Abstract
We studied the genetic pattern of 21 Ethiopian populations of Prunus africana by using six nuclear and five plastid microsatellites. In total, 89 alleles were found in the nuclear and 16 haplotypes in the plastid genome. High levels of diversity both in cpSSRs (h T = 0.703) and nSSR (H T = 0.725) were detected. Genetic differentiation among populations at the nuclear and plastid level was moderate (F ST = 0.122 vs. G ST = 0.478). While Ethiopian populations harbored the highest plastid haplotype diversity throughout Africa, the level of nuclear diversity was lower than in the remaining part of the species’ range. Ten of the observed 16 plastid haplotypes were unique to Ethiopia, suggesting an isolated plastid evolution. Remarkably, all plastid haplotypes found in Ethiopia belonged to one single lineage, while other populations from East Africa and Madagascar contain haplotypes from up to four more divergent lineages. This suggests that in contrast to previous expectations, the Horn of Africa is a hot spot of plastid diversity but not the ancestral origin for present populations of P. africana. The ratio between pollen to seed flow was estimated to be 7.1, indicating predominant gene flow by pollen. The exhaustive pollen flow also facilitated gene exchange with West African nuclear lineages probably in the early Holocene. The Ethiopian rift formed a genetic barrier resulting in population differentiation east and west of the rift; however, it was less effective in disrupting gene flow than the Eastern Rift in more southern parts of the East African range.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The population history of most plant species in East Africa is still in the dark, but accumulated evidence suggests that the variable climate in the region over the millennia has resulted in plastic and complex patterns of diversity (Livingstone 1975; Hamilton 1982). In particular, Afromontane forest species have been subject to a number of population expansion and retraction dynamics, following changing precipitation conditions (Street-Perrott and Perrott 1993). Among the better studied species is Prunus africana (Hook. f.) Kalkman (synonym Pygeum africanum Hook. f.), a keystone species of the Afromontane forests. It is a medium to large multipurpose tree of 30–40 m height, commonly known as African cherry. P. africana is hermaphroditic, insect pollinated, and bird dispersed. It mainly occurs in montane forests but also along creeks at lower elevation. The species is an economically important and endangered member of the Afromontane flora and occurs geographically widespread in mainland Africa and outlying islands (Bioko, Grande Comore, Sao Tome, and Madagascar) (Kalkman 1965). In Ethiopia, P. africana is one of the victims of omnipresent deforestation (Reusing 2000). During the last century, most of the Afromontane forests have been cleared and only a very small proportion of the original vegetation remains (Teketay and Granström 1995). Besides timber and numerous traditional medicinal uses of this species, its bark extracts are commercially used to treat benign prostatic hyperplasia (Kadu et al. 2012, and references therein). Bark is especially exploited in Cameroon and Madagascar (Cunningham and Mbenkum 1993; Cunningham et al. 1997). In addition to this, overexploitation for pharmacological use, deforestation, habitat fragmentation, wildfires, and invasive alien plants threaten this species at different levels throughout its native range (Jimu 2011). Moreover, the modeled future distribution of P. africana indicates that it will be negatively affected by climate change (Vinceti et al. 2013). Consequently, the “Convention on International Trade in Endangered Species of Wild Fauna and Flora” includes African cherry as a vulnerable species that may become threatened by extinction unless its trade is regulated (http://www.cites.org/eng/app/appendices.php accessed 8/2015; Dawson et al. 2000). At the national level, policies have been established in various African countries aiming to ensure sustainable utilization and management of P. africana, although enforcement problems persist (Vinceti et al. 2013). It is very likely that valuable genetic resources have already been lost as some of the populations have been overexploited or gone extinct.
A high level of molecular diversity is still maintained in the species despite its heavy exploitation. Previous studies using randomly amplified polymorphic DNA (RAPD) markers indicated high diversity levels within certain regions of its native range (Barker et al. 1994; Dawson and Powell 1999; Muchugi et al. 2006). More recent studies using microsatellite markers also pointed to high genetic diversity (Farwig et al. 2008; Kadu et al. 2011, 2013).
In regard to its phylogeography, Aubréville (1976) suggested that P. africana or its progenitor migrated from its possible origin in the Middle East into the northeast of Africa. Based on DNA findings, it is likely that this assumed southward migration route split into a western and eastern lineage, which finally contributed to the colonization of southern Africa (Muchugi et al. 2006; Kadu et al. 2011). Populations in Madagascar are highly diverged and were isolated for an extensive time period (Kadu et al. 2011, 2013). The Eastern Rift Valley (ERV) was identified as a major barrier to gene flow between population groups (Kadu et al. 2011, 2013). The Ethiopian highlands, which form the northern part of the ERV, are of particular importance, because they represent the suggested primary immigration and spreading route of the species or its predecessors into Africa. Unfortunately, Ethiopian samples were unavailable at the time of the genetic inventory of P. africana across its native range in the previous analysis conducted by Kadu and her colleagues (2011, 2012, 2013). Six Ethiopian populations have already been studied using RAPDs by Atnafu (2007), resulting into two main clusters geographically separated by the main Ethiopian Rift. Nevertheless, an extensive sampling was desirable to elucidate the phylogeography and population history of the species in this important region and in particular the integration of Ethiopian data with common methods into the available data set to provide an overall understanding of the phylogeography of this important species. In the present study, we investigate the population genetic structure of this species in Ethiopia and study whether Ethiopia is an ancestral region for the continental diversity of this species, by adding chloroplast and microsatellite analysis of Ethiopian populations to the data sets of Kadu et al. (2011, 2013). The results of our study are expected to increase our understanding of the phylogeography of this Afromontane tree and should also increase the efficiency of conservation efforts toward the species. The data set presented here is one of the most complete concerning any African tree species with a wide-ranging distribution.
Material and methods
Sampling
Twenty-one populations were sampled in Ethiopia from 35° 32′ (Gore) to 41° 49′ (GaraMuleta) longitude east and 5° 53′ (Kibre Mengist) to 11° 50′ (Debre Tabor) latitude north covering an altitudinal range from 1584 m (Harenna) and 2859 m (Debre Tabor) above sea level (Table 1). The geographic distance between populations ranged from 26 km (Addis Ababa—Menagesha) to 700 km (Gara Muleta—Gore). From each population, young leaves from 10 trees were collected and dried in ziplock plastic bags containing silica gel. To minimize the collection of closely related individuals, sampled trees were at least separated by a distance of 100 m. Due to the very small population size in Denkoro (DE) and Wof Washa (WW), this minimum distance was abandoned and every available tree was sampled. Compared to the sampling scheme of Kadu et al. (2011, 2013), the average sampled population size was smaller but more populations were sampled over a comparatively restricted geographic area.
DNA isolation
Total genomic DNA was isolated from 40 to 60 mg of dried leaf samples using a modified triple cetyltrimethyl ammonium bromide (CTAB) extraction technique (Borsch et al. 2003). DNA samples were run in 1 % agarose gelelectrophoresis in order to check their quality and quantity. Concentration and purity of the DNA samples were further determined using a ND-1000 spectrophotometer (NanoDrop, USA).
DNA amplification and genotyping
In total, 11 microsatellites originally developed for other species were used to genotype all samples. Nuclear DNA was studied using six microsatellite loci (nuclear simple sequence repeat (nSSR)) originally characterized in peach [Prunus persica, primer pairs U3 (UDP9-403) and U5 (UDP96-018) (Cipriani et al. 1999) and P2 (PS12A02) Sosinski et al. 2000] and wild cherry (Prunus avium, primer pairs EMPaS01, EMPaS06, and EMPaS10; Vaughan and Russell 2004). These microsatellites were previously transferred to P. africana (Cavers et al. 2009; Kadu et al. 2013). For the investigation of plastid DNA variation, five microsatellites originally developed for Japanese plum (Prunus salicina, primer pairs TPSCP1, TPSCP5, and TPSCP10; Ohta et al. 2005) and Sorbus aucuparia (primer pairs rps16pm2 and trnT-Lpm1; Chester et al. 2007) were used. These chloroplast simple sequence repeat (cpSSR) markers were also previously transferred to P. africana, and for rps16pm2 and trnT-Lpm1, two indels were detected by Kadu et al. (2011). Indels were treated as four additional loci, i.e., rps16pm2a, rps16pm2b, trnT-Lpm1a, and trnT-Lpm1b.
PCR reactions were performed in a 10 μL volume containing 1 μL (10–70 ng) total genomic DNA, 2.3 μL KAPA2G™ Buffer A (Kapa Biosystems), 0.05 mM of each dNTP, 0.21 mM of each primer, one of the two being 5′ labeled with a fluorescence dye, 0.45 U KAPA2G™ Fast DNA Polymerase (Kapa Biosystems), and autoclaved deionized water. Amplifications were run on a PTC-100 thermocycler using the following heating profile: a first step initial denaturing at 95 °C for 3 min followed by 35 cycles, each consisting of 30 s denaturing at 94 °C, 30 s annealing at a specific temperature (54 °C for all nSSR primers, 49 °C for trnT-Lpm1, 50 °C for TPSCP5 and rps16pm2, 53 °C for TPSCP1, and 55 °C for TPSCP10), and 5 s extension at 72 °C. The last cycle was ended by additional 30 s at 72 °C to complete extension.
PCR-amplified DNA fragment size was determined by capillary gel electrophoresis using a CEQ8000 sequencer (Beckman Coulter, USA). Selected DNA samples from the previous studies of Kadu et al. (2011, 2013) were used as reference to standardize genotyping and to link the results with those of previous studies.
Data analyses—Ethiopia
In a first step, the Ethiopian nSSR data set was checked for the presence of genotyping errors and null alleles using the program Micro-Checker (van Oosterhout et al. 2004). Thereafter, the number of observed alleles (N a ), the number of effective alleles (N e ), the observed heterozygosity (H o ) and unbiased expected heterozygosity (H e ), the fixation index (F), the number of private alleles (A p ), and the percentage of polymorphic loci (PPL) per population were calculated, and deviations from Hardy–Weinberg expectations (HWEs) were assessed using GenAlEx 6.501 (Peakall and Smouse 2012). The number of alleles corrected for equal sample size (allelic richness, R s ) was calculated with FSTAT (Goudet 1995). The presence of a phylogeographic pattern within the Ethiopian populations was tested using SPAGeDi1.4b (Hardy and Vekemans 2002) by evaluating the contribution of the stepwise mutation to the differentiation patterns at the nSSRs. Here, the observed differentiation R ST is compared to the differentiation obtained after 1000 allele size permutations (pRST) as an approximate for F ST. If stepwise mutations do not contribute to differentiation, R ST should be equal to F ST, but if stepwise mutations have contributed to differentiation, R ST is expected to be significantly higher than pRST, respectively, F ST.
The model-based Bayesian clustering method implemented in STRUCTURE 2.3 (Pritchard et al. 2000) was used to infer clusters by assigning individual multilocus genotypes probabilistically to a user-defined number of K clusters. The admixture model without incorporation of population information was used, assuming correlated allele frequencies applying K values ranging between 1 and 21 as well as run lengths of 800,000 iterations with a burn-in period of 200,000. Five runs per K were performed for the Ethiopian populations separately. The most likely number of clusters was inferred using the ΔK statistic of Evanno et al. (2005) implemented in STRUCTURE HARVESTER 0.6.93 (Earl and von Holdt 2012). Analysis of molecular variance (AMOVA) was carried out in Arlequin 3.5 (Excoffier and Lischer 2010) to investigate population differentiation at various subdivisions based on geographical units, tree seed zones, and individual-based population clusters from STRUCTURE using both F ST and R ST in separate analyses based on 10,000 permutations. Isolation by distance was evaluated by a Mantel test (Mantel 1967) using Nei’s unbiased genetic distance (Nei 1978).
The chloroplast data were transformed into multilocus haplotypes and for a unique identification of haplotypes combined with the range-wide data set of Kadu et al. (2011). The number of haplotypes per population (N a ), the effective number of haplotypes (N e ), the number of private haplotypes (N p ), and haplotype diversity (H e ) were calculated using GenAlEx 6.501 (Peakall and Smouse 2012). Total haplotype diversity (h T ) and average within population haplotype diversity (h S ) were calculated according to Pons and Petit (1995, 1996) using the software PerMut (http://www.pierroton.inra.fr/genetics/labo/Software). To test for the existence of phylogeographic pattern in haplotype diversity, we compared the two measures of differentiation N ST and G ST, which are based on ordered and unordered alleles, respectively. Significance was tested based on 1000 permutations.
The data sets of both the nSSRs and the cpSSRs were split into two groups (north-northeastern vs. south-southwestern populations, low vs. high elevated population, and farmland vs. non-farmland) in order to test for differences by t- tests. The results of these tests were corrected for multiple comparisons by Bonferroni corrections (one-sided or two-sided). Estimates of diversity derived for nuclear and plastid genomes, respectively, were rank-correlated.
Both AMOVA and Mantel test were computed using GenAlEx 6.501 (Peakall and Smouse 2012), and significances were tested based on 9999 permutations. The pollen-to-seed migration proportion was calculated after Ennos (1994) by \( r=\frac{\left[\left(\frac{1}{F_{\mathrm{ST}}}-1\right)\left(1+{F}_{\mathrm{IS}}\right)-2\left(\frac{1}{N_{\mathrm{st}}}-1\right)\right]}{\left(\frac{1}{N_{\mathrm{ST}}}-1\right)} \)
Data analysis—range wide
To analyze the relationships between the Ethiopian populations and the range-wide collection of Kadu et al. (2011, 2013), an unweighted pair group arithmetic mean dendrogram (UPGMA) based on Nei’s standard genetic distances (Nei 1972) was computed for the nSSR data using 1000 bootstrapped distance matrices created in MSA (Dieringer and Schlotterer 2003). The computer programs NEIGHBOR and CONSENSE in the PHYLIP 3.63 package (Felsenstein 1989) were used for tree construction. As an additional method to identify population clusters, principal co-ordinate analysis was conducted using GenAlEx 6.501 (Peakall and Smouse 2012) to explore multivariate relationships among inter-individual Nei’s standard genetic distance (Nei 1972). Also, the STRUCTURE analysis as described above was performed for the total data set (including the data from Kadu et al. 2013).
Haplotype relationships of all chloroplast polymorphisms were inferred by constructing a statistical parsimony network following a two-step strategy according to Bänfer et al. (2006). This procedure accounts for the (presumably) different mutation rates underlying indel and microsatellite variation. First, haplotype data from the indel variation were employed to construct a backbone network using TCS 1.18 (Clement et al. 2000). Second, the network was enlarged by adding the variation at microsatellite loci manually at the respective positions of the backbone.
Results
Genetic structure within Ethiopia
In total, 89 alleles were detected at six nuclear microsatellites within the Ethiopian populations (mean number of alleles per locus was 14.83). The most variable locus was EMPAS10 with 27 alleles, while the least variable one was U3 at which only two alleles were detected (Suppl. Table S1). Total gene diversity for all populations (H T ) was 0.725, and overall observed heterozygosity (H o ) was 0.678 resulting into a positive fixation index (F) of 0.056. Observed number of alleles (N a ), number of effective alleles (N e ), fixation index (F), and number of private alleles (A p ) are shown in Table 2.
Very few populations showed heterozygote deficiency or excess at single loci. Population DE is remarkable, because here, for five out of six markers, significant deviations from HWE were found (Suppl. Table S2). Genetic comparisons based on the effective number of alleles between cohorts [low vs. high altitude (number of population equally split) and farmland vs. non-farmland] were not significant. Overall, differentiation among populations was highly significant (P < 0.001) both based on unordered alleles (F ST = 0.122) and ordered alleles (R ST = 0.186). Comparison of R ST and pRST values (pRST = 0.125) revealed a weak phylogeographic signal as R ST > pRST (P > 0.05).
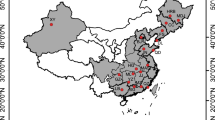
STRUCTURE analysis for the Ethiopian populations resulted in two biologically meaningful groups; also, STRUCTURE HARVESTER 0.6.93 (Earl and von Holdt 2012) identified the most likely group structure to be two (Suppl. Fig. S1 and Suppl. Table S3). These two population groups were largely located east and west of the Ethiopian rift, and an extensive admixture gradient between the populations was evident (Fig. 1), showing ongoing exchange of migrants between both sides of this gene flow barrier. AMOVA partitioned the variation based on geographical units (north, southwest, central, and southeast), tree seed zones, and STRUCTURE-based groups (east and west of the Ethiopian Rift). Although significant differentiation was found, the proportion of the variation due to variation between the assumed regions (geographic units, seed zones, and genetic clusters) was generally small (Table 3). The pairwise population matrix of Nei’s unbiased genetic distances and geographic distances for the Mantel test is given in Suppl. Table S4. Isolation by distance has not significantly contributed to the nuclear population structure as only a very weak positive correlation between genetic and geographic distance matrices was found within Ethiopia (Rxy = 0.20, P = 0.057) (Fig. 2).
Results of individual population assignment (admixture model) performed with STRUCTURE for the combined data set (Ethiopian populations and data set of Kadu et al. (2013) using K = 6 and separate for the 21 Prunus africana populations from Ethiopia (K = 2)). Stars indicate populations east of the Ethiopian rift
Even though five chloroplast DNA (cpDNA) and four cpDNA indels (the same as used in Kadu et al. 2011) were considered in generating data from 210 individuals from Ethiopia, only three of these microsatellite loci were polymorphic (Suppl. Table S1). At TPSCP5 and TPSCP10, four haplotypic variants were detected at each locus, while at TPSCP1, two variants were found resulting in a total of 14 haplotypes in the Ethiopian samples. The monomorphic indel loci were not excluded from further analysis for the sake of consistency in comparing the present data with previously published results (Kadu et al. 2011). Haplotypic variation strongly differed among populations (Suppl. Table S5). Sixteen of the 32 multilocus haplotypes, which were constructed from cpSSRs of the range-wide samples, were found in Ethiopia. Total haplotype diversity within the Ethiopian sample amounted to h T = 0.703, which could be significantly partitioned into 47.8 % among populations and 52.2 % within populations. The mean within population plastid haplotype diversity was estimated to be h S = 0.367. Comparison of observed N ST (0.483) with mean-permuted N ST (0.473) did not reveal a significant phylogeographic pattern. A Mantel test showed only a very weak positive correlation between geographic and Nei’s genetic distance matrices (Rxy = 0.107, P < 0.001).
The ratio between pollen flow to seed flow was estimated as r = 7.1, indicating a several fold more extensive gene flow by pollen compared to seed dispersal. A test for correlations between the effective number of nuclear alleles and the number of plastid haplotypes amounted to low and insignificant rank correlation (Spearman’s r 2 = −0.15).
Genetic structure across Africa
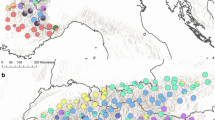
The relationship among populations from the range-wide sample (including populations from Kadu et al. 2011, 2013) based on a bootstrapped Nei’s standard genetic distance of the nSSRs is shown in the UPGMA tree (Fig. 3). With one exception (population DE), all Ethiopian populations fell within one major branch of the tree. Ethiopian populations east of the Ethiopian Rift also grouped close together on a sub-branch. Interestingly, all Ethiopian populations clustered with populations of the western lineage from the range-wide sample (Kadu et al. 2013). In the principal co-ordinate analysis, the following six groups were identified: (1) Ethiopia, (2) Cameroon and Equatorial Guinea, (3) East Africa west of the ERV (Uganda and western Kenya), (4) East Africa east of the ERV (Tanzania and eastern Kenya), (5) Zimbabwe and South Africa, and (6) Madagascar (Fig. 4). Consistent with the UPGMA, the Ethiopian cluster was more closely related to the western lineage populations than to populations originating east of the ERV. A similar pattern was obtained in the STRUCTURE analysis of the combined nuclear data set (Fig. 1): the results of K = 6 show the grouping already reported by Kadu et al. (2013) plus a well-defined Ethiopian group that exhibits some admixture with the western lineages of the range-wide sample.
UPGMA dendrogram for the combined data set of P. Africana from Ethiopia and Kadu et al. (2013). Populations located east of the Ethiopian Rift Valley are marked by a star
Principal co-ordinate analysis showing the multivariate relationships of 46 P. africana populations including the data set of Kadu et al. (2013). Population acronyms for non-Ethiopian populations are from GQ1 Moka (Equ. Guinea), CM1 Lower Man’s Spring (Cameroon), CM2 Ngashie Mountains (Cameroon), CM3 Mount Danoua (Cameroon), UG1 Bwindi (Uganda), UG2 Kalinzu (Uganda), UG3 Kibale (Udana), UG4 Mabira (Uganda), KE1 Kibiri (Kenya), KE2 Kakamega (Kenya), KE3 Kapcherop (Kenya), KE4 Londiani (Kenya), KE5 Chuka (Kenya), KE6 Lari (Kenya), KE7 Ol Danyo Sambuk (Kenya), KE8 Taita Hills (Kenya), TZ1 Kilimanjaro Catchment Forest Reserve (Tanzania), TZ2 Kindoro Catchment Reserve (Tanzania), TZ3 Shume Mamba Forest Reserve (Tanzania), TZ4 Meru Catchment Reserve (Tanzania), TZ5 Udzungwa (Tanzania), TZ6 Kidabaga (Tanzania), ZW1 Nyanga (Zimbabwe), ZW2 Cashel Valley Chimanimani (Zimbabwe), ZW3 Chirinda Forest Reserve Chipinge (Zimbabwe), SA1 Mpumalanga (South Africa), SA2 Kwazulu-Natal (South Africa), and MG1 Marovoay (Madagascar); for more information on non-Ethiopian populations, see Kadu et al. (2013). For Ethiopian population acronyms, see Table 1
The relationship between the 32 haplotypes is demonstrated by the genealogical network in Fig. 5. The backbone of the network consists of five main haplotypes (denoted HT1–HT5). Ten of the Ethiopian haplotypes (HT1p–HT1y) were not found in other countries. Five of the sixteen haplotypes (HT1a, HT1c, HT1g, HT1h, and HT1i) were shared with Eastern (excluding Uganda) and southern African populations, but only one haplotype (HT1k) was shared with a West African population (Equatorial Guinea) (Fig. 6). All haplotypes found in Ethiopia were members of the HT1 family, which is the most commonly detected haplotype in Kadu et al. (2011). In the Ethiopian samples, the predominant haplotype HT1h mainly was found on the northwestern side of the Ethiopian Rift. This haplotype occurred in 16 Ethiopian populations, while it was rare in the non-Ethiopian populations occurring only within one Kenyan and one Tanzanian population, respectively.
Genealogical network of 32 haplotypes (HTs) of 53 P. africana populations from 10 African countries (including 32 populations of Kadu et al. 2011). Non-white colors represent haplotypes found in Ethiopia, and haplotypes shared with populations of other African countries are represented by concentric circles. The size of the haplotypes is shown proportional to their frequencies. Very small white circles represent hypothetical intermediate haplotypes
Distribution of 16 “Ethiopian” haplotypes of P. africana found in Africa. Codes for non-Ethiopian populations are according to Kadu et al. (2011) GQ Moka, KA Chuka, KB Kinale, KC Kapcherop, KD Kakamega, KE Londiani, KG Taita, KS Kibiri Forest, SA Mpumalanga, SB KwaZulu-Natal, TB Kilimanjaro, TD Shume Magamba, TE Kidabaga, TT Udzungwa, ZWA Nyanga, ZWB Chimanimani, and ZWC Chipinge, and for Ethiopian populations, see Table 1 (map adapted from Kadu et al. 2011)
Discussion
While the phylogeographic patterns of northern hemisphere tree species are well analyzed and found to be mainly affected by repeated poleward expansion-contraction cycles in the Quaternary (e.g., Hewitt 1999), the population history of trees in the low-latitude zone is less studied and seems to be shaped by interaction dynamics between topography and changing climate patterns (e.g., Hamilton and Taylor 1991). A frequent difficulty in developing likely scenarios of migration and population expansion at low latitudes is the identification of the founder populations and origins of spread. The present study makes use of one of the most complete data sets of an African tree species that—although restricted to Afromontane forests—is widespread across the continent. Until today, populations from the Ethiopian highland were thought to be the founder populations in Africa, because it was suggested that Afroalpine and Afromontane species colonized Africa through the coastal mountain ranges of the Arabian Peninsula and the Ethiopian highlands (Koch et al. 2006; Aubréville 1976). However, our joint analysis of Ethiopian populations with data derived from the remaining species’ range reported by Kadu et al. (2011, 2013) suggests that the Horn of Africa is a hot spot of plastid diversity and evolution but not the ancestral origin for a majority of populations across Africa. Ethiopian populations seem to form an additional distinct entity with independent plastid evolution for an extended period of time resulting in the highest values of plastid diversity across the species’ range. Total plastid haplotypic diversity (h T ) for all Ethiopian populations nearly reached the estimate calculated for 32 sub-Saharan populations from nine countries and within populations diversity (h s ) averaged to 0.367 throughout Ethiopia but only to 0.234 for the rest of the species’ range (Kadu et al. 2011). Besides this remarkable diversity, the genetic relatedness among the cpDNA haplotypes provides new answers to the questions whether P. africana has initiated its migration from the Horn of Africa or from locations further south; all haplotypes found in Ethiopia were members of the HT1 lineage, while East African populations from Kenya, Tanzania, and Madagascar also contained haplotypes from the more diverged and probably “older” lineages HT2–HT5. Thus, the initial diversification very likely did not start in Ethiopia but probably in the Kenyan or Tanzanian Mountains. This model would also be supported by one of the oldest continuous pollen records of Eastern Africa retrieved from Sacred Lake, north-east of Mount Kenya spanning more than 40,000 years before present (BP). It is remarkable that since approximately 30,000 years BP, Prunus pollen was constantly found here with a slight increase in the mid-Holocene (Coetzee 1967). Very old pollen records of approximately 11,000 years BP were also reported close to Lake Victoria in eastern Kenya (Hamilton 1982; Vincens 1986), as well as in Burundi, Rwanda, and western Uganda (Jolly et al. 1997). It is therefore tempting to speculate that from this region, P. africana had spread to north and south after the Last Glacial Maximum. However, maximum time scales covered among different palynological studies vary erratically. But to the best of the authors’ knowledge, pollen of P. africana has been only reported during the last 3000 years in the Arsi Mountains in southeastern Ethiopia (Bonnefille and Umer 1994). Since all major occurrences now have been sampled with inconclusive results, it may be that the primary immigration route into Africa and the origin of the species stay dubious. Several climate oscillations have taken place in East Africa over an extensive time period, and it is possible that original patterns have been blurred by several cycles of population expansion and contraction or that ancestral populations went extinct or lost the majority of its genetic diversity by drift.
At present, Ethiopian populations are well isolated from populations further south (Uganda, Tanzania, and Kenya), since the Turkana Basin forms a strong gene flow barrier. At least by the middle of the Holocene (around 5000 years BP), the climatic conditions had become similar to those found today (Owen et al. 1982) and the arid environment in this basin inevitably has disrupted gene flow.
Six out of 16 haplotypes found in Ethiopia were also present in Kenya and Tanzania, Zimbabwe, and South Africa (Kadu et al. 2011), while populations in Cameroon, Uganda, Nigeria, and Madagascar did not share haplotypes with the Ethiopian samples. Only HT1k was shared between one Ethiopian and one Equatorial Guinean population, maybe as a result of homoplasy which may have influence haplotype patterns in closely related variants (Kadu et al. 2011). But the absence of “Ethiopian” haplotypes in West Africa and in populations west of the Albertine Rift rejects certain migration scenarios of P. africana proposed by Kadu et al. (2011), such as (i) the southward migration of P. africana from the southern fringe of the Ethiopian highlands to the Albertine Rift Valley (Uganda) and then to West Africa and (ii) the independent colonization of West Africa directly from the Ethiopian highlands using the Marrah Mountains (Sudan) as stepping stones via a northwestern migration corridor and the subsequent colonization of western Uganda from West Africa. Instead, it seems more likely that West Africa was colonized via the southern migration tract (White 1993) or that populations from west of the Albertine Rift, which so far showed the highest similarities to West African populations, are early descendents of Kenyan or Tanzanian populations. An analysis of populations in southwestern Africa (e.g., Angola) is strongly needed to clarify this issue. The time period of the western immigration is difficult to determine, but pollen records of P. africana obtained from central Cameroon imply that the species has already been present in western Africa 7000 years BP (Vincens et al. 2010).
Despite missing evidence for migration of P. africana from the Ethiopian highland to West Africa, the analysis of the nuclear markers revealed a closer clustering between populations from West Africa and populations west of the ERV than populations east of the ERV. A possible explanation is that gene flow via pollen between Ethiopian and West African populations took place during a comparatively recent climatic episode when the species may have been distributed also at lower altitudes. Such favorable conditions existed between 9000 and 6000 years BP in the area (Street-Perrott and Perrott 1993). After this humid period, precipitation in the region decreased and climate changed to drier conditions leading to retraction into higher altitudes and the strong isolation of present populations.
The high diversity found in the plastid genomes was not reflected in the nuclear data, where we obtained only moderate estimates of molecular diversity. For instance, the range of expected heterozygosities (0.452–0.732) in our study was similar to those obtained for 25 P. africana populations across the remaining native distribution area of sub-Saharan Africa (H e = 0.430–0.827; Kadu et al. 2013). Similar results were found for eight cohorts of an Ethiopian population in the southern Amhara region (H e = 0.77) by Yineger et al. (2014) who employed identical microsatellites as we did. Therefore, the highest nuclear diversity across the species’ range was found in Ugandan and Kenyan populations west of the Eastern Rift Valley averaging to 6.723 for R s (N = 10) and 0.755 for H e (eight populations studied; Kadu et al. 2013), while our respective Ethiopian values amounted to only 4.989 (arithmetic mean across regions) and 0.678, respectively. One might speculate that landscape fragmentation and overexploitation might have caused the low nuclear genetic diversity, since populations of P. africana are especially fragmented on farmland. However, our comparison of farmland versus non-farmland population did not result in significant differences and suggests that the genetic structure is so far not or at least not heavily affected by overexploitation.
Haplotypic and nuclear diversities were not significantly correlated in Ethiopian populations. However, there are only very few reports to our knowledge which tackle the correlation between nuclear and plastid data in forest trees (Vriesea gigantea, r rank = 0.47, not significant, calculated based on data in Palma-Silva et al. 2009; Sorbus domestica, r rank = 0.077, not significant, George 2015), and all of them are not significant. The levels of genetic differentiation found in the Ethiopian populations both for nuclear (F ST = 0.122) and plastid markers (N ST = 0.483) were expectedly lower than in the remaining species’ distribution area ranging from Madagascar to Equatorial Guinea and from South Africa to Uganda (F ST = 0.27, Kadu et al. 2013; N ST = 0.735, Kadu et al. 2011), likely due to the much smaller region covered. Moreover, when Afromontane forests were widely distributed in Ethiopia between 9000 and 6000 years BP (Street-Perrott and Perrott 1993), many populations were presumably connected genetically, resulting in low among-population differentiation. Today, most Ethiopian populations of P. africana are genetically isolated due to anthropogenic fragmentation of the forests (Teketay and Granström 1995), which probably started approximately 2500 years ago and has accelerated strongly during the twentieth century (Hurni 1988). But some remnant trees still exist and may function as stepping stones for gene flow, e.g., between populations like Bonga-Bedele and Chilimo-Nekemte, as relatively small genetic distances (for both populations pairs 0.048) across moderate geographic distances (134 and 168 km, respectively) indicate.
The genetic differentiation among populations in cpDNA was expectedly higher than in nuclear DNA (nDNA), which could be explained (1) by the fact that gene flow via seeds and pollen is generally higher than by seeds alone and (2) by genetic drift which acts stronger on cpDNA with smaller effective populations sizes than on nDNA (Birky et al. 1983). In support of the first explanation, Berens et al. (2013) reported different effective gene flow ratios of pollen over seed dispersal derived from propagule samples at different ontogenetic stages (from seeds to older seedlings) that varied from 3.4 to 24.8 in P. africana, while our indirect measures of this ratio resulted in a 7-fold preponderance of pollen flow.
In the Kenyan and Tanzanian parts of the African Rift Valley, eastern and western populations were strongly diverged both at nuclear and plastid markers (Kadu et al. 2011, 2013). Such a gene flow barrier was also present at the Ethiopian rift, although the pattern was weaker and included a strong admixture cline with the STRUCTURE analysis. We presume that during humid periods (interglacials) of the Pleistocene, the Afromontane vegetation belt shifted to lower altitudes and covered the highlands, while the valley bottom in Ethiopia (between 1000 and 1500 m asl) provided probably ample growing conditions for P. africana and thus may have enabled effective gene flow between eastern and western populations. Further south (main ERV), montane forests probably did not expand to the valley floor due to dryer conditions.
AMOVA revealed weak genetic differentiation for R ST and a lack of differentiation if stepwise mutation were neglected and F ST was used. Expectedly, also the seed zones (Aalbæk 1993), which had been delineated based on ecological conditions (Aalbæk 1993), were not supported by the molecular data. In our study, isolation by distance was very weak and probably of little biological significance. Environmental barriers, historical processes, and life history traits may shape the genetic structure of populations (e.g., Poelchau and Hamrick 2012) and eventually lead to isolation by distance due to limited pollen and seed dispersal; e.g., in a strongly fragmented Ethiopian P. africana population (Yineger et al. 2014), where both for adults (r = 0.68) and young seedlings (r = 0.43), a significant Mantel correlation each has been reported.
Although the Ethiopian highland was not confirmed to be the point of origin for continental genetic diversity, the present study demonstrates that Ethiopian populations harbor unique genetic variation, which requires urgent conservation measures to counteract deforestation and climate change. Our data suggest that P. africana in Ethiopia has been isolated for an extensive period of time evidenced by independent plastid evolution, although probably, extensive gene flow via pollen in more recent millennia facilitated gene exchange with West African nuclear lineages. The contrasting relatedness of Ethiopian populations to East African populations on the chloroplast genome, but to populations of West African and west of the Rift Valley on the nuclear genome, corroborate the importance of the complex interaction between the topographical structures (i.e., the Rift Valley) and climate oscillations as drivers of population structure and diversity. Further studies are needed to investigate local adaptation but also differences in bark compounds that could be utilized for medicine and would create a further stimulus to plant and conserve the species in the region.
References
Aalbæk A (1993) Tree seed zones for Ethiopia. Forestry Research Center/National Tree Seed Project, Addis Ababa
Atnafu H (2007) Genetic variation in some natural population of Prunus africana from Ethiopia as revealed by randomly amplified polymorphic DNA (RAPD). MSc Thesis, Addis Ababa University
Aubréville A (1976) Centres tertiaires d’origine, radiations et migrations des flores angiospermiques tropicales. Andansonia 16:297–354
Bänfer G, Moog U, Fiala B, Mohamed M, Weising K, Blattner FR (2006) A chloroplast genealogy of myrmecophytic Macaranga species (Euphorbiaceae) in Southeast Asia reveals hybridization, vicariance and long-distance dispersals. Mol Ecol 15:4409–4424
Barker N, Cunningham AB, Morrow C, Harley EH (1994) A preliminary investigation into the use of RAPD to assess the genetic diversity of a threatened African tree species: Prunus africana. Strelitzia 1:221–230
Berens DG, Griebeler EM, Braun C, Chituy BB, Nathan R, Böhning-Gaese K (2013) Changes of effective gene dispersal distances by pollen and seeds across successive life stages in a tropical tree. Oikos 122:1616–1625
Birky CW, Maruyama T, Fuerst P (1983) An approach to population and evolutionary genetic theory for genes in mitochondria and chloroplasts, and some results. Genetics 103:513–527
Bonnefille R, Umer M (1994) Pollen-inferred climate fluctuations in Ethiopia during the last 3000 years. Palaeogeogr Palaeoclimatol Palaeoecol 109:331–343
Borsch T, Hilu KW, Quandt D, Wilde V, Neinhuis C, Barthlott W (2003) Non-coding plastid trnT-trnF sequences reveal a well resolved phylogeny of basal angiosperms. J Evol Biol 16:558–576
Cavers S, Munro RC, Kadu CAC, Konrad H (2009) Transfer of microsatellite loci for the tropical tree Prunus africana (Hook.f.) Kalkman. Silvae Genet 58:276–279
Chester M, Cowan RS, Fay MF, Rich TCG (2007) Parentage of endemic Sorbus L. (Rosaceae) species in the British Isles: evidence from plastid DNA. Bot J Linn Soc 154:291–304
Cipriani G, Lot G, Huang WG, Marrazzo MT, Peterlunger E, Testolin R (1999) AC/GT and AG/CT microsatellite repeats in peach [Prunus persica (L) Batsch]: isolation, characterization and cross-species amplification in Prunus. Theor Appl Genet 99:65–72
Clement M, Posada D, Crandall KA (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9:1657–1660
Coetzee JA (1967) Pollen analytical studies in East and southern Africa. Palaeoecol Afr 3:1–146
Cunningham AB, Mbenkum FT (1993) Sustainability of harvesting prunus Africana bark in Cameroon: a medicinal plant in international trade. UNESCO, People and Plants Working Paper 2, Paris
Cunningham M, Cunningham AB, Schippmann U (1997) Trade in Prunus africana and the implementation of CITES. German Federal Agency for Nature Conservation, Bonn
Dawson I, Powell W (1999) Genetic variation in the Afromontane tree Prunus africana, an endangered medicinal species. Mol Ecol 8:151–156
Dawson I, Were J, Lengkeek A (2000) Conservation of Prunus africana, an over-exploited African medicinal tree. In: Palmberg-Lerche C, Hald S, Sigaud P (eds) Forest genetic resources, no. 28. FAO, Rome, pp 27–33
Dieringer D, Schlotterer C (2003) Microsatellite analyser (MSA): a platform independent analysis tool for large microsatellite data sets. Mol Ecol Notes 3:167–169
Earl DA, von Holdt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Ennos RA (1994) Estimating the relative rates of pollen and seed migration among plant populations. Heredity 72:250–259
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567
Farwig N, Braun C, Bohning-Gaese K (2008) Human disturbance reduces genetic diversity of an endangered tropical tree, Prunus africana (Rosaceae). Conserv Genet 9:317–326
Felsenstein J (1989) PHYLIP—Phylogeny Inference Package (version 3.2). Cladistics 5:164–166
George JP, Konrad H, Collin E, Thevenet J, Ballian D, Idzojtic M, Kamm U, Zhelev P, Geburek T (2015) High molecular diversity in the true service tree (Sorbus domestica) despite rareness: data from Europe with special reference to the Austrian occurence. Ann Bot 115:1105-1115
Goudet J (1995) FSTAT (version 1.2): a computer program to calculate F-statistics. J Hered 86:485–486
Hamilton AC (1982) Environmental history of East Africa—a study of the Quaternary. Academic Press, London
Hamilton AC, Taylor D (1991) History of climate and forests in tropical Africa during the last 8 million years. In: Myers N (ed) Tropical Forests and Climate. Springer, Dordrecht, pp 65–78
Hardy O, Vekemans X (2002) SPAGEDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes 2:618–620
Hewitt GM (1999) Post-glacial re-colonization of European biota. Biol J Linn Soc 68:87–112
Hurni H (1988) Degradation and conservation of the resources in the Ethiopian highlands. Mt Res Dev 8:123–130
Jimu LN (2011) Threats and conservation strategies for the African cherry (Prunus africana) in its natural range—a review. J Ecol Nat Environ 3:118–130
Jolly D, Taylor D, Marchant R, Hamilton A, Bonnefille R, Buchet G, Riollet G (1997) Vegetation dynamics in central Africa since 18,000 yr BP: pollen records from the interlacustrine highlands of Burundi, Rwanda and western Uganda. J Biogeogr 24:495–512
Kadu CAC, Schueler S, Konrad H, Muluvi GM, Eyog-Matig O, Muchugi A, Williams VL, Ramamonjisoa L, Kapinga C, Foahom B, Katsvanga C, Hafashimana D, Obama C, Geburek T (2011) Phylogeography of the Afromontane Prunus africana reveals a former migration corridor between East and West African highlands. Mol Ecol 20:165–178
Kadu CAC, Parich A, Schueler S, Konrad H, Muluvi GM, Eyog-Matig O, Muchugi A, Williams VL, Ramamonjisoa L, Kapinga C, Foahom B, Katsvanga C, Hafashimana D, Obama C, Vinceti B, Schumacher R, Geburek T (2012) Bioactive constituents in Prunus africana: geographical variation throughout Africa and associations with environmental and genetic parameters. Phytochemistry 83:70–78
Kadu CAC, Konrad H, Schueler S, Muluvi GM, Eyog-Matig O, Muchugi A, Williams VL, Ramamonjisoa L, Kapinga C, Foahom B, Katsvanga C, Hafashimana D, Obama C, Geburek T (2013) Divergent pattern of nuclear genetic diversity across the range of the Afromontane Prunus africana mirrors variable Pleistocene climate of African highlands. Ann Bot 111:47–60
Kalkman C (1965) The old world species of Prunus sub-genus Laurocerasus. Blumea 13:33–35
Koch MA, Kiefer C, Ehrich D, Vogel J, Brochmann C, Mummenhoff K (2006) Three times out of Asia Minor: the phylogeography of Arabis alpina L. (Brassicaceae). Mol Ecol 15:825–839
Livingstone DA (1975) Late Quaternary climatic change in Africa. Ann Rev Ecol Syst 6:249–280
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
Muchugi A, Lengkeek G, Agufa C, Muluvi G, Njagi E, Dawson I (2006) Genetic variation in the threatened medicinal tree Prunus africana in Cameroon and Kenya: implications for current management and evolutionary history. S Afr J Bot 72:498–506
Nei M (1972) Genetic distance between populations. Am Nat 106:283–292
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590
Ohta S, Nishitani C, Yamamoto T (2005) Chloroplast microsatellites in Prunus, Rosaceae. Mol Ecol Notes 5:837–840
Owen RP, Barthelme JW, Renault RW, Vincens A (1982) Palaeolimnology and archaeology of Holocene deposits north-east of Lake Turkana, Kenya. Nature 298:523–529
Palma-Silva C, Lexer C, Paggi GM, Barbara T, Bered F, Bodanese-Zanettini MH (2009) Range-wide patterns of nuclear and chloroplast DNA diversity in Vriesea gigantea (Bromeliaceae), a neotropical forest species. Heredity 103:600–612
Peakall R, Smouse PE (2012) GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28:2537–2539
Poelchau MF, Hamrick JL (2012) Differential effects of landscape-level environmental features on genetic strcutre in three co-distributed tree species in Central America. Mol Ecol 21:4970–4982
Pons O, Petit RJ (1995) Estimation, variance and optimal sampling of gene diversity. I. Haploid locus. Theor Appl Genet 90:462–470
Pons O, Petit RJ (1996) Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics 144:1237–1245
Pritchard JK, Stephens P, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Reusing M (2000) Change detection of natural high forests in Ethiopia using remote sensing and GIS techniques. International Archives of Photogrammetry Remote Sensing XXXIII (Part B7):1253–1257
Sosinski B, Gannavarapu M, Hager LD, Beck LE, King GJ, Ryder CD, Rajapakse S, Baird WV, Ballard RE, Abbott AG (2000) Characterization of microsatellite markers in peach Prunus persica (L.) Batsch. Theor Appl Genet 101:421–428
Street-Perrott FA, Perrott RA (1993) Holocene vegetation, lake levels and climate of Africa. In: Wright HE, Kutzbach JE, Webb T, Ruddiman WF, Street-Perrott FA, Bartlein PJ (eds) Global climate since the Last Glacial Maximum. University of Minnesota Press, Minneapolis, pp 318–356
Teketay D, Granström A (1995) Soil seed banks in dry Afromontane forests of Ethiopia. J Veg Sci 6:777–786
van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) Micro-Checker: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Vaughan SP, Russell K (2004) Characterization of novel microsatellites and development of multiplex PCR for large-scale population studies in wild cherry, Prunus avium. Mol Ecol Notes 4:429–431
Vincens A (1986) Diagramme pollinique d’un sondage Pleistocene superieur—Holocene du Lac Bogoria (Kenya). Rev Palaeobot Palynol 47:169–192
Vincens A, Buchet G, Servant M, Barton C, Ngos S, Nguetsop F, Servant-Vildary S, Tchotsoua M (2010) Vegetation response to the “African Humid Period” termination in Central Cameroon (7°N)—new pollen insight from Lake Mbalang. Clim Past 6:281–294
Vinceti B, Loo J, Gaisberger H, van Zonneveld MJ, Schueler S, Konrad H, Kadu CAC, Geburek T (2013) Conservation priorities for Prunus africana defined with the aid of spatial analysis of genetic data and climatic variables. PLoS One 8:e59987
White F (1993) Refuge theory, ice-age aridity and the history of tropical biotas: an essay in plant geography. Fragm Florist Geobot Suppl 2:385–409
Yineger H, Schmidt DJ, Hughes JM (2014) Genetic structuring of remnant forest patches in an endangered medicinal tree in north-western Ethiopia. BMC Genet 15:31
Acknowledgments
We thank all persons involved in carrying out the sample collections and Thomas Thalmayr for his technical assistance during the laboratory analysis and with creating figures. This research was done with financial contribution of the following institutions: Addis Ababa University and Bahir Dar University, Ethiopia; BFW: Federal Research and Training Centre for Forests, Natural Hazards and Landscape, Department of Forest Genetics, Austria; and Bioversity International, Italy.
Data archiving statement
All data will be deposited in DRYAD on acceptance.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by W. Ratnam
Rights and permissions
About this article
Cite this article
Mihretie, Z., Schueler, S., Konrad, H. et al. Patterns of genetic diversity of Prunus africana in Ethiopia: hot spot but not point of origin for range-wide diversity. Tree Genetics & Genomes 11, 118 (2015). https://doi.org/10.1007/s11295-015-0945-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-015-0945-z