and group 2 (acquired hepatopathy). Eleven of the 15 samples in groups 1 and 2 were correctly classified (probability p>0.10), 2 were borderline to being correctly predicted (0.10>p>0.05) and samples 3 and 10 were wrongly classified. Sample 10 was again misclassified as congenital PVA as was observed in negative ion mode.
The intensity of ions at m/z 496.3400 (16:0-lysophosphatidylcholine, C24H50NO7P, error=−5.0 mDa, −10.1 ppm) and 524.3700 (18:0-lysophosphatidylcholine, C26H54NO7P, error=−2–2 mDa, −4.2 ppm) was reduced. The identities have been confirmed by exact mass LC-MS/MS product ion analysis against authentic standards (table 3). On LC-MS/MS analysis, the lysophospholipids readily fragmented to produce an ion at m/z 184 (figure 6), which is attributable to the phosphocholine head group (Lehmann et al., 1997). In addition, ions at m/z 758.5676 (postulated as [M+H]+ of C42H80NO8P theoretical mass=758.5700, 786.5939 ([M+H]+ of C44H84NO8P theoretical mass=786.6013) and 810.5992 (postulated as the [M+H]+ of C46H84NO8P theoretical mass 810.6013, error=−2.1 mDa, −2.6 ppm), showed an increased intensity in the PVA samples. LC-MS/MS analysis indicated that these ions correspond to phosphatidylcholine-34:2, phosphatidylcholine-36:2 and phosphatidylcholine-38:4 respectively.
Although preliminary, our results suggest that this experimental approach might be able to distinguish different forms of liver disease in dogs. The numbers of dogs in each study group were small but, despite this, the metabolomic strategy could separate the three groups with ease. Metabolomic strategies have successfully differentiated groups of similar size in a recent experimental model of infections with Schistosoma infection (Wang et al., 2004). This would suggest that metabolomics will prove to be more powerful than conventional laboratory diagnostics, a supposition which is supported by the fact that other clinical parameters could not be used to separate the groups in the current study.
The PCA studies showed overlap of the dogs with acquired liver disease. These disorders may have different aetiologies and therefore comprise a phenotypically diverse group of animals. Nonetheless, using chemometric tools it was possible to distinguish fully this group of dogs. The three-dimensional PLS-DA with Pareto scaling of the negative ion data gave the best separation between classes. The loadings plot indicated that there was a significant increase in specific bile acids of dogs with PVA, as might have been expected because elevations in the concentrations of plasma bile acid commonly occur in a broad spectrum of general liver diseases and are routinely used as part of diagnostic investigations of PVA. However, no single biomarker for PVA was identified; rather a combination of metabolites seemed to be responsible, although this requires further investigation. This finding supports profiling of multiple low molecular weight metabolites to diagnose liver disorders (Mustaq et al., 1999). The identification of specific biochemical profiles, directly related to the disease states may also provide a greater insight into the pathogenesis of certain forms of hepatic dysfunction. These results demonstrate the ‘power’ of metabolomics in being able to detect subtle changes in the profile of other endogenous low molecular weight metabolites in body fluids and tissues.
One concern with the results of the current study is that the control population might not have been truly representative of normal individuals. The choice of appropriate control group for clinical studies is a contentious issue in both human and veterinary medicine. A group of normal individuals has the advantage that the true effect, of a particular disease can be seen. However, when attempting to develop a diagnostic test, normal animals are not necessarily the best control population because the clinician is attempting to differentiate a particular disease (e.g. hepatic disease) from a range of ‘differential diagnoses’ (e.g. alternative diseases which present with similar clinical signs). For canine hepatic disease, differential diagnoses include neurological or neuromuscular disorders (which produce signs similar to hepatic encephalopathy e.g. seizures), and gastrointestinal disorders (which can cause vomiting and diarrhoea). Therefore, for clinical studies the use of normal animals as controls is criticised and a ‘hospital control’ population is preferred, which would include patients from the same hospital population, ideally, with diseases that cause clinical signs mimicking the disease of interest. This latter strategy was chosen for the current study, and it is of note that five of 10 individuals in the control group had diseases which were potential differential diagnoses for liver disease (e.g. Campylobacter infection, idiopathic epilepsy, myasthenia gravis, leukoencephalopathy, eosinophilic enteritis). Nevertheless, the current study is small and preliminary, and further validation with a larger population is now required.
Concluding remarks
This preliminary investigation supports the concept of a global approach for characterizing the metabolite disturbances associated with liver disease. Further studies are still needed to analyze samples from larger cohorts of patients. In time, this should enable the technique not only to diagnose diseases in a similar manner and differentiate conditions with similar clinical presentations, but to provide prognostic information. This will allow the most appropriate treatment to be given at an early stage. This work suggests that metabolomic-based strategies may play an invaluable role in the future diagnosis and management of liver disease in humans and in veterinary species.
Conflicts of interest
The study was a collaboration between the University of Liverpool and Waters Corporation (Hilary Major), who manufactured the equipment used in the study.
References
Allen, J., Davey, H.M., Broadhurst, D., et al. (2003) High-throughput classification of yeast mutants for functional genomics using metabolic footprinting. Nat. Biotechnol. 21, 92–696
Brindle, J.T., Antti, H., Holmes, E., et al. (2002) Rapid and noninvasive diagnosis of the presence and severity of coronary heart disease using 1H-NMR-based metabolomics. Nat. Med. 8, 1439–1444
Center, S.A. (1996) Hepatic vascular diseases in Guilford, W.G., Center, S.A., Strombeck, D.R., Williams, D.A. and Meyer, D.J. (Eds), Strombeck’s Small Animal Gastroenterology, 3rd edn. WB Saunders, Philadelphia, pp. 802–846
Dunn, W.B., Bailey, N.J. and Johnson, H.E. (2005) Measuring the metabolome: current analytical technologies. Analyst 130, 606–625
Fang, N., Yu, S. and Badger, T.M. (2003) LC-MS/MS analysis of lysophospholipids associated with soy protein isolate. J. Agric. Food Chem. 51, 6676–6682
German, J.B., Roberts, M.A. and Watkins, S.M. (2003) Genomics and metabolomics as markers for the interaction of diet and health: lessons from lipids. J. Nutr. 133, 2078S–2083S
Griffin, J.L. (2003) Metabonomics: NMR spectroscopy and pattern recognition analysis of body fluids and tissues for characterisation of xenobiotic toxicity and disease diagnosis. Curr. Opin. Chem. Biol. 7, 648–654
Griffin, J.L. and Shockcor, J.P. (2004) Metabolic profiles of cancer cells. Nat. Rev. Cancer 7, 551–561
Lamers, R.J., DeGroot, J., Spies-Farber, E.J., et al. (2003) Identification of disease- and nutrient-related metabolic fingerprints in osteoarthritic guinea pigs. J. Nutr. 133, 1776–1780
Lehmann, W.D., Koester, M., Erben, G. and Keppler, D. (1997) Characterization and quantification of rat bile phosphatidylcholine by electrospray-tandem mass spectrometry. Anal. Biochem. 246, 102–110
Lenz, E.M., Bright, J., Knight, R., Wilson, I.D. and Major, H. (2004a). A metabonomic investigation of the biochemical effects of mercuric chloride in the rat using (1)H NMR and HPLC-TOF/MS: time dependant changes in the urinary profile of endogenous metabolites as a result of nephrotoxicity. Analyst 129, 535–541
Lenz, E.M., Bright, J., Knight, R., Wilson, I.D. and Major, H. (2004b) Cyclosporin A-induced changes in endogenous metabolites in rat urine: a metabonomic investigation using high field 1H NMR spectroscopy, HPLC-TOF/MS and chemometrics. J. Pharm. Biomed. Anal. 35, 599–608
Lindon, J.C., Nicholson, J.K., Holmes, E., et al. (2003). The role of metabonomics in toxicology and its evaluation by the COMET project. Toxicol. Appl. Pharmacol. 187, 137–146
Mills, K.A., Mustaq, I., Johnson, A.W., Whitfield, P.D. and Clayton, P.T. (1998) A method for the quantitation of conjugated bile acids in dried blood spots using electrospray ionization-tandem mass spectrometry. Pediatr. Res. 43, 361–368
Mustaq, I., Logan, S., Morris, M., et al. (1999) Screening of newborn infants for cholestatic hepatobilary disease with tandem mass spectrometry. Brit. Med. J. 319, 471–477
Nicholson, J.K., Connelly, J., Lindon, J.C. and Holmes, E. (2002). Metabonomics: a platform for studying drug toxicity and gene function. Nat. Rev. Drug Discov. 1, 153–161
Nicholsonm, J.K. and Wilson, I.D. (2003) Understanding ’global’ systems biology: metabonomics and the continuum of metabolism. Nat. Rev. Drug Discov. 2, 668–676
Plumb, R.S., Stumpf, C.L., Granger, J.H., Castro-Perez, J., Haselden, J.N. and Dear, G.J. (2003). Use of liquid chromatography/time-of-flight mass spectrometry and multivariate statistical analysis shows promise for the detection of drug metabolites in biological fluids. Rapid Commun. Mass Spectrom. 17, 2632–2638
Raamsdonk, L.M., Teusink, B., Broadhurst, D., et al. (2000). A functional genomics strategy that uses metabolome data to reveal the phenotype of silent mutations. Nat. Biotechnol. 19, 45–50
Roessner, U., Luedemann, A., Brust, D., et al. (2001) Metabolite profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell. 13, 11–29
Rosado, B. and Kamath, P.S. (2003). Transjugular intrahepatic portosystemic shunts: an update. Liver Transpl. 9, 207–217
Schafer, K., Ukida, M., Steffen, C., Bode, C., Zelder, O. and Bode, J.C. (1985) Effect of ammonia on plasma and cerebrospinal amino acids in dogs with and without portacaval anastomoses. Res. Exp. Med. 185, 35–44
Wang, Y., Holmes, E., Nicholson, J.K., et al. (2004). Metabonomic investigations in mice infected with Schistosoma mansoni: an approach for biomarker identification. Proc. Natl. Acad. Sci. USA 101, 12676–12681
Watanabe, A. (2000). Portal-systemic encephalopathy in non-cirrhotic patients: classification of clinical types, diagnosis and treatment. J. Gastroenterol. Hepatol. 15, 969–979
Waters, N.J., Holmes, E., Waterfield, C.J., Farrant, R.D. and Nicholson, J.K. (2002). NMR and pattern recognition studies on liver extracts and in livers from rats treated with alpha-naphthylisothiocyanate. Biochem. Pharmacol. 64, 67–77
Wilson, I.D., Plumb, R., Granger, J., Major, H., Williams, R. and Lenz, E.M. (2005). HPLC-MS-based methods for the study of metabonomics. J. Chromatogr. B 817, 67–76
Yang, J., Xu, G., Zheng, et al. (2004). Diagnosis of liver cancer using HPLC-based metabonomics avoiding false-positive result from hepatitisis and hepatocirrhosis diseases. J. Chromatogr. B 813, 59–65
Acknowledgments
We wish to thank the clinicians, support staff and laboratory technicians of the Small Animal Hospital, University of Liverpool for their assistance with the study. Alex German’s lectureship, at the University of Liverpool, is currently funded by Royal Canin.
Author information
Authors and Affiliations
Corresponding author
Additional information
This work was supported in part by grants from The Royal Society, Petplan Charitable Trust and The Waltham Foundation.
Rights and permissions
About this article
Cite this article
Whitfield, P.D., Noble, PJ.M., Major, H. et al. Metabolomics as a diagnostic tool for hepatology: validation in a naturally occurring canine model. Metabolomics 1, 215–225 (2005). https://doi.org/10.1007/s11306-005-0001-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11306-005-0001-3