Abstract
Understanding the genetic diversity and phylogenetic relationship of citron (Citrus medica L.) is of great importance for making conservation and utilization strategies. Southeast Asia and its vicinity are usually regarded as one of the centers of origin for Citrus. The principal aims of this work were to evaluate the genetic diversity of 56 accessions of citron and its relatives mainly from southwest China, to investigate the genetic structures of citron populations, and to construct a phylogenetic tree to understand the evolution of some hybrids among citrons and other citrus types. SSR analysis detected a total of 387 alleles ranging from 2 to 12 alleles per locus, and nearly all accessions identified could be unequivocally distinguished. The observed and expected heterozygosities averaged 0.36 and 0.49, respectively. From a Bayesian cluster analysis, citrons were characterized by two distinguished genetic structures, which corresponded to the geographical distribution in southwest China. Citron and fingered citron might derive from a common ancestor. Based on the chloroplast sequences, the phylogenetic trees were constructed with congruent topologies and similar levels of statistical support for relationships among citron and its relatives using both maximum parsimony and Bayesian inference methods. Citron formed a monophyletic clade, which was completely different from mandarin (C. reticulata) and pummelo (C. grandis). The genetic contributors of some hybrid species, such as C. limonia, C. aurantifolia, and C. limon, were also discussed, and citron appeared to be a primary contributor to the speciation of these secondary citrus species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The citron (Citrus medica L.), an ancient species of the genus Citrus, plays a crucial role in the economic and environmental context in mountainous areas. Citron is native to northeastern India, Burma, and southwest China where it is found in the valleys at the foot of Himalaya Mountain and its adjacent zone (Lim 2012). Before and after the third century BC, citron was the first citrus fruit introduced to Mediterranean basin and remained the only representative of citrus there until the seventh century. Not only had its medical value attracted the attention of Theophrastus and Roman Naturalists, but also, the citron is still used for worship by Jews during the feast of Tabernacles, which is supported by ample archeological as well as literature records (Reuther et al. 1968; Nicolosi et al. 2005).
In China, the citron is extensively distributed across a large area from tropical to sub-tropical regions, such as Yunnan, Guizhou, Sichuan, Guangxi, and Tibet Autonomous Regions. It is widespread in Yunnan and can be found on both sides of the “Tanaka Line” (Gmitter and Hu 1990). Both cultivated and wild types of citron exist there, such as “Gou cheng” citron, “Muli” citron, “Xiangyangguo” citron, fragrant citron, sour citron, sweet citron, fingered citron, and so on, which display a wide variety of genetic characteristics. Since ancient times, local people have made full use of C. medica as ornamental plants and later as rootstocks of citrus. They have started to trade the fruits for the pleasant fragrant rinds or uniquely flavored juice in recent years. Just like other citrus species, the fruits of citron and its relatives are regarded as a kind of hesperidium varied in size with a different quantity of albedo. Typically, the surface is not only bumpy but also somewhat ribbed, with medium-sized oil bubbles scattered randomly and emitting charming fragrance. Most citron varieties usually lack juice or just contain somewhat dry juice vesicles and impart an acidic flavor instead of a sweet one. Citrons are mono-embryonic, a characteristic that could cause abundant genetic diversity through long-term evolution.
The success of breeding program is based on the knowledge and availability of genetic variability for efficient selection. Morphological and agronomic characteristics of citrus species are greatly subject to environmental modifications and cannot always unambiguously distinguish or correctly cluster closely related taxa. It is difficult to reach an agreement on the species status, and the apparent hybrids among naturally occurring types of citron and its relatives are also confusing. Based on the results from Scora (1975) and Barrett and Rhodes (1976), citron along with mandarin (C. reticulata) and pummelo (C. grandis) constitute the three ancestral species within the genus Citrus. In addition, other genotypes are derived from hybridization between these three basic species, which has gained further support from various biochemical and molecular studies (Nicolosi et al. 2000; Barkley et al. 2006; Garcia-Lor et al. 2013; Liu et al. 2013). However, Citrus phylogeny and taxonomy are still complicated and obscure, mainly due to a wide range of factors including the large degree of morphological diversity, sexual compatibility between the species, apomixis in many genotypes, high frequency of bud mutations, long history of cultivation, and wide dispersion (Nicolosi et al. 2000; Garcia-Lor et al. 2013). Some taxonomists hold the view that C. medica has made a contribution to the development of several important genotypes. Despite the fact that lemon (C. limon) is accepted as a species by the Swingle and Tanaka systems (Tanaka 1961; Swingle and Reece 1967), there is a prevailing hypothesis that lemon is of a hybrid origin, with sour orange (C. aurantium) as the maternal parent and citron as the paternal parent (Nicolosi et al. 2000; Garcia-Lor et al. 2013). Along with C. micrantha or C. hystrix, C. medica contributed to the development of C. aurantifolia (Ollitrault et al. 2012a). C. limonia is reported to be closely related with citron in the previous studies, and it could be a result of a cross between citron and rough lemon (C. limon) or mandarin (Nicolosi et al. 2000; Li et al. 2010). Although citron is a typical member of the genus Citrus, the genetic diversity, population structure, and the phylogenetic relationships of itself and its relatives have not been extensively studied so far.
Various molecular marker systems are used to analyze the genetic diversity and phylogenetic relationship of Citrus, such as RFLP, RAPD, AFLP, SSR, indel, SNP, and chloroplast DNA sequence (Federici et al. 1998; Barkley et al. 2006; Pang et al. 2006; Morton 2009; Li et al. 2010; Garcia-Lor et al. 2012, 2013). Among these, SSR markers have been very successfully utilized for genotyping and genetic diversity studies of Citrus (Barkley et al. 2006; Biswas et al. 2012; Garcia-Lor et al. 2012, 2013; Snoussi et al. 2012; Liang et al. 2015), due to its desirable genetic attributes, including high polymorphism, codominant inheritance, and chromosome-specific location. There is a relatively high level of genetic diversity in citron as evidenced by SSR markers studies (Luro et al. 2012; Ramadugu et al. 2015). In the majority of angiosperms, chloroplast DNA (cpDNA) is characterized by its evolutionary conservatism, matrilineal inheritance, and lack of recombination (Wolfe et al. 1987). Thus, it can be used to elucidate phylogenetic relationship at low taxonomic levels and identify the potential hybrid lineages. Chloroplast DNA has been successfully used to study the molecular phylogeny and the taxonomy of Aurantioideae subfamily and hybrid identification. After a study on the sequences of the chloroplast DNA from two regions (trnL-trnF and trnT-trnL), de Araujo et al. (2003) conclude that C. medica is distinct from other citrus species because of its position relative to other non-citrus genera. In studies including more genera and several chloroplast genes, the Aurantioideae is identified to be monophyletic within Rutaceae, and the two tribes, Citreae and Clauseneae, are not monophyletic (Morton et al. 2003; Bayer et al. 2009; Morton 2009; Penjor et al. 2010, 2013). More importantly, this conclusion has partly shaken the traditional view about the classification of the tribes and subtribes of Aurantioideae widely used by Swingle and Reece (1967). All these controversies are triggered by the complexity of the phylogenetic relationship of Citrus.
Specifically, this work was aimed at assessing the genetic diversity and population structure of 56 accessions of citron and its relatives mainly from southwest China by using a substantial number of nSSR markers. Meanwhile, based on the combined analysis of coding and non-coding chloroplast regions, we established robust phylogenetic relationships between the members of this group, laid a foundation for clarifying the origin of some secondary species, such as C. limonia, C. aurantifolia, and C. limon, and ascertained the identity of their putative parents.
Materials and methods
Plant materials
A total of 74 genotypes were used for the diversity analysis with nSSR markers, and 39 of the genotypes were selected for the phylogenetic analysis with coding and non-coding chloroplast sequence. A total of 56 cultivated, wild, and dooryard accessions were surveyed and sampled from Yunnan, Tibet, and surrounding areas in southwest China, including part of the range of natural distribution of citron and its relatives (Fig. 1). The typical flowers and fruits of a few accessions are also depicted (Fig. 2). Others were collected from the Institute of Citriculture of Huazhong Agriculture University (HZAU, Wuhan, China) and Citrus Research Institute of Chinese Academy of Agriculture Sciences (CRI, Chongqing, China). Almost all of them were chosen from the genus Citrus and Murraya paniculata (L.) Jack was selected as an outgroup. Details of sample codes, origin, and further information were summarized in Supplementary Table S1.
DNA extraction and microsatellite amplification
Total genomic DNA was extracted from leaf samples as previously described (Cheng et al. 2003). All accessions were genotyped with 77 selected nSSR markers obtained from different research groups containing genomic-SSR and EST-SSR markers with clear and repeated amplification patterns, randomly distributed in the genome (Kijas et al. 1997; Barkley et al. 2006; Chen et al. 2006; Froelicher et al. 2008; Luro et al. 2008; Ollitrault et al. 2010; Xu et al. 2013). More information on each pair of primers and annealing temperature was listed in the Supplementary Table S2. The SSR amplification reactions were conducted according to the protocol described by Chai et al. (2013). PCR products were separated by polyacrylamide gel electrophoresis and visualized by silver staining following the protocol developed by Ruiz et al. (2000).
Amplification and sequencing of chloroplast DNA
Six chloroplast regions, five non-coding regions (trnS-trnG, rps16, rpl16, atpB-rbcL, accD-psaI), and one coding region (matK) were amplified by PCR, and the primer pairs for each region were designed based on the chloroplast genome of sweet orange (Bausher et al. 2006), and related information was presented in the Supplementary Table S3. Amplification of all PCR products was performed in a total volume of 20 μl containing 50 ng of genomic DNA, 0.4 mM dNTPs, 0.4 U NovoStar FastPfu DNA Polymerase (NovoGene), a corresponding 2× reaction buffer and 0.4 μM of each primer pair. Amplification was carried out as follows: 5 min at 94 °C, followed by 32 cycles of 30 s at 94 °C, 30 s at 55–60 °C, 45–60 s at 72 °C (depending on the annealing temperature of each primer and length of the amplified regions; Supplementary Table S3) and a final extension at 72 °C for 10 min in a MJ-PTC_200 thermal controller (MJ Research, Waltham Mass). Products were first separated by 1 % agarose gel stained with ethidium bromide. The expected size bands were excised and gel-purified with a DNA Purification kit (Dingguo, Beijing, China). Both strands of the resulting products were sequenced directly by using an ABI 3700XL DNA Analyzer (Applied Biosystem, Foster City, USA). In the case of matK, primers matKiF and matKiR were required for internal sequencing.
Genetic diversity assessment, population structure and principal coordinates analysis for nSSR data set
The polymorphism bands were recorded as 1 (present) or 0 (absent) for the same amplified fragments. Data format could be changed according to the requirement of different analysis softwares. The resulting binary matrix was converted to the required data input format according to the instructions for NTSYS-pc version 2.1 (Rohlf 2000). A similarity matrix was constructed using nSSR data, based on the Dice’s coefficient (Nei and Li 1979). From the similarity matrix of nSSR data set, a dendrogram was constructed using the UPGMA clustering method. The robustness of the phylogenetic tree was evaluated by bootstrap analysis with 1000 replicates using the bootstrap function of the FreeTree program (Hampl et al. 2001). The level of genetic diversity was estimated using POPGENE version 1.32 (Yeh et al. 1997) with the following statistics: number of alleles per locus (Na), effective number of alleles (Ne), observed heterozygosity (Ho), and expected heterozygosity (He). Polymorphism information content (PIC) was calculated using Power Marker version 3.25 (Liu and Muse 2005).
The model-based software program STRUCTURE version 2.3 (Pritchard et al. 2000) was used to infer population structure by a Bayesian approach using nSSR data set. The optimal value of K (the number of clusters) was deduced by evaluating K = 1–10, under the admixture model with correlated allele frequencies. For each K value, at least ten independent runs of 100,000 iterations were processed following a burn-in period of 10,000 iterations. The optimal K value was identified as previously described (Evanno et al. 2005). To confirm the results obtained from UPGMA and population structure analysis, we also carried out an additional analysis for visualizing the distribution of individual genotypes. The data from nSSR markers was also used to obtain the pairwise genetic distance matrix, which was standardized and used to perform the principal coordinate analysis (PCoA) using GenALEx version 6.5 (Peakall and Smouse 2012).
Chloroplast coding and non-coding sequences alignment and phylogenetic analyses
All the nucleotide sequences were imported into ClustalX 2.0 (Larkin et al. 2007), and alignments were adjusted manually. Gaps were inserted to maintain sequence homology. Indels were aligned and scored as previously reported (Simmons and Ochoterena 2000). The sequences from all six chloroplast DNA regions were analyzed separately and then combined into a single alignment matrix containing all data for a final analysis. Chloroplast DNA datasets were analyzed using maximum parsimony and Bayesian inference methods. For the maximum parsimony analysis, all concatenated data sets were implemented in PAUP version 4.0b10 (Swofford 2002). All characters were weighted equally and gaps treated as missing data. A heuristic search was implemented employing 1000 random addition sequence replicates with tree bisection and reconnection branch swapping. The strict consensus tree was then calculated from the most parsimonious trees. Bootstrapping was performed with 1000 replicates to establish support for relationships inferred.
Bayesian inference of phylogeny was performed in MrBayes version 3.1.2 (Huelsenbeck and Ronquist 2001; Ronquist and Huelsenbeck 2003) with the most suitable choice of nucleotide-substitution models determined by the corrected Akaike Information Criterion (AIC) calculated using ModelTest version 3.7 (Posada and Buckley 2004). Subsequently, the commands “lset NST = 6, RATES = gamma” and “lset coding = variable” were entered in MrBayes version 3.1.2 for the nucleotide and gap characters, respectively. Two independent runs of Metropolis-coupled Markov Chain Monte Carlo (MCMC) were performed simultaneously, with each run consisting of one cold chain and three incrementally heated chains and all started randomly in the parameters space. One million generations were run, and every 100 generations were sampled with the first 25 % of samples discarded as burn-in. Tracer version 1.4 was used to check further whether the chains converged (Rambaut and Drummond 2007). Finally, the tree was visualized using Treeview Version 1.6.6 (Page 2001).
Results
nSSR polymorphism and genetic diversity
The diversity pattern of the 56 accessions for the 77 SSR loci revealed a highly variable number of alleles (here, we only included C. medica, C. medica var. sarcodactylis, C. limon, C. limonia, and C. aurantifolia). Almost all the loci evaluated in this study were multi-allelic. The frequencies of alleles obtained in different loci and the relevant parameters of variability analyzed were presented in Table 1. A total of 387 alleles were detected, with an average of 5 alleles per locus and the number of alleles ranging from 2 to 12. The highest number of polymorphic alleles was detected at locus TAA41 with a total of 12 alleles. The effective number of alleles per locus (2.26), which minimized the input of alleles with low frequencies, was less than half of the mean of total alleles. Across the accessions, Ho and He varied from 0.02 to 0.96 and 0.12 to 0.86, averaging 0.36 and 0.49, respectively. The mean Ho was slightly lower than the mean He at most loci. Consequently, the fixation index (F) values of most of loci were positive, with an average of 0.25. The heterozygosity observed in citron and finger citron accessions ranged from 0.09 to 0.34, and the mean value was 0.23 (here, we excluded two putative hybrids, CIT-X and CIT-AF; Supplementary Table S4). As a measurement of the genetic diversity of the species, the PIC value for the SSR loci ranged from 0.12 to 0.83, with an average of 0.45.
Phylogenetic relationships of citrons and its relatives based on nSSR
Based on the nSSR data set, a similarity matrix was calculated according to Dice’s coefficient. A total of 74 accessions (here we included all the taxa) were clustered using UPGMA method (Fig. 3). The similarity values among the studied genotypes ranged from 0.16 to 1.00. Individual genotype matching (pairwise comparisons) based on multi-locus SSR profiles did not detect match pairs, except for some of C. limon and C. sinensis. The dendrogram clearly revealed that almost all of the studied accessions were of a different genotype and their relationships were organized into seven distinct clusters. The largest identified group, cluster I, included 36 members of C. medica and C. medica var. sarcodactylis with a similarity value of 0.72. Meanwhile, cluster II, C. aurantifolia and C. hystrix, were grouped together, showing a close relationship with C. medica. Cluster III consisted of three species, including C. aurantium, C. sinensis, as well as cultivated C. reticulata. Cluster IV was reportedly formed by a number of accessions with a hybrid origin, such as C. aurantifolia “Bergamot”, C. limon, and C. limonia. All of the members from Cluster III and Cluster IV showed a slightly distant phylogenetic relationship with wild mandarins which were grouped alone in Cluster V. All the C. grandis formed an independent cluster VI, but two kinds of hybrid citron also fell into this group. In cluster VII, there was only one C. hongheensis that was grouped very closely to C. grandis. M. paniculata (L.) jack as an out group was not grouped with any accession and finally placed in a single distinct cluster VIII.
UPGMA dendrogram of all the analyzed genotypes, including 56 accessions of citron and its relatives, mandarin, pummelo, several other hybrid species and one outgroup, Murraya paniculata (L.) Jack based on 77 nSSR markers. Numbers at the nodes are bootstrap percentages out of 5000. Only values of ≥50 are indicated. The abbreviated accession names of corresponding samples are listed in the brackets. Clusters defined as I, II, III, IV, V, VI, VII, and VIII represent different groups
The genetic divergence between citron and its relatives based on Nei’s genetic distance (Nei and Li 1979) was used to generate the PCoA scatter plot. There were seven groups obviously displayed by the PCoA plot in Fig. 4. PCoA was very applicable in describing the organization of genetic diversity especially when potential hybrids were included in different groups. The distant groups on the positive X- and Y-axis were formed by individuals of all the citron and fingered citron accessions across the entire range, and they were strongly differentiated from C. reticulata and C. grandis. However, C. grandis was differentiated from C. reticulata by the second axis. Two hybrid citrons and C. hongheensis showed a close relationship with C. grandis. C. aurantium and C. sinensis were clearly positioned between C. reticulata and C. grandis. Nearly all members of C. limonia clustered together with C. reticulata instead of C. grandis. Accessions of C. aurantifolia and C. hystrix were grouped together and formed one distinct cluster.
Principal coordinates analysis of all the analyzed genotypes, including 56 accessions of citron and its relatives, mandarin, pummelo, papeda, and several other hybrid species based on genetic distance estimates. Clustered groups are represented by different icons. The plane of the first three PCoA axes accounts for 47.12 % of the total variation (first axis = 31.79 %, second = 9.75 %, and third = 5.57 %)
Genetic structure of citron population
Genotype data from 77 SSR markers was used to determine population structure among 36 citron and fingered citron accessions (here, we excluded two putative hybrids, CIT-X and CIT-AF). A Bayesian clustering approach was used to make a probabilistic assignment of individuals based on genotypes. The Evanno’s test indicated that a sharp signal was found at K = 2, implying that two gene pools shaped the genetic structure of the population analyzed (Supplementary Fig. 1). The final proportion of each of the two hypothetical gene pools present in each accession was obtained, and the results were shown in Supplementary Fig. 2. The assignment of a cultivar to a specific gene pool was provided by a membership probability of q i (the mean proportion of ancestry). Genotypes with a membership probability lower than 70 % were considered to belong to more than one gene pool. Thirty-four genotypes (94.44 %) showed a strong component derived from one specific gene pool. Only two genotypes (5.56 %) were considered ambiguous. Analysis of the geographic origin of each individual revealed that the red gene pool mainly included fingered citrons and wild citrons collected from Tibet, while the green gene pool included most of the other citrons presumed to be from Yunnan.
Alignments, phylogenetic analysis based on the coding and non-coding sequences
Sequences of the six chloroplast loci were generated for all the 39 investigated accessions (Supplementary Table S1). The final alignments consisted of 1573 aligned positions of the matK gene, 674 of the trnS-trnG spacer, 772 of the rps16 intron, 920 of the rpl16 intron, 793 of the atpB-rbcL spacer, and 839 of the accD-psaI spacer. Altogether, the concatenated sequence matrix contained 5571 characters, of which 228 (4.09 %) were polymorphic sites, and 76 (1.36 %) were parsimony informative. A total of 128 phylogenetically informative indels were coded as present/absent characters and added to the sequence matrix in the maximum parsimony and Bayesian analyses.
Tree topologies resulting from separate analysis of the six individual loci slightly differed from each other, but no well-supported conflicting nodes were observed. All trees resulting from the combined data sets generally showed higher resolution than any of the single-locus trees and displayed similar topologies. Therefore, all data sets were combined and subjected to maximum parsimony and Bayesian analyses. The combined parsimony analysis recovered a single most parsimonious tree with a tree length (L) = 292. Consistency and retention indices were high, indicating a low level of homoplasy within the dataset (CI = 0.863, RI = 0.939). The strict consensus maximum parsimony tree was shown in Fig. 5, with a robust statistical support for many internal nodes given above the branches, suggesting an unambiguous relationship among these accessions. Bayesian inference from the combined data set generated a topology identical to the strict consensus parsimony tree with slightly better clade support. Posterior probabilities for the Bayesian analysis were calculated from all post-burn-in generations and presented in Supplementary Fig. 3.
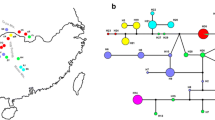
Strict consensus of the single most parsimonious tree recovered by PAUP* 4.0b10 from the alignment of the matk, trnS-trnG, rps16, rpl16, atpB-rbcL, and accD-psaI chloroplast DNA regions from 39 Citrus taxa. Tree statistics are tree length (L) = 292, consistency index (CI) = 0.863, and retention index (RI) = 0.939. Numbers above the branches are bootstrap values derived from 1000 heuristic replicates. Clades defined as I, II, III, and IV represent different groups
With successive branching of several clades that received moderate to high bootstrap support, a grade was formed within the ingroup, which not only included bootstrap (BS) in the maximum parsimony trees but also posterior probability (PP) in the Bayesian analysis. The first branch, clade I, comprised all investigated C. medica, and C. medica var. sarcodactylis and the sister relationship between them was rather strongly supported (BS 100; PP 99). The second branch splitted into a dichotomy with C. limon “Rough lemon”, C. reticulata, and C. limonia, which formed clade II (BS 100; PP 100). Within clade II, several taxa including C. limonia “Yellow limonia”, C. limonia “Rangpur”, C. limonia “Guangxi local lemon”, C. limonia “Red limonia”, and C. limon “Rough lemon” formed a subclade and clustered with different accessions of mandarin. Clade III comprised C. grandis, C. aurantium, C. sinensis, C. hongheensis, C. limon, C. aurantifolia “Bergamot” and one hybrid citron C. medica “Xiangyangguo” (BS 85; PP 100). Within clade III, the hybrid “Xiangyangguo” citron was branched first, followed by C. hongheensis and C. aurantium “Goutou” in the Maximum parsimony tree. On one hand, lemon from Mediterranean such as C. limon “Eureka”, C. limon “Villafranca”, C. aurantifolia “Bergamot”, C. limon “Femminello” had an affinity with C. aurantium “Daidai”, a species that originated from southwest China. The relationship between cultivated lemon and C. aurantium “Daidai” was moderately supported in the maximum parsimony analysis (BS 77) but received a high posteriori probability in the Bayesian analysis (PP 100). On the other hand, the ancient and wild lemon from China, C. limon “Muli” and C. limon “Meyer”, were grouped with sweet orange (BS 83; PP 99). Although C. aurantium “Goutou” was grouped with Chinese ancient lemon in Bayesian tree, the posteriori probability was relatively low (PP 51). The two accessions, C. aurantifolia “Mexican” and C. hystrix formed a subclade (BS 100; PP 100), which acted as a sister to the remainder of clade III. All of the aforementioned species in clade III were also clustered with C. grandis. M. paniculata (L.) Jack as a distinct species was separated from all the accessions used.
Discussion
Fruit improvement program usually starts with identification of genetic variation among the genotypes which is the first step towards rational conservation and efficient use of natural resources. The user-friendly nature of SSR marker is successfully exploited in some fruit species, such as chestnut (Castanea sativa Mill.), Pyrus and guava (Psidium guajava L.) for better understanding of the genetic diversity, geographic divergence, and distribution (Sitther et al. 2014; Quintana et al. 2015; Rana et al. 2015). Previous studies on the genetic diversity of citron and its relatives by SSR markers are mainly focused on citron and its relatives in Europe such as the Mediterranean region (Barkley et al. 2006; Luro et al. 2012). In the present study, the citron and its relatives studied encompassed many precious cultivated, dooryard, and wild genotypes, which represented a significant portion of genetic variation in southwest China. Furthermore, most of the accessions used in our investigation came from Yunnan and Tibet, which were supposed to be the places of origin of Citrus (Gmitter and Hu 1990).
A number of useful SSR markers, scored for over 387 alleles, provided a fairly broad coverage of the genome of citron and its relatives. The average PIC value of the 77 nSSR markers was 0.54, indicating that all the loci selected were highly informative and suitable for distinguishing ambiguous citrons and their relatives. An average of five alleles per locus and the expected mean heterozygosity of 0.49 illustrated that SSR markers could provide unique molecular profiles for individual genotypes. The average F (0.40) indicated the significant relative genetic differentiation among the citron accessions studied. For most loci, the high level of polymorphism indicated an excess of alleles in the homozygous state that could be expected for advanced cultivars and breeding genotypes. These useful markers also provided us with abundant informative SSR loci for further research and reduced the number of loci necessary to characterize a citron collection.
The high level of genetic differentiation detected among citron accessions could be attributed to either the different geographical regional adaptation of accessions, greatest diversity of wide species tested, or long evolutionary history. Another key factor for the genetic diversity level was the difference of species in the reproductive mode. A few studies demonstrated that mandarin and pummelo usually harbor high levels of genetic diversity, due to the production of citrus cultivars through sexual hybridization (Barkley et al. 2006). Conversely, citron is believed to be a purer species compared with the other two, since it tends to be fertilized by self-pollination. This hypothesis was supported, at least to a degree, by the result from the analysis through nSSR markers that the mean value of the observed heterozygosity of citron is 0.23, which is lower than that of pummelo (0.33) or mandarin (0.39). According to the opinion from Ramadugu et al. (2015), the true citrons also had a low level of heterozygosity. Citron is generally considered to be a male rather than a female parent of some hybrid citrus species (Luro et al. 2012). The inclination to pollinate itself instead of adopting the mode of reproduction of cross-pollination is the most probable reason for the relatively low genetic diversity of C. medica compared to C. reticulata and C. grandis.
The results of both PCoA and UPGMA clustering were partially congruent and revealed a clear delimitation among all the analyzed accessions. The results showed three main clusters within Citrus, but mandarin and pummelo shared each clade and both of them were separated from citron, which is consistent with previous investigations using various markers, such as SSR, SNP, indel, chloroplast sequence (Barkley et al. 2006; Bayer et al. 2009; Garcia-Lor et al. 2012, 2013; Ollitrault et al. 2012b). The structure analysis, with a K value to 2, facilitated our understanding of the genetic relationships between citron populations at the micro-scale level. Citrons from Yunnan and Tibet were separated completely from each other. It was noteworthy that, in cluster I, fingered citron was clustered closely with citron originating from Tibet rather than Yunnan. After a deep analysis of cpDNA sequences, fingered citron was just closely related with citron, which coincided with the results of nSSR data analysis. Yunnan and Tibet are in southwest China and are known as “the Kingdom of Plants” in China. The number of native species of higher plants in these two areas accounts for more than half of the number of plant species native to China (Long et al. 2003). Plants of the Rutaceae are abundant there, such as wild Honghe papeda (C. hongheensis), Ichang papeda (C. ichangensis), and some other treasure troves of Citrus (Chen et al. 2012; Li et al. 2015). In the present study, the close genetic relationship observed among several fingered citron species with a specific geographic origin supported the hypothesis that fingered citron was not only a variety of citron but might have evolved with citron from the same common ancestor.
In a dry-hot valley region, wild and semi-wild citrus species are widely scattered in southwest China and “Xiangyangguo” citron is one member of these species. Local people traditionally recognized it as a variety of citron, because of its development from endogenous carpel to fruit, which produced an intermediate appearance between citron and fingered citron. Although molecular analysis showed its similarity with citron, the chloroplast gene analyses indicated that its chloroplast might come from pummelo. It phylogenetically clustered in clade III based on both maximum parsimony and Bayesian analyses. Based on these results, we could rule out the possibility that it is a variety of citron and accept the probability that it is just a hybrid progeny. Consequently, there are numerous citron species with a complex evolutionary mechanism in southwest China, which is consistent with the prevailing opinion that southwest China is one of the important centers of origin for citron.
It is particularly evident that most of species of Citrus have been derived from hybridization. Combining the nuclear SSR data with chloroplast sequence information, we could assess the probable parentage of these species initially. Nearly all accessions of the same species formed a clade with high branch support values in both maximum parsimony and Bayesian analyses. A significant observation of the maximum parsimony trees constructed from chloroplast gene sequences was that all the mandarin, C. limonia “Rangpur”, C. limon “Rough lemon”, C. limonia “Guangxi local lemon”, C. limonia “Red lemon”, and C. limonia “Yellow lemon” formed a monophyletic group, suggesting that mandarin was most likely a maternal parent of these hybrids. However, there was no direct relationship between C. limonia and C. medica at first glance of the nSSR data sets. One of the possible reasons was that none of the nSSR markers selected was mined from citron, which caused a bias effect. Another conceivable reason might be that C. limonia was not a direct interspecific hybrid species but underwent a backcross with C. reticulata in the history of speciation. Even based on the current nSSR data sets containing many different accessions, C. limonia was grouped with C. reticulata, which coincided with the result from chloroplast gene analyses. Our results also matched the observation from the analysis of nSSR markers, and chloroplast data sets, especially about the hybrid origin of C. limonia “Rangpur lime”, an emblematic species in Mediterranean (Nicolosi et al. 2000; Gulsen and Roose 2001; Barkley et al. 2006; Morton 2009). There was a great possibility that both C. reticulata and C. medica participated in the formation of C. limonia, with the former as a female parent and the later as male parent.
Several hypotheses have been proposed for the origin of C. aurantifolia “Mexican”. According to Barrett and Rhodes (1976), it probably arose from a trihybrid cross involving C. medica, C. grandis, and Microcitrus species, whereas Nicolosi et al. (2000) inferred that it would be a direct interspecific cross between C. medica and C. micrantha, which gained support from Ollitrault et al. (2012a, b) and Garcia-Lor et al. (2012, 2013), and they also proposed that either C. micrantha or C. hystrix might be another potential female parent. In the present study, C. aurantifolia “Mexican” was clustered with the accession C. hystrix, rather than C. hongheensis, which was supported by both maximum parsimony and Bayesian analyses. It was thus speculated that not all the papeda species, but a few, such as C. hystrix, were mostly likely a maternal parent of C. aurantifolia “Mexican”. Moreover, the nSSR data sets indicated that C. aurantifolia “Mexican” had a close relationship with C. hystrix and both of them had a close affiliation with C. medica.
Lemon (C. limon) is conventionally accepted as a species by the two most widely cited taxonomic systems (Tanaka 1961; Swingle and Reece 1967). In the initial phase, Nicolosi et al. (2000) were one of the first to propose that lemon arose from hybridization between C. aurantium and C. medica by molecular markers. From then on, more and more studies have suggested that lemon is likely to be of a hybrid origin, with sour orange being the maternal parent and citron being the paternal parent (Gulsen and Roose 2001; Ollitrault et al. 2012a, b; Garcia-Lor et al. 2012, 2013). It was obvious that “Eureka” lemon, “Lisbon” lemon, “Femminello” lemon, and “Villafranca” lemon were highly heterozygous and were very similar to each other. We could not distinguish these taxa through 77 nSSR markers, which shared a common outlook that they originated from a single clonal parent via a series of mutations (Gulsen and Roose 2001). All the lemon species from Mediterranean were clustered with C. aurantium “Daidai” within clade III based on maximum parsimony and Bayesian analyses. This result clearly demonstrated that sour orange was probably a maternal parent of lemon. The nSSR clustering analysis indicated that all accessions of C. limon were grouped with C. limonia and appeared to be closely related with C. reticulata. Furthermore, the results of PCoA showed that their position was between the C. aurantium and C. medica group in each factorial axis. These results suggested that citron was most likely a parent of lemon. The ancient and wild C. limon “Meyer” and C. limon “Muli”, which are native to China, were identified by nSSR data sets and chloroplast gene analyses to have a similar genetic background as the typical lemons from the Mediterranean, such as “Eureka”, “Lisbon”, “Femminello”, and “Villafranca”. Interestingly, lemon that originated in the East (China) appeared to inherit its chloroplast genome from sweet orange. However, lemon that originated in the West (Mediterranean) probably obtained the chloroplast genome from sour orange. Although mandarin and pummelo participate in the origin of sour orange and sweet orange, they are different from each other (Nicolosi et al. 2000; Li et al. 2010; Uzun et al. 2010; Xu et al. 2013; Wu et al. 2014). The lemon accessions from different geographic locations varied in matrilineal inheritance patterns. Obviously, if more wild types were included in the analysis, and the whole genome sequence information of lemon, citron, sour orange, and sweet orange were analyzed, the precise contours of the origin of lemon would be clear. To some extent, our data provided further evidence to support the hybrid origin of lemon.
Conclusions
This work provides detailed information on genetic variation of citron and its relatives in southwest China. Some of the well-described accessions will provide a solid base for future selection and breeding. The distribution of genetic variation verified the existence of two diversified regions for citron, which could be attributed to the specific adaptation to different regional environments or experience of a different evolutionary mechanism. By incorporating well-characterized hybrid species assumed to belong to this group, we also provide a phylogenetic evolution pattern of citron and its relatives to uncover some of their potential parents. This result supported that C. reticulata was probably a maternal parent of C. limonia and C. medica was a paternal parent. C. hystrix, one kind of papeda, was more likely to be the female parent of C. aurantifolia, and C. medica was the male parent. C. medica and C. aurantium or C. sinensis might be the parents of C. limon, with the former acting as male parent and the latter as female one. Coincidentally, all of these three typical hybrid citrus species happened to inherit the pedigree of C. medica.
References
Barkley NA, Roose ML, Krueger RR, Federici CT (2006) Assessing genetic diversity and population structure in a citrus germplasm collection utilizing simple sequence repeat markers (SSRs). Theor Appl Genet 112:1519–1531
Barrett H, Rhodes A (1976) A numerical taxonomic study of affinity relationships in cultivated Citrus and its close relatives. Syst Bot 1:105–136
Bausher MG, Singh ND, Lee S-B, Jansen RK, Daniell H (2006) The complete chloroplast genome sequence of Citrus sinensis (L.) Osbeck var ‘Ridge Pineapple’: organization and phylogenetic relationships to other angiosperms. BMC Plant Biol 6:21
Bayer RJ, Mabberley DJ, Morton C, Miller CH, Sharma IK, Pfeil BE, Rich S, Hitchcock R, Sykes S (2009) A molecular phylogeny of the orange subfamily (Rutaceae: Aurantioideae) using nine cpDNA sequences. Am J Bot 96:668–685
Biswas MK, Chai LJ, Mayer C, Xu Q, Guo WW, Deng XX (2012) Exploiting BAC-end sequences for the mining, characterization and utility of new short sequences repeat (SSR) markers in citrus. Mol Biol Rep 39:5373–5386
Chai LJ, Biswas MK, Yi HL, Guo WW, Deng XX (2013) Transferability, polymorphism and effectiveness for genetic mapping of the pummelo (Citrus grandis Osbeck) EST-SSR markers. Sci Hortic 155:85–91
Chen CX, Zhou P, Choi YA, Huang S, Gmitter FG Jr (2006) Mining and characterizing microsatellites from citrus ESTs. Theor Appl Genet 112:1248–1257
Chen HM, Jiang D, Hu ZR, Li KM, He YR, Chen SC, Li LH, Zheng DS, Liu X (2012) A newly discovered wild Ichang papeda population in Yuanjiang county, Yunnan province. J Plant Genet Res 13:929–935 (in Chinese with Engnish abstract)
Cheng YJ, Guo WW, Yi HL, Pang XM, Deng XX (2003) An efficient protocol for genomic DNA extraction from citrus species. Plant Mol Biol Report 21:177–178
de Araujo EF, de Queiroz LP, Machado MA (2003) What is citrus? Taxonomic implications from a study of cp-DNA evolution in the tribe Citreae (Rutaceae subfamily Aurantioideae). Org Divers Evol 3:55–62
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Federici C, Fang D, Scora R, Roose M (1998) Phylogenetic relationships within the genus Citrus (Rutaceae) and related genera as revealed by RFLP and RAPD analysis. Theor Appl Genet 96:812–822
Froelicher Y, Dambier D, Bassene JB, Costantino G, Lotfy S, Didout C, Beaumont V, Brottier P, Risterucci AM, Luro F, Ollitrault P (2008) Characterization of microsatellite markers in mandarin orange (Citrus reticulata Blanco). Mol Ecol Resour 8:119–122
Garcia-Lor A, Luro F, Navarro L, Ollitrault P (2012) Comparative use of InDel and SSR markers in deciphering the interspecific structure of cultivated citrus genetic diversity: a perspective for genetic association studies. Mol Gen Genet 287:77–94
Garcia-Lor A, Curk F, Snoussi-Trifa H, Morillon R, Ancillo G, Luro F, Navarro L, Ollitrault P (2013) A nuclear phylogenetic analysis: SNPs, indels and SSRs deliver new insights into the relationships in the ‘true citrus fruit trees’ group (Citrinae, Rutaceae) and the origin of cultivated species. Ann Bot 111:1–19
Gmitter FG, Hu X (1990) The possible role of Yunnan, China, in the origin of contemporary Citrus species (Rutaceae). Econ Bot 44:267–277
Gulsen O, Roose M (2001) Lemons: diversity and relationships with selected Citrus genotypes as measured with nuclear genome markers. J Am Soc Hortic Sci 126:309–317
Hampl V, Pavlícek A, Flegr J (2001) Construction and bootstrap analysis of DNA fingerprinting-based phylogenetic trees with the freeware program FreeTree: application to trichomonad parasites. Int J Syst Evol Microbiol 51:731–735
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755
Kijas J, Thomas M, Fowler J, Roose M (1997) Integration of trinucleotide microsatellites into a linkage map of Citrus. Theor Appl Genet 94:701–706
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) ClustalW and ClustalX version 2.0. Bioinformatics 23:2947–2948
Li XM, Xie RJ, Lu ZH, Zhou ZQ (2010) The origin of cultivated citrus as inferred from internal transcribed spacer and chloroplast DNA sequence and amplified fragment length polymorphism fingerprints. J Am Soc Hortic Sci 135:341–350
Li H, Yang XM, Zhu LS, Yi HL, Chai LJ, Deng XX (2015) Parentage analysis of natural citrus hybrid ‘Zhelong Zhoupigan’ based on nuclear and chloroplast SSR markers. Sci Hortic 186:24–30
Liang M, Yang XM, Li H, Su SY, Yi HL, Chai LJ, Deng XX (2015) De novo transcriptome assembly of pummelo and molecular marker development. PLoS One 10(3):e0120615
Lim TK (2012) Citrus medica. Edible medicinal and non-medicinal plants. Springer, Netherlands, pp 682–689
Liu K, Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21:2128–2129
Liu CH, Jiang D, Cheng YJ, Deng XX, Chen F, Fang L, Ma ZC, Xu J (2013) Chemotaxonomic study of Citrus, Poncirus and Fortunella genotypes based on peel oil volatile compounds-deciphering the genetic origin of Mangshanyegan (Citrus nobilis Lauriro). PLoS One 8(3):e58411
Long CL, Li H, Ouyang ZQ, Yang XY, Li Q, Trangmar B (2003) Strategies for agrobiodiversity conservation and promotion: a case from Yunnan, China. Biodivers Conserv 12:1145–1156
Luro FL, Costantino G, Terol J, Argout X, Allario T, Wincker P, Talon M, Ollitrault P, Morillon R (2008) Transferability of the EST-SSRs developed on Nules clementine (Citrus clementina Hort ex Tan) to other Citrus species and their effectiveness for genetic mapping. BMC Genomics 9:287
Luro F, Venturini N, Costantino G, Paolini J, Ollitrault P, Costa J (2012) Genetic and chemical diversity of citron (Citrus medica L.) based on nuclear and cytoplasmic markers and leaf essential oil composition. Phytochemistry 77:186–196
Morton CM (2009) Phylogenetic relationships of the Aurantioideae (Rutaceae) based on the nuclear ribosomal DNA ITS region and three noncoding chloroplast DNA regions, atpB-rbcL spacer, rps16, and trnL-trnF. Org Divers Evol 9:52–68
Morton CM, Grant M, Blackmore S (2003) Phylogenetic relationships of the Aurantioideae inferred from chloroplast DNA sequence data. Am J Bot 90:1463–1469
Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci U S A 76:5269–5273
Nicolosi E, Deng Z, Gentile A, La Malfa S, Continella G, Tribulato E (2000) Citrus phylogeny and genetic origin of important species as investigated by molecular markers. Theor Appl Genet 100:1155–1166
Nicolosi E, La Malfa S, El-Otmani M, Negbi M, Goldschmidt EE (2005) The search for the authentic citron (Citrus medica L.): historic and genetic analysis. HortSci 40:1963–1968
Ollitrault F, Terol J, Pina JA, Navarro L, Talon M, Ollitrault P (2010) Development of SSR markers from Citrus clementina (Rutaceae) BAC end sequences and interspecific transferability in Citrus. Am J Bot 97:124–129
Ollitrault P, Terol J, Chen C, Federici CT, Lotfy S, Hippolyte I, Ollitrault F, Berard A, Chauveau A, Cuenca J, Costantino G, Kacar Y, Mu L, Garcia-Lor A, Froelicher Y, Aleza P, Boland A, Billot C, Navarro L, Luro F, Roose ML, Gmitter FG, Talon M, Brunel D (2012a) A reference genetic map of C. clementina hort. ex Tan.; citrus evolution inferences from comparative mapping. BMC Genomics 13:593
Ollitrault P, Terol J, Garcia-Lor A, Bérard A, Chauveau A, Froelicher Y, Belzile C, Morillon R, Navarro L, Brunel D (2012b) SNP mining in C. clementina BAC end sequences; transferability in the Citrus genus (Rutaceae), phylogenetic inferences and perspectives for genetic mapping. BMC Genomics 13:13
Page R (2001) Treeview (Win32) version 1.6.6. Available at. http://taxonomy.zoology.gla.ac.uk/rod/treeview.html
Pang XM, Hu CG, Deng XX (2006) Phylogenetic relationships within Citrus and its related genera as inferred from AFLP markers. Genet Resour Crop Evol 54:429–436
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in excel. Population genetic software for teaching and research-an update. Bioinformatics 28:2537–2539
Penjor T, Anai T, Nagano Y, Matsumoto R, Yamamoto M (2010) Phylogenetic relationships of Citrus and its relatives based on rbcL gene sequences. Tree Genet Genomes 6:931–939
Penjor T, Yamamoto M, Uehara M, Ide M, Matsumoto N, Matsumoto R, Nagano Y (2013) Phylogenetic relationships of Citrus and its relatives based on matK gene sequences. PLoS One 8(4):e62574
Posada D, Buckley TR (2004) Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and bayesian approaches over likelihood ratio tests. Syst Biol 53:793–808
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Quintana J, Contreras A, Merino I, Vinuesa A, Orozco G, Ovalle F, Gomez L (2015) Genetic characterization of chestnut (Castanea sativa Mill.) orchards and traditional nut varieties in El Bierzo, a glacial refuge and major cultivation site in northwestern Spain. Tree Genet Genomes 11:1–12
Ramadugu C, Keremane ML, Hu X, Karp D, Federici CT, Kahn T, Roose ML, Lee RF (2015) Genetic analysis of citron (Citrus medica L.) using simple sequence repeats and single nucleotide polymorphisms. Sci Hortic 195:124–137
Rambaut A, Drummond AJ (2007) Tracer version 1.4. Available at: http://beast.bio.ed.ac.uk/Tracer
Rana JC, Chahota RK, Sharma V, Rana M, Verma N, Verma B, Sharma TR (2015) Genetic diversity and structure of Pyrus accessions of Indian Himalayan region based on morphological and SSR markers. Tree Genet Genomes 11:1–14
Reuther W, Batchelor L, Webber H (1968) The citrus industry. University of California, USA
Rohlf FJ (2000) NTSYS-pc: numerical taxonomy and multivariate analysis system version 2.1. Exeter Software, Setauket
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Ruiz C, Breto MP, Asins M (2000) A quick methodology to identify sexual seedlings in citrus breeding programs using SSR markers. Euphytica 112:89–94
Scora RW (1975) On the history and origin of Citrus. Bull Torr Bot Club 102:369–375
Simmons MP, Ochoterena H (2000) Gaps as characters in sequence-based phylogenetic analyses. Syst Biol 49:369–381
Sitther V, Zhang D, Harris DL, Yadav AK, Zee FT, Meinhardt LW, Dhekney SA (2014) Genetic characterization of guava (Psidium guajava L.) germplasm in the United States using microsatellite markers. Genet Resour Crop Evol 61:829–839
Snoussi H, Duval MF, Garcia-Lor A, Belfalah Z, Froelicher Y, Risterucci AM, Perrier X, Jacquemoud-Collet JP, Navarro L, Harrabi M, Ollitrault P (2012) Assessment of the genetic diversity of the Tunisian citrus rootstock germplasm. BMC Genet 13:16
Swingle WT, Reece PC (1967) The botany of Citrus and its wild relatives. In: Reuther W, Webber HJ, Batchelor LD (eds) The Citrus industry, vol 1. University of California, Berkeley, pp 190–430
Swofford D (2002) PAUP: phylogenetic analysis using parsimony, version 4.0 b10. Sinauer Associates, Sunderland
Tanaka T (1961) Citologia: semi-centennial commemoration papers on citrus studies. Citrologia Supporting Foundation, Osaka, p 114
Uzun A, Yesiloglu T, Polat I, Aka-Kacar Y, Gulsen O, Yildirim B, Tuzcu O, Tepe S, Canan I, Anil S (2010) Evaluation of genetic diversity in lemons and some of their relatives based on SRAP and SSR markers. Plant Mol Biol Report 29:693–701
Wolfe KH, Li WH, Sharp PM (1987) Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci U S A 84:9054–9058
Wu GA, Prochnik S, Jenkins J, Salse J, Hellsten U, Murat F, Perrier X, Ruiz M, Scalabrin S, Terol J, Takita MA, Labadie K, Poulain J, Couloux A, Jabbari K, Cattonaro F, Del Fabbro C, Pinosio S, Zuccolo A, Chapman J, Grimwood J, Tadeo FR, Estornell LH, Munoz-Sanz JV, Ibanez V, Herrero-Ortega A, Aleza P, Perez-Perez J, Ramon D, Brunel D, Luro F, Chen C, Farmerie WG, Desany B, Kodira C, Mohiuddin M, Harkins T, Fredrikson K, Burns P, Lomsadze A, Borodovsky M, Reforgiato G, Freitas-Astua J, Quetier F, Navarro L, Roose M, Wincker P, Schmutz J, Morgante M, Machado MA, Talon M, Jaillon O, Ollitrault P, Gmitter F, Rokhsar D (2014) Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nat Biotechnol 32:656–662
Xu Q, Chen LL, Ruan X, Chen D, Zhu A, Chen C, Bertrand D, Jiao WB, Hao BH, Lyon MP, Chen J, Gao S, Xing F, Lan H, Chang JW, Ge X, Lei Y, Hu Q, Miao Y, Wang L, Xiao S, Biswas MK, Zeng W, Guo F, Cao H, Yang X, Xu XW, Cheng YJ, Xu J, Liu JH, Luo OJ, Tang Z, Guo WW, Kuang H, Zhang HY, Roose ML, Nagarajan N, Deng XX, Ruan Y (2013) The draft genome of sweet orange (Citrus sinensis). Nat Genet 45:59–66
Yeh FC, Yang R-C, Boyle TB, Ye Z, Mao JX (1997) POPGENE, the user-friendly shareware for population genetic analysis. Molecular Biology and Biotechnology centre, University of Alberta, Canada
Acknowledgments
This work was funded by the National Natural Science Foundation of China (31330066 and 31221062), the Special Fund for Agro-scientific Research in the Public Interest (201303093), and the Ministry of Agriculture (CARS-27). The authors are grateful to Dr. Xiuli Zeng for collecting plant materials.
Data archiving statement
We follow standard Tree Genetics and Genomes policy. All the chloroplast gene sequences will be submitted to GenBank, and accession numbers will be provided once available.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Communicated by W.-W. Guo
This article is part of the Topical Collection on Germplasm Diversity
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1
Plot of Delta K (filled circles, solid line) calculated as the mean of the second-order rate of change in likelihood of K divided by the standard deviation of the likelihood of K, m(|L"(K)|)/s [L(K)] (GIF 73 kb)
Supplementary Fig. 2
Inferred clusters based on Bayesian analysis for K = 2 among 36 citron and fingered citron accessions. Each vertical line represents an individual genotype. Different colors stand for the most likely ancestry of the cluster. Individuals with multiple colors have admixed genotypes. The height of each bar represents the probability of a variety belonging to a different subgroup. The identified abbreviated accession names of corresponding samples are listed in Supplementary Table S1 (GIF 28 kb)
Supplementary Fig. 3
The 50 % majority rule consensus tree of 1502 trees obtained from four rounds of Bayesian analysis of the combined data set (matk, trnS-trnG, rps16, rpl16, atpB-rbcL, and accD-psaI chloroplast DNA regions), implementing the GTR model from 39 Citrus taxa. Branch lengths reflect changes per site. Posterior probabilities are given above branches. Clades defined as I, II, III, and IV represent different groups (GIF 54 kb)
Supplementary Table S1
Classification, origin, group, and list of studied accessions (DOCX 27 kb)
Supplementary Table S2
Summary information of the 77 nSSR primer pairs and the value of annealing temperature (T) (DOCX 20 kb)
Supplementary Table S3
The six chloroplast DNA regions used in the phylogenetic analysis, the marker names and sequences used to amplify them, the type of DNA each region contains and their annealing temperature (T) (DOCX 17 kb)
Supplementary Table S4
Heterozygosity observed in all the analyzed accessions by the 77 nSSR markers (DOCX 17 kb)
Rights and permissions
About this article
Cite this article
Yang, X., Li, H., Liang, M. et al. Genetic diversity and phylogenetic relationships of citron (Citrus medica L.) and its relatives in southwest China. Tree Genetics & Genomes 11, 129 (2015). https://doi.org/10.1007/s11295-015-0955-x
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-015-0955-x