Abstract
Human activities such as fragmentation and selective logging of forests can threaten population viability by modification of ecological and genetic processes. Using six microsatellite markers, we examined the effects of forest fragmentation and local disturbance on the genetic diversity and structure of adult trees (N = 110) and seedlings (N = 110) of Prunus africana in Kakamega Forest, western Kenya. Taking samples of adults and seedlings allowed for study of changes in genetic diversity and structure between generations. Thereby, adults reflect the pattern before and seedlings after intensive human impact. Overall, we found 105 different alleles in the six loci examined, 97 in adults and 88 in seedlings. Allelic richness and heterozygosity were significantly lower in seedlings than in adults. Inbreeding increased from adult tree to seedling populations. Genetic differentiation of adult trees was low (overall F ST = 0.032), reflecting large population sizes and extensive gene flow in the past. Genetic differentiation of seedlings was slightly higher (overall F ST = 0.044) with all of the 28 pairwise F ST-values being significantly different from zero. These results suggest that human disturbance in Kakamega Forest has significantly reduced allelic richness and heterozygosity, increased inbreeding and slightly reduced gene flow in P. africana in the past 80–100 years.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Tropical ecosystems suffer profoundly as a result of human over-exploitation (Chapin et al. 2000). Extensive deforestation, habitat fragmentation and local disturbance, for example through selective logging, threaten the survival of species (Aldrich and Hamrick 1998). Moreover, fragmentation and local disturbance have been shown to modify ecological and genetic processes (Young et al. 1996; Chapin et al. 2000; Keller and Waller 2002; Luck et al. 2003).
Fragmentation and local disturbance, i.e. selective logging, reduce the local population size of tree species and therefore, their genetic diversity. If the populations become subdivided and gene flow is restricted, genetic drift and inbreeding may lead to genetic differentiation (Young et al. 1996; Keller and Waller 2002). Decreased genetic diversity may imperil the adaptive potential of populations and, as a consequence, their fitness (McLaughlin et al. 2002). To understand the effects of human activity on genetic diversity as well as to develop conservation strategies it is crucial to analyse the levels and patterns of genetic structure in fragmented and locally disturbed populations (Rossetto et al. 2004).
Studies on effects of fragmentation for the genetic structure of trees often revealed decreasing allelic richness and heterozygosity as well as increasing differentiation among populations as a result of fragmentation or local disturbance (reviewed in Lowe et al. 2005). Most of these studies have been conducted across space comparing differently fragmented populations of adult trees. For example, Hall et al. (1994) found limited gene flow in the tree Pentaclethra macroloba among sites in a fragmented terrain compared to sites within a continuous forest reserve. Prober and Brown (1994) were able to link levels of genetic variation to patch size in Eucalyptus albens.
Due to the longevity of trees, we can gain a better understanding of patterns in gene flow by including not only adults but also seedlings or saplings into the study. In case fragmentation and local disturbance had taken place in between the time periods when adult trees and seedlings got established, patterns of genetic variation in adults might reflect genetic diversity and gene flow before human impact on the forest and in seedlings after human impact. In this case allelic richness and gene flow in adult trees might have been high, whereas genetic diversity and gene flow in the seedling generation could already be reduced.
The studies investigating the genetic structure of tropical tree populations from a multistage perspective including adults and seedlings show ambiguous results. For example, significant inbreeding and genetic differentiation among patches were recorded for the tree Symphonia globulifera only in the seedling stage and in fragmented forest patches but not for adults and not in a continuous rain forest area in Costa Rica (Aldrich et al. 1998). Also, a study on Carapa guianensis revealed lower allelic richness and greater genetic distances among the sapling cohort than the adult population in an isolated managed forest (Dayanandan et al. 1999). Particularly for juveniles, fragmented forests tend to lose allelic richness and heterozygosity (Rossetto et al. 2004; Fernandez-M and Sork 2007). In contrast to these findings, high levels of allelic richness and heterozygosity were observed in all stage classes and study sites despite intensive logging in the past for Swietenia macrophylla (Cespedes et al. 2003). Also, Cloutier et al. (2007) recorded similar levels of gene diversity and allelic richness before and after logging in both the adult and the seed generations of C. guianensis.
Most of the studies that compared the genetic structure of adults and seedlings were based on unequal number of samples per stage class, i.e. several seedlings per adult tree. It is therefore difficult to compare these stage classes as the measure used to describe the genetic diversity and structure of populations depends on the number and spatial pattern of the sampled individuals (Lowe et al. 2005). Thus, taking a larger number of seedlings than of adults and calculating allelic richness is expected to yield higher values for seedlings, because a larger number of sampled individuals is expected to sample just by chance also more alleles.
To avoid an unequal number of samples per stage class, we collected adult trees and seedlings in pairs to obtain a spatially matched random sample of each generation. Our study species was P. africana (Rosaceae), a widespread commercially used tree species in the Afrotropics. Wild populations of P. africana have been declining over much of their geographical range due to over-exploitation of their medicinally valuable bark (Cunningham and Mbenkum 1993). P. africana showed low levels of heterozygosity in Kenya compared for instance to Ethiopia (H E = 0.067, Dawson and Powell 1999). The only rainforest in Kenya that still harbours high-population densities of P. africana is Kakamega Forest, but in the recent past the species has declined rapidly (Fashing 2004). Kakamega Forest consists of a continuous forest block and several fragments and its disturbance history is well documented (Mitchell 2004). Therefore, Kakamega Forest is an ideal study area to examine whether forest fragmentation and local disturbance through selective logging affect the genetic diversity and gene flow of adult trees and seedlings of P. africana. We assume that adults represent the historical genetic structure in Kakamega Forest, possibly only slightly influenced by starting forest fragmentation and local disturbance, whereas the genetic structure of seedlings should already be affected by human modification of the forest (Mitchell 2004). We compared the genetic diversity and structure of adult trees and seedlings using six microsatellite markers.
Methods
Study species
Prunus africana Hook f. (Rosaceae) is an evergreen tree species native to Africa and Madagascar typical of mature climax forests (Kalkman 1965). The monoecious tree grows up to 40 m in height producing small white flowers in elongated clusters, which are basically insect pollinated (Hall et al. 2000). It has purple fleshy one-seeded fruits (seeds (mean ± SD): diameter: 6.6 ± 0.5 mm, mass: 0.15 ± 0.05 g, N = 30), which are dispersed by birds and monkeys (Farwig et al. 2006). P. africana belongs to the group of species which has been intensively logged in all parts of Kakamega Forest (Mitchell 2004, see below).
Study site
The study was conducted in Kakamega Forest, western Kenya (between latitudes 0°14′ and 0°21′N and longitudes 34°47′ and 34°48′E). Kakamega Forest is the eastern most relict of the Guineo-Congolian rainforest belt and lies at an altitude of 1,500–1,700 m asl (Kokwaro 1988). It is a highly fragmented and locally disturbed montane tropical rainforest (Tsingalia 1990; Mitchell 2004). Average monthly temperature of the forest lies between 10.6 and 27.7°C (Tsingalia 1990). Annual precipitation averages 1,960 mm and is highly seasonal with a rainy season from April to November and a short dry season from December to March (averaged from Forest Department records at Isecheno Forest Station from 1982 to 2005).
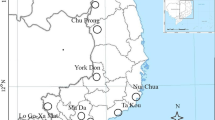
The indigenous forest cover of Kakamega has been reduced from 23,785 ha in 1933 to 13,990 ha in about 1990 (Blackett 1994). Most clear-felling was conducted in the southern part of the forest and along the western fringe close to the city of Kakamega (Mitchell 2004). Clear-felling has not only destroyed a quarter of the forest but also resulted in fragmentation of the forest (Fig. 1). Presently, the forest covers an area of 8,537 ha of a main forest block surrounded by five forest fragments of various sizes (132–1,370 ha). Malava and Kisere are the northern fragments and Ikuywa, Yala and Kaimosi are the southern fragments (Fig. 1). The fragments Malava and Kisere in the north have most likely been connected by riparian forest corridors with the other parts of the forest in historical times (Mitchell 2004, but see Brooks et al. 1999). In contrast, the fragment Kaimosi in the south was separated from the main forest between 1913 and 1959 and the fragments Yala and Ikuywa have been separated since the early 1960s (Mitchell 2004). Selective logging of trees has taken place in all parts of the forest, which has reduced the local population size of P. africana (Fashing 2004; Mitchell 2004). The forest is surrounded by a densely settled agricultural area (600 people/km2; KIFCON 1994) which is composed of small patches of fields planted with sugarcane, maize, beans or tea interspersed by single large forest trees and small shrubs. Further information on the fragmentation and disturbance history of Kakamega Forest is given in Tsingalia (1990), Mitchell (2004) and Bleher et al. (2006).
Plant material
In June 2002, we collected leaf material of adult trees and seedlings of P. africana in eight different sites of Kakamega Forest, four sites in the main forest block (two in the north and two in the south) and four in fragments (Malava, Kisere, Ikuywa and Kaimosi) (Fig. 1). In each site we randomly sampled on average 14 pairs of adult trees (range 9–17; 80–100 years old) and seedlings (1–5 years old). For each adult tree we collected one seedling within a radius of 10 m from the tree trunk. By sampling adults and seedlings in pairs we obtained spatially matched, random samples of each generation per site. In total, 110 adult and 110 seedling individuals were sampled. Considering the age of the adult trees, we assume that adult trees had been established before the two southern fragments Ikuywa and Kaimosi were separated among themselves and from the main forest. Note that the northern fragments, Malava and Kisere and the main forest most likely have been connected via riparian forest when the adult trees were established as seedlings. In contrast, the seedlings became established during the present distribution of forest (Fig. 1). Plant material was silica-dried in the field. For statistical analyses, each site was defined as a single population.
Molecular analysis
Leaves were ground to a fine powder in liquid nitrogen using a mortar and pestle. DNA was extracted from the powder of ∼1.0 g leaf material of P. africana, following the protocol described in DNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA). The standard protocol was slightly modified by using 500 μl buffer AP1 and 160 μl buffer AP2 which handled the amount of leaf material better. DNA was stored at −20°C in AE elution buffer.
We studied the levels of genetic variation using six microsatellite marker loci, four listed by Cipriani et al. (1999) and two listed by Sosinski et al. (2000). The forward and reverse primers for each microsatellite locus are presented in Table 1. Instead of radioactive labelling we used fluorescent 5′end-labelled primers (6-FAM, NED, HEX, Applied Biosystems, ABI, Foster City, CA, USA).
Polymerase chain reactions (PCR) were performed in a volume of 25 μl containing 10 mM Tris–HCl (pH 9.0), 50 mM KCl, 1.5–2.5 mM MgCl2, 4 × 0.2 mM2 dNTP, ∼1.5 U Taq polymerase (Ready-To-Go™ PCR-Beads, Amersham Pharmacia Biotech Inc., Chalfont, UK), primer ranging from 0.04 to 0.28 μM, respectively (Table 1), and 25 ng genomic DNA using the following temperature profile: 95°C for 5 min, then 35 cycles of (94°C for 45 s, 60–62°C for 45 s and 72°C for 45 s), finishing with 72°C for 8 min.
The labelled PCR products were separated on a 4.5%-polyacrylamide gel on an ABI 377 automated sequencer, and allele sizes were determined using genescan 2.1. and Genotyper 3.1 (ABI).
Data analysis
The mean number of alleles per locus and site were estimated from 9 to 17 individuals and therefore corrected for variation in sample size by using the rarefaction method implemented in hp-rare (Kalinowski 2005). Allele frequencies, the mean observed heterozygosity HO and the mean expected heterozygosity HE (Nei 1978) over all loci in both stage classes (adults, seedlings) were computed using tfpga (Tools for population genetic analysis, Miller 1997). Inbreeding was estimated as f = 1—HO/HE per population. Genotypic linkage disequilibria between pairs of loci and departures from Hardy–Weinberg equilibrium at each locus were tested using genepop 3.4 software (Raymond and Rousset 1995a). F-statistics for all loci was calculated using arlequin Version 3.11 (Excoffier et al. 2005).
To examine the relative partitioning of genetic variation within and among populations an analysis of molecular variance (AMOVA) was calculated with arlequin Version 3.11 (Excoffier et al. 2005). For each pair of populations we estimated pairwise F ST-values for population differentiation determined after 110 permutations (Raymond and Rousset 1995b). To test for isolation by distance (IBD) at the scale of populations, we regressed F ST/1-F ST, based on pairwise F ST-values among all sampled populations, against the log-transformed geographic distance separating the populations (Slatkin 1993; Rousset 1997). Statistical significance was evaluated with a Mantel-test using 110 permutations. Percentage of rare alleles, observed heterozygosity H O, expected heterozygosity H E and pairwise F ST-values were arcsine square root transformed prior to statistical analyses. As locus P1 caused most population-locus Hardy–Weinberg disequilibria we also performed all analyses without this locus. Excluding locus P1 from the analyses led to similar results. We therefore report only the results using all six loci.
Results
Descriptive population genetics of adults and seedlings
All of the 110 adult trees and 110 seedlings were unambiguously genotyped at the six microsatellites loci. The loci were highly polymorphic with 105 alleles in the six loci, 97 of them in adults and 88 of them in seedlings (mean number of alleles per locus over all populations 8.0 and 7.1, respectively). The number of alleles varied between populations ranging from 40 to 56 over six loci in adults and from 35 to 50 over six loci in seedlings. The mean number of alleles per population was significantly higher for adults than for seedlings (adults: 41.0, seedlings: 36.6, Wilcoxon Signed-Rank: Z 7 = 18, p = 0.008) (Table 2). The frequencies for individual alleles per population ranged from 0.029 to 0.640 in adults and from 0.029 to 0.750 in seedlings. Between 0 and 34% of the alleles per population in adults and 0 and 31% of the alleles in seedlings were rare alleles below the 5% threshold. The percentage of rare alleles per population did not differ between the stage classes (rare alleles: adults: mean 20%, seedlings: mean 18%, paired t-test: t 7 = 0.62, p = 0.55). Mean observed heterozygosity H O and mean expected heterozygosity H E per population were always higher in adults than in seedlings (H O: adults: mean 0.69, range = 0.53–0.83, seedlings: mean 0.54, range = 0.46–0.65, paired t-test: t 7 = 5.24, p = 0.0008; H E: adults: mean 0.80, range = 0.73–0.83, seedlings: mean 0.75, range = 0.71–0.80; paired t-test: t 7 = 6.11, p = 0.0003) (Table 2). Inbreeding per population increased significantly from the adult to the seedlings stage class (f: adults: mean 0.14, range = 0.01–0.35, seedlings: mean 0.29, range = 0.14–0.42; paired t-test: t 7 = 4.91, p = 0.0017) (Table 2).
Linkage and Hardy–Weinberg equilibrium of adults and seedlings
No linkage disequilibrium was detected between any pair of loci (non-significant after sequential Bonferroni correction, Rice 1989). Significant deviations from Hardy–Weinberg equilibrium were detected at the following population locus combinations (significant after sequential Bonferroni correction, Rice 1989): Adults: Malava-P1, Kisere-U3, Kisere-P1, Mukangu-P1, IsechenoB-P1, IsechenoA-P1, Kaimosi-P1; Seedlings: Malava-P2, Malava-P1, Kisere-U5, Kisere-P1, Mukangu-P2, Mukangu-P1, Buyangu-P2, Buyangu-U2, Buyangu-P1, IsechenoA-P1, Kaimosi-P1 (all heterozygosity deficits). These heterozygosity deficits might be caused by inbreeding but then a deficit would be expected for all loci which was not the case in our species. Alternatively the deficit could be explained by the lack of amplification of some alleles (null alleles) particularly in locus P1. This locus caused most population-locus Hardy–Weinberg disequilibria and showed the highest F IS-values (Table 3). Nevertheless, excluding locus P1 from the analyses led to similar results (see Methods).
Genetic structure of adults and seedlings
The results of the AMOVA revealed for adults as well as for seedlings that most of the genetic variation was found within populations (96.8 and 95.5%, respectively, Table 4). Variation among populations was 3.2% for adults and 4.4% for seedlings. The variance component among populations in both stage classes was highly significant after the permutation test (1,023 permutations) in the amova.
In adults significant differentiation was found in 23 of the 28 pairwise comparisons, and mainly between the southern populations (Table 5). In seedlings, all of the 28 pairs of populations were significantly different from zero. The frequency of significant pairwise F ST-values was significantly larger in seedlings than in adults (χ2-test: χ2-value = 7.42, df = 1, p = 0.006, Table 5).
The pairwise genetic variance among populations increased significantly from adults to seedlings (paired t-test: t 27 = -3.51, p = 0.0016) (Table 5, Fig. 2a, b). However, a test for IBD resulted in no correlation between the geographic and the genetic distance neither for adults (Pearson correlation: r = 0.13, Mantel p = 0.28, n = 8, Fig. 2a) nor for seedlings (Pearson correlation: r = 0.23, Mantel p = 0.12, n = 8, Fig. 2b).
Discussion
Genetic structure of adults
Our results showed high levels of allelic richness in populations of P. africana adults combined with high levels of heterozygosity indicating a predominantly outcrossing species. This corresponds to field observations demonstrating that self-pollinated flowers did not develop fruits (N. Farwig, own observations). The majority of tropical trees investigated to date appeared to be outcrossers with extensive gene flow (e.g. Doligez and Joly 1997; Cespedes et al. 2003; Fernandez-M. and Sork 2007). Prunus africana showed higher values of heterozygosity in the present study (H E = 0.73–0.83) than in an African-wide P. africana study (H E = 0.067 for Kenya; Dawson and Powell 1999). The lower value for Kenya could be explained by the fact that Dawson and Powell (1999) studied isolated, remnant trees in farmland around Mt. Kenya, which might have lost genetic diversity. Our results, therefore, suggest that P. africana populations in Kakamega Forest comprise a high amount of genetic diversity given its severe fragmentation and disturbance history.
Prunus africana exhibited weak genetic structure in adults (F ST = 0.032). Also other studies on tropical trees revealed rather low F ST-values ranging between 0.003 and 0.063 (e.g. Pacheco and Simonetti 2000; Cespedes et al. 2003). Low F ST-values are found for most tree species (Hamrick and Godt 1997). This can be explained by a combination of the life history traits (long-lived, woody plants) and breeding system (outcrossing insect-pollinated flowers, animal-dispersed seeds) of most studied trees (Hamrick and Godt 1997; Jordano and Godoy 2000). Trees have long generation cycles and overlapping generations fostering little or no genetic structure among populations (Knowles 1991; Xie and Knowles 1991). The majority of tropical rainforest tree species that are outcrossers showed extensive gene flow in terms of pollen and seed movement (Doligez and Joly 1997; Nason and Hamrick 1997). Extensive gene flow via seeds has, for instance, been demonstrated in Syzygium nervosum (Shapcott 1999) and P. mahaleb (Godoy and Jordano 2001).
The lack of genetic differentiation among populations in adults is congruent with the fragmentation and disturbance history of Kakamega Forest, which started about 100 years ago (Mitchell 2004). We assume that the establishment of adults took place when the forest was still contiguous, population size of P. africana was higher than today and extensive gene exchange could take place. Historic data suggest that the area to the north of Kakamega main forest was only thinly populated by humans about 100 years ago (Mitchell 2004). Furthermore, in these times, the northern parts of the main forest and the fragments Malava and Kisere were probably connected via riparian forest corridors and a multitude of trees (Mitchell 2004) that might have been stepping stones especially for birds acting as seed dispersers. In contrast, populations of the southern fragments are very different from the main forest, in particular the most southern population of Kaimosi, which differs from all other populations. This could be due to rigorous selective logging of P. africana trees in Kaimosi, which has removed the commercially most valuable individuals from the population (Fashing 2004; Mitchell 2004). This is supported by a low-mean number of alleles in the sampled adult trees (Table 2).
Comparison between adults and seedlings
Comparing genetic diversity between adults and seedlings showed that mean allelic richness, mean observed heterozygosity H O and mean expected heterozygosity H E decreased significantly from adults to seedlings. Moreover, inbreeding increased from the adult to the seedling stage class. These results could be caused by modifications in population sizes and genetic exchange between the two stage classes. Prunus africana has been heavily logged in the past 70 years (Mitchell 2004) and populations have further declined in the recent past (Fashing 2004). Loss of individual reproductive trees and subdivision of adult tree populations might have reduced genetic diversity in the seedling stage class through random genetic drift and inbreeding within subpopulations. Genetic drift and inbreeding can result in genetic differentiation among subpopulations if gene flow between these populations is restricted. Reduced gene flow between our studied populations may have increased the effects of genetic drift and therefore, the loss of genetic diversity in the seedling stage class.
High densities of conspecifics in the past might have promoted extensive gene flow since distances between tree individuals were smaller and might have been easily covered by pollinators and seed dispersers (Murawski and Hamrick 1992; Young et al. 1996 but see White et al. 2002). Reduced population densities due to human influence might have led to limited pollen and seed flow within populations of P. africana and therefore contributed to loss of genetic diversity. Moreover, distances of 4.5–10 km between the fragments in our study area might have reduced gene flow among populations and slightly increased the genetic differentiation in the seedling stage class. A relationship between limited seed dispersal and increased population genetic structure has been shown in a number of studies (Perry and Knowles 1991; Pacheco and Simonetti 2000; Ueno et al. 2000). For example, a comparative study on the tropical tree Inga ingoides revealed more differentiation among subpopulations where the spider monkey as seed disperser was absent than where it was present (Pacheco and Simonetti 2000).
The comparison between adults and seedlings revealed slightly higher genetic differentiation among populations in seedlings than in adults (F ST = 0.044 versus 0.032). Such a pattern has also been recorded for other tree species (Hamrick et al. 1993; Alvarez-Buylla et al. 1996). Genetic structure is likely to persist through the adult stage if mortality with age is random (Pacheco and Simonetti 2000). However it may become less evident if balancing selection favouring heterozygote survival takes place (Hamrick et al. 1993; Alvarez-Buylla et al. 1996). As we collected adults and seedlings in pairs and not a larger number of seedlings per adult we obtained a spatially matched random sample of each generation per site. Thus, with our sampling design it is rather likely that the higher genetic structure of seedlings will persist through the adult stage if mortality with age is random. If selection favouring heterozygote survival takes place the populations might become even more structured as thinning will most likely further reduce genetic diversity per site. We found neither for adult nor for seedling populations a significant correlation between the geographic and the genetic distance. However, pairwise genetic variance among populations increased significantly from adults to seedlings.
Our results showed that P. africana populations in Kakamega Forest still contain high levels of genetic diversity but recent forest fragmentation and selective logging led to reduction of allelic richness and heterozygosity in seedlings. This loss of genetic diversity within the younger generation could be caused by a combination of reduced population density of adult P. africana trees and restricted gene flow. Further studies measuring gene flow within and among populations directly will improve our understanding of the extent of gene flow in P. africana. This knowledge is essential for developing sound conservation and management strategies for tree populations at the landscape scale.
References
Aldrich PR, Hamrick JL (1998) Reproductive dominance of pasture trees in a fragmented tropical forest mosaic. Science 281:103–105
Aldrich PR, Hamrick JL, Chavarriaga P, Kochert G (1998) Microsatellite analysis of demographic genetic structure in fragmented populations of the tropical tree Symphonia globulifera. Mol Ecol 7:933–944
Alvarez-Buylla ER, Chaos A, Pinero D, Garay AA (1996) Demographic genetics of a pioneer tropical tree species: patch dynamics, seed dispersal and seed banks. Evolution 50:1155–1166
Blackett HL (1994) Forest inventory report no. 3: Kakamega Kenya indigenous forest conservation programme. Nairobi, Kenya
Bleher B, Uster D, Bergsdorf T (2006) Assessment of threat status and management effectiveness in Kakamega Forest, Kenya. Biodivers Conserv 15:1159–1177
Brooks TM, Pimm SL, Oyugi JO (1999) Time lag between deforestation and bird extinction in tropical forest fragments. Conserv Biol 13:1140–1150
Cespedes M, Gutierrez MV, Holbrook NM, Rocha OJ (2003) Restoration of genetic diversity in the dry forest tree Swietenia macrophylla (Meliaceae) after pasture abandonment in Costa Rica. Mol Ecol 12:3201–3212
Chapin FS, Zavaleta ES, Eviner VT et al (2000) Consequences of changing biodiversity. Nature 405:234–242
Cipriani G, Lot G, Huang WG, Marrazzo MT, Peterlunger E, Testolin R (1999) AC/GT and AG/CT microsatellite repeats in peach Prunus persica (L) Batsch: isolation, characterisation and cross-species amplification in Prunus. Theor App Genet 99:65–72
Cloutier D, Kanashiro M, Ciampi AY, Schoen DJ (2007) Impact of selective logging on inbreeding and gene dispersal in an Amazonian tree population of Carapa guianensis Aubl. Mol Ecol 16:797–809
Cunningham AB, Mbenkum FT (1993) Sustainability of harvesting Prunus africana bark in Cameroon. People and Plants Working Paper, 2. UNESCO, Paris
Dawson IK, Powell W (1999) Genetic variation in the Afromontane tree Prunus africana, an endangered medicinal species. Mol Ecol 8:151–156
Dayanandan S, Dole J, Bawa K, Kesseli R (1999) Population structure delineated with microsatellite markers in fragmented populations of a tropical tree, Carapa guianensis (Meliaceae). Mol Ecol 8:1585–1592
Doligez A, Joly HI (1997) Genetic diversity and spatial structure within a natural stand of a tropical forest tree species, Carapa procera (Meliaceae), in French Guiana. Heredity 79:72–82
Excoffier L, Laval G, Schneider S (2005) arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evol Bioinfor Online 1:47–50
Farwig N, Böhning-Gaese K, Bleher B (2006) Enhanced seed dispersal of Prunus africana in fragmented and disturbed forests? Oecologia 147:238–252
Fashing PJ (2004) Mortality trends in the African cherry (Prunus africana) and the implications for colobus monkeys (Colobus guereza) in Kakamega Forest, Kenya. Biol Conserv 120:449–459
Fernandez-M JF, Sork VL (2007) Genetic variation in fragmented forests stands of the Andean oak Quercus humboldtii Bonpl. (Fagaceae). Biotropica 39:72–78
Godoy JA, Jordano P (2001) Seed dispersal by animals: exact identification of source trees with endocarp DNA microsatellites. Mol Ecol 10:2275–2283
Hall JB, O’Brien EM, Sinclair FL (2000) Prunus africana: a monograph. School of Agricultural and Forest Sciences Publication Number 18. University of Wales, Bangor, pp 104
Hall P, Chase MR, Bawa KS (1994) Low genetic-variation but high population differentiation in a common tropical forest tree species. Conserv Biol 8:471–482
Hamrick JL, Godt MJW (1997) Effects of life history traits on genetic diversity in plant species. In: Silverton J, Franco M, Harper JL (eds) Plant life histories: ecology, phylogeny and evolution. Cambridge Royal Society, Cambridge, pp 102–118
Hamrick JL, Murawski DA, Nason JD (1993) The influence of seed dispersal mechanisms on the genetic structure of tropical tree populations. Vegetatio 107/108:281–297
Jordano P, Godoy JA (2000) RAPD variation and population genetic structure in Prunus mahaleb (Rosaceae), an animal-dispersed tree. Mol Ecol 9:1293–1305
Kalinowski ST (2005) hp-rare: a computer program for performing rarefaction on measures of allelic diversity. Mol Ecol Notes 5:187–189
Kalkman C (1965) The Old World species of Prunus subg. Laurocerasus, including those formerly referred to Pygeum. J. J. Groen & Zoon, Leiden
Keller LF, Waller DM (2002) Inbreeding effects in wild populations. Trends Ecol Evol 17:230–241
KIFCON (1994) Kakamega Forest. The official guide. (KIFCON) Kenya Indigenous Forest Conservation Programme. Nairobi, Kenya
Knowles P (1991) Spatial genetic structure within two natural stands of black spruce (Picea mariana (Mill.) B.S.P). Silvae Genet 40:13–19
Kokwaro JO (1988) Conservation status of the Kakamega Forest in Kenya: the easternmost relict of the equatorial rain forests of Africa. Monographs in Systematic Botany from the Missouri Botanical Garden 25:471–489
Lowe AJ, Boshier D, Ward M, Bacles CFE, Navarro C (2005) Genetic resource impacts of habitat loss and degradation; reconciling empirical evidence and predicted theory for neotropical trees, Heredity 95:255–273
Luck GW, Daily GC, Ehrlich PR (2003) Population diversity and ecosystem services. Trends Ecol Evol 18:331–336
McLaughlin JF, Hellman JJ, Boggs CL, Ehrlich PR (2002) Climate change hastens population extinctions. Proc Natl Acad Sci USA 99:6070–6074
Miller MP (1997) Tools for populations genetics analyses (TFPGA) 1.3. a Windows program for the analysis of allozyme and molecular population genetic data. Computer software distributed by author; http: //bioweb.usu.edu/mpmbio/tfpga.htm
Mitchell N (2004) The exploitation, disturbance history of Kakamega Forest, western Kenya. In: Bleher B, Dalitz H (eds) BIOTA-East Report No 1, vol 20. Bielefelder Ökologische Beiträge, Bielefeld
Murawski DA, Hamrick JL (1992) The mating system of Cavanillesia platanifolia under extremes of flowering-tree density: a test of predictions. Biotropica 26:141–159
Nason JD, Hamrick JL (1997) Reproductive and genetic consequences of forest fragmentation: Two case studies of neotropical canopy trees. J Hered 88:264–276
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590
Pacheco LF, Simonetti JA (2000) Genetic structure of a mimosoid tree deprived of its seed disperser, the spider monkey. Conserv Biol 14:1766–1775
Perry DJ, Knowles P (1991) Spatial genetic structure within three sugar maple (Acer saccharum Marsh.) stands. Heredity 66:137–142
Prober SM, Brown AHD (1994) Conservation of the grassy white box Woodlands - population- genetics and fragmentation of Eucalyptus albens. Conserv Biol 8:1003–1013
Raymond M, Rousset F (1995a) Genepop (version 1.2), population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Raymond M, Rousset F (1995b) An exact test for population differentiation. Evolution 49:1280–1283
Rice WR (1989) Analyzing tables of statistical tests. Evolution 43:223–225
Rossetto M, Jones R, Hunter J (2004) Genetic effects of rainforest fragmentation in an early successional tree (Elaeocarpus grandis). Heredity 93:610–618
Rousset F (1997) Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 145:1219–1228
Shapcott A (1999) Vagility and the monsoon rain forest archipelago of northern Australia: Patterns of genetic diversity in Syzygium nervosum (Myrtaceae). Biotropica 31:579–590
Slatkin M (1993) Isolation by distance in equilibrium and non-equilibrium populations. Evolution 47:264–279
Sosinski B, Gannavarapu M, Hager LD et al (2000) Characterization of microsatellite markers in peach Prunus persica (L.) Batsch. Theor App Genet 101:421–428
Tsingalia MH (1990) Habitat disturbance, severity and patterns of abundance in Kakamega Forest, Western Kenya. Afr J Ecol 28 213–226
Ueno S, Tomaru N, Yoshimaru H, Manabe T, Yamamoto S (2000) Genetic structure of Camellia japonica L. in an old-growth evergreen forest, Tsushima, Japan. Mol Ecol 9:647–656
White GM, Boshier DH, Powell W (2002) Increased pollen flow counteracts fragmentation in a tropical dry forest: an example from Swietenia humilis Zuccarini. Proc Natl Acad Sci USA 99:2038–2042
Xie C-Y, Knowles P (1991) Genetic substructure within natural populations of jack pine (Pinus banksiana Lamb.). Can J Bot 69:547–551
Young A, Boyle T, Brown T (1996) The population genetic consequences of habitat fragmentation for plants. Trends Ecol Evol 11:413–418
Acknowledgements
We thank the Kenyan Ministry for Education & Research for research permission, the KWS and FD for granting us access to their reserves, Jasper M. Kirika for help with field work and Bärbel Bleher for support. We are grateful to D. G. Berens, T. Diegisser, E. M. Griebeler, J. Johannesen, F. A. Voigt and two anonymous reviewers for comments on earlier drafts of the manuscript. Financial support was provided by the BMBF (Biota East Africa 01LC0025/01LC0405).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Farwig, N., Braun, C. & Böhning-Gaese, K. Human disturbance reduces genetic diversity of an endangered tropical tree, Prunus africana (Rosaceae). Conserv Genet 9, 317–326 (2008). https://doi.org/10.1007/s10592-007-9343-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-007-9343-x