Abstract
A network of 92 pedigreed ex situ conservation plantings of Pinus tecunumanii, established as replicated progeny within provenance trials, is used to present a principal components-based analysis that illustrates the climatic preferences of 23 populations from the species’ native range. This meta-analysis quantifies changes in the relative productivity, assessed as individual-tree volume, of populations across climatic gradients and associates the preference of a population with increased volume production along the climatic gradient. Clustering and ordination on the matrix containing estimates of change in productivity for each population summarise differentials in productivity associated with climatic gradients. The preference of populations along principal components therefore reflects the adaptive profiles of populations, which may be used with breeding-value estimates from routine genetic evaluations to assist with the development of deployment populations targeting different environments. As well, the approach may be used to test whether the preference of a population, estimated as population loadings for growth differentials, is affected by the climate in the native range of the population. This relationship may be interpreted as an estimate of how much local climate shapes the adaptive profiles of populations. The amount and seasonality of precipitation most clearly differentiate the adaptive profiles of populations, with less variation in the population responses explained by temperature differentiation. As expected from type-B correlation estimates, most populations exhibited small changes in relative productivity across climatic gradients. However, patterns of similarities in adaptive profiles among populations were evident using spatial orientation to display population responses to the climatic variables experienced in the provenance trials. Clustering and ordination of population responses derived from empirical data served to identify populations that responded positively or negatively to climatic variables; this information may help guide conservation genetics efforts, direct the deployment of germplasm, or identify seed sources that are sensitive to changes in climatic variables. Linking response patterns to the climatic data from the native range of each population indicated little effect of local climate shaping adaptive profiles.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pinus tecunumanii is a medium to large tree that is native to most of Central America and Chiapas, Mexico (Perry 1991). Seeds collected from more than 1,500 individual trees within the species’ native range by the Camcore program (Dvorak et al. 2000) have been established in ex situ genetic conservation areas in the form of pedigreed provenance trials in Argentina, Brazil, Colombia, Kenya, Mozambique, South Africa and Venezuela. These trials are used to evaluate the productivity of different populations, families and individuals within populations. Results of provenance performance as related to overall productivity were summarised by Hodge and Dvorak (1999; 2012) in order to guide conservation and breeding strategies. In the present study, productivity data from a subset of well-tested populations were extracted and analysed to identify patterns of productivity change across the climate gradients sampled by the various field trials in order to develop adaptation profiles for populations of this pine.
The species is widely distributed and includes populations that are vulnerable to over-exploitation or loss from natural disasters. These populations have been evaluated in field trials established across a wide range of environments; however, there is uncertainty about which population will perform best when planted into production forests on contrasting sites or when reintroduced into the species’ native range. This uncertainty arises because of changes in the performance of populations and families across trials due to genotype by environment interactions (GxE). While traditional genetic analyses are used to identify which populations or families are best across sites, understanding changes in performance across environmental gradients is of increasing interest (Hodge and Dvorak 2004; Costa e Silva et al. 2006; Baltunis and Brawner 2010; Cullis et al. 2010; Brawner et al. 2012; Dutkowski and Potts 2012).
The paper describes a process for identifying the environmental variables that are associated with changes in the productivity of P. tecunumanii that may be useful for directing the deployment of germplasm, identifying patterns in population responses to climatic variables, informing the development of species distribution models and guiding gene conservation efforts. An important feature of the method is that it abstracts performance away from the specific trials that have sampled the target planting environment so that changes in productivity are associated with the environmental variables used to classify the trials. This allows for the portrayal of changes in productivity across climatic variables that classify the landscape rather than changes in productivity across a specific to sets of trials. This facilitates an alternative portrayal of GxE that is associated with the environmental variables used to characterise all planting locations rather than individual trial environments. Presentation of results may have to be carried out across grossly imbalanced data sets, and care must therefore be taken in the interpretation of the results of this type of meta-analysis. A clear understanding of the distribution of populations across trial networks and genetic connectivity across trials is required if results are used to assist with the interpretation of species distribution models (Estes et al. 2013) or understanding how the impact of environmental drivers differs within physiological models of plant growth (Almeida et al. 2010). Concisely describing similarities in how populations perform across climatic gradients relative to one another using a set of P. tecunumanii provenance trials is the primary objective of this work.
A highly condensed summary of analysis results typically generated for comparison of populations across environments, such as for average productivity, is produced with clustering and ordination in the form of a preference analysis (Carroll 1972). Climate data of trials included in this study is used with estimates of the relative productivity of populations in these trials to produce an alternative form of the traditional biplot that is typically used to classify patterns of GxE in terms of differences in performance across trials (Gabriel 1971; DeLacy et al. 1996; Hardner et al. 2010). Data from many trials is used to place well-tested populations within the space of the environmental variables driving the interactions rather than the space of the trial network describing the environmental variables. The main contrast with GxE biplots that place field trials within the space of variable populations is the use of environmental variables to define the space that classifies changes in population productivity. The clustering and ordination methodology described below has been detailed in Brawner et al. (2013) along with the strengths and weaknesses of the process.
Materials and methods
This study extends the results derived from the provenance trial network described by Hodge and Dvorak (2012), which provides detailed descriptions of the populations sampled from these provenances, the trials involved in the network, data standardisation processes and the statistical methods. Briefly, data were standardised by dividing all observations by the individual tree phenotypic standard deviation, followed by adding a constant (100 %) and multiplying by another constant (coefficient of variation) prior to generating least-square estimates for each population in each trial where effects were estimable (Eisen and Saxton 1983). Estimates from an across-sites analysis of standardised trial data removed the environmental main effects in order to isolate the genetic and GxE effects used to illustrate productivity differentials among populations with respect to climatic variables. Species distribution models with maps encompassing the testing environments presented by Leibing et al. (2009) provide background for the discussion of potential productivity in present and alternative climate scenarios.
Populations and field trials
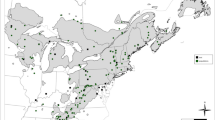
Of the 48 P. tecunumanii populations established in field trials, the 23 populations described in Fig. 1 were assessed for diameter and height 5 years after planting and were also present in eight or more of the field trials where at least three native-range population samples were evaluated in each trial. Particulars of these populations are shown in Table 1. Data from all P. tecunumanii trials represented in Fig. 2 were combined to produce the least-square estimates of individual tree volume for each population at each location as detailed in Hodge and Dvorak (2012). Subsequent to the original collections of P. tecunumanii in Mexico, the Juquila source was re-classified as an atypical form of Pinus pringleii/Pinus herrerae possibly resulting from historic introgression of several closed-cone pines (Dvorak et al. 2001) but was still included in the study for demonstrative purposes.
Location of well tested P. tecunumanii provenances established in the trial network with numbers referring to Table 1 codes
P. tecunumanii may be subdivided into broad ecotypes based on morphological and molecular information which is apparently influenced by the altitude of their occurrence in Central America and Southern Mexico (Dvorak 1986), i.e. high- and low-elevation populations. The two ecotypes are conceptually treated as separate breeding populations with high- and low-elevation ecotypes described in Dvorak et al. (2000). These populations were planted in disconnected sets of field trials. Differences in the testing environment experienced by high- and low-elevation ecotypes are demonstrated in Supplementary Figs. 1, 2, 3, 4 and 5, and the impact on inferences that may be made across high and low populations are discussed below.
In this study, climatic data for the native-range populations described in Fig. 1 and trials described in Fig. 2 were derived from the Bioclim global climate data set (Hijmans et al. 2005). No environmental data on soil, aspect, or pest or disease occurrence were used, largely due to the difficulty of obtaining consistent information. Environmental variables were compiled from the high resolution (30″ or approximately 1-km resolution) Bioclim dataset using R (Bivand et al. 2013). The Bioclim dataset, generated by interpolation of long-term climate data, is described with the environmental variables listed in Table 2 to clarify abbreviations used in figures.
Preference analysis, methodology and interpretation
Climatic variables associated with trials were used to examine changes in the productivity of each population across climate gradients. For any specific experiment, the estimate of productivity for any population is interpreted as a deviation about the overall mean, related to population (G) and population by environment interactions (GxE). The extent of change in each population’s estimate of productivity across the range of the variables sampled by that population in these trials is defined as the correlation (r) between the population estimates and the climate variables that were used to describe the growing conditions of each trial environment. A separate correlation was estimated for each population using the population’s productivity estimate as the dependent variable and the Bioclim variables associated with each trial as independent variables. These correlations (Supplementary Table 1) were then subjected to clustering and ordination to describe patterns in change across the Bioclim-derived environmental gradients. Cluster analyses used Ward’s minimum variance method, while ordination used conventional principal component analysis of unscaled and centred variables to produce rotated variables for presentation of loadings and scores. Clustering and ordination was completed using the matrix comprised of the population preferences (Supplementary Table 1) using the hclust and prcomp function, respectively, in the Stats package of the R statistical software environment (R Development Core Team 2013). Clusters were considered to be different if the percent of variation explained by an additional cluster accounted for more than 50 % of the unexplained variance using the k-means algorithm. Clustering results are included in the preference analysis-based biplot by altering font and colour for clusters of environmental scores and altering line types for clusters of population loadings.
These changes in productivity across climate gradients reflect preference in the analysis of Carroll (1972), and preference is herein associated with increased productivity along a climate gradient. The preference analysis approach differs from those typically used for the depiction of GxE as estimated from multi-environment trials (Basford et al. 1996; DeLacy et al. 1996; Yan and Hunt 2002). The biplot presented herein provides an indication of preference for higher levels of the certain environmental variables with populations spanning different environmental gradients depending on what environments were experienced in field trials (see Supplementary Figs. 1, 2, 3, 4 and 5). The method uses replicated field trials to isolate genetic responses rather than using phenotypic responses in natural stands so that the environmental main effect may then be removed from the population comparisons. Classification based on correlations \( \left(r=\frac{ dy}{ dx}\right) \) that indicate change in productivity (y) across a climate gradient (x) makes references to changes in population productivity across climate gradients possible. However, this also means no information on the absolute genetic merit of the populations or the effect of environmental variables is conveyed in the resulting biplot; other types of genetic analyses readily provide that information.
Within the biplot, objects (populations) are compared with respect to variables (Bioclim environmental variables) so that provenance loadings are displayed as vectors from the origin to the population code, and scores are displayed as Bioclim variable names (Kroonenberg 2008). The relative proximity of populations and environmental variables in this presentation provides an intuitive way to associate descriptors with population loadings located near each other implying similarity of preferences or tightly clustered environmental scores representing variables that elicit similar preference patterns across populations. Using this categorisation of scores and loadings, the length of the species’ vectors indicates the extent of principal component (PC) fit, and the direction of the vector indicates the preference of a given population for the environmental variables associated with this PC. Preference (as reflected by increased individual tree volume) increases as the vector moves from the origin towards any given environmental score. The score for each environmental variable may be orthogonally projected onto any population vector, and the environmental variable that projects furthest along a vector in the direction in which it points is the variable which the population prefers most.
Quantifying among-population comparisons in terms of productivity changes across climatic variables from this principal component analysis provides greater precision in describing adaptive patterns. For among-population comparisons, the cosine of the angle between two vectors approximates the correlation between the response patterns of the two populations across all environmental gradients. The dot product of the PC1 and PC2 loadings for two populations provides an estimate of correlation with the cosine of the angle between the two vectors in degrees equal to the correlation coefficient (r). An acute angle between two vectors implies similar preferences while a 90° angle implies a correlation of zero or no preference. To facilitate presentation, scores and loadings were rescaled by multiplying each point by the reciprocal of the maximum score or loading so that each point is presented as a percentage of the maximum score or loading.
To gain insight into the ability to predict populations’ sensitivity to changes in environmental variables using climate data from their native habitat, linear models were used to relate populations’ loadings for growth differentials to the climatic conditions in that populations’ native range. The significance of the correlation (r i where i indexes 19 Bioclim variables) between the native range climate, and the PC loadings for growth differentials may be formally tested. This was undertaken for both PC1 and PC2 axes represented in the biplot as PC1 was primarily associated with precipitation variables, and PC2 was primarily associated with temperature variables.
Results
Interpretations of the preference analysis are described to clarify the inferences that may be made using the cluster diagrams of Fig. 3 and the biplot of Fig. 4, which depict similarities of the regression of population relative productivity across Bioclim variables. The cluster diagram presented in the right of Fig. 3 identifies Jocón (8) and Cabricán (1) as the least dissimilar populations, followed by a larger cluster containing Mountain Pine Ridge (14) and El Carrizal (5). Interestingly, Jocón, Cabricán and Mountain Pine Ridge appear to have higher levels of introgression with Pinus oocarpa than the other populations evaluated in this study (Dvorak and Raymond 1991). Similar inferences may be made using the cluster diagram for environmental variables presented on the left of Fig. 3. In the case of climate variables, isothermality and the precipitation of the wettest month and wettest quarter are readily partitioned.
Clustering of changes of P. tecunumanii productivity elicited by changes in Bioclim-derived environmental variables showing similarities in environmental variable (Table 2 code) effects on populations (left) and similarities in population (Table 1 code) responses induced by environmental variables (right). The green line delineates clusters of populations (numbered in Table 1) differentiated by line type and clusters of environmental variables differentiated by font and greyscale in Fig. 4 (Colour figure online)
Biplot depicting relationships between P. tecunumanii populations’ (Table 1 codes) responses (vectors) elicited by changes in Bioclim-derived environmental variables (Table 2 codes) (centre of climate variable name) with four clusters of population scores distinguished by style of line, populations from the low-elevation ecotype in boldface and three clusters of environmental loadings distinguished by greyscale
The groupings in these cluster diagrams are annotated in the biplot (Fig. 4), with shades of grey differentiating climatic variable clusters and line types differentiating population clusters. As 84 % of the variation in productivity changes across the climatic variables explained by the first two principal components, little is to be gained from interrogating additional principal components (PC3 explained 9 % of the variation, details not shown). Differences in results from Ward’s clustering algorithm and the PCA analysis are evident; for example, Jocon and Cabrican cluster closely together but occupy different spaces in the biplot.
Figure 4 allows for inference about the impact of climatic variables on changes in population performance as well as the responses of populations to climatic variables. The clearest finding is the separation of environmental scores related to precipitation along the first principal component axis, showing the strongest contrast between seasonality of precipitation and precipitation in the driest month. The proximity of the environmental variables that explain the greatest changes in rankings (precipitation in the driest quarter and precipitation in the driest month) demonstrates the similarity in their impact on changes in population performance and the ability of PCA to display co-linearity in descriptors. The climatic scores related to temperature variables are somewhat less clearly differentiated along the second principal component axis. As well, a relationship between the temperature and precipitation variables is apparent, where variables associated with lower rainfall are also associated with higher temperatures. In addition to the proximity of climatic variables to one another, the proximity of climatic variables to the origin is informative. Climatic variables near the origin produce smaller changes in the relative performance of population than those that are located at a greater distance from the origin, such as precipitation in the wettest quarter or precipitation in the wettest month.
Inferences about the importance of environmental variables for particular populations may be quantified by projecting climatic variables onto a population vector. The relative importance of each climatic variable in bringing about changes in relative performance may be demonstrated using the Juquila (9) vector, where the projection of precipitation in the driest month (the most influential environmental variable) is slightly further along the vector than precipitation in the driest quarter. This vector represents Juquila’s adaptation profile with respect to all climatic variables.
Further inferences that may be made about populations relate to the nearness of populations, which reflects both similarities in their pattern of change and stability with proximity to the origin reflecting a lack of predictable change in relative performance, i.e. an average response due to the regression slopes being near to zero. Populations close to the origin are relatively stable and do not have predictable response patterns. For example, San Esteban (16) is located closest to the origin and may therefore be regarded as a stable population with little change in performance induced by climatic variables. The opposite may be inferred for Juquila (9), which is the population with the greatest vector length and therefore the population that exhibits the greatest predictable changes in relative productivity induced by climatic variables. Quantifying the similarities in population responses to environmental variables is possible using the angle between population vectors. For example, the Juquila and (9) Mountain Pine Ridge (14) responses to environmental variables are negatively correlated (r = −0.92) while the El Carrizal (5) and Mountain Pine Ridge responses are quite similar with respect to all variables (r = 0.94).
While identifying populations that respond positively or negatively to changes in climate variables may be used to guide deployment of a priori tested populations, it would be useful if this approach could be extended to identify the climatic variables that shape populations’ adaptive profiles in the species’ native range. Significant relationships between Bioclim climate data from the native range and the loadings of these populations in trials may be interpreted as adaptation associated with local climate. When quantified as the correlation (r i ) between population loadings for growth differentials and the climate experience in the native range, formal tests of significance may be used to identify climate variables that are associated with preference. For PC1, a positive regression may be interpreted as an association between native-range climatic variables and preferences for higher precipitation or lower seasonality of precipitation and lower temperatures. None of the regressions indicated that there was a significant association between responsiveness to environmental variables and native range climate (details not shown). While the preference of populations was not significantly associated with native range climate, the climate variables that provide a contrast along PC1 showed weak associations with population preference (annual precipitation p = 0.14 and precipitation seasonality p = 0.16). These associations may be interpreted as follows: (1) populations derived from areas of high annual precipitation present a negative response pattern when rainfall increases, and (2) populations derived from areas with high precipitation seasonality respond positively to increased rainfall.
Discussion
A selection of well-tested populations from a network of P. tecunumanii field trials was analysed so that patterns of genetic response to changes in environmental variables could be visualised. Similar to findings for eucalypt trials (Brawner et al. 2013), pine population changes over precipitation gradients accounted for a much greater proportion of the variation in productivity differentials than changes over temperature gradients. This indicates rainfall is the principal driver of change in the relative performance of populations. For example, a population such as Juquila responds positively as precipitation increases and negatively as precipitation decreases, demonstrating a positive preference for higher precipitation. In contrast, populations like Chanal and El Carrizal are relatively less productive as precipitation increases demonstrating an aversion to higher precipitation. This specific example emphasises that comparisons among population loadings for change across climatic gradients are made relative to one another with environmental main effects removed; it is not suggested that the absolute productivity of these populations would increase as precipitation decreases. Similar examples may be developed for temperature gradients using the second principal component.
Interpretations of the influence of a climatic variable on the relative productivity of P. tecunumanii populations should be visually apparent in the biplot as it spatially represents similarities in climatic variables and population responses to these climatic variables. This allows for the interpretation of all environmental variables and populations simultaneously. For example, the climatic variables associated with precipitation were tightly clustered to the right of the biplot indicating they have similar effects on the populations’ performance. Conversely, the Jocón population is spatially separated from other populations, indicating that Jocón is distinct in its response to the environmental variables. Verifying the genetic differences among populations with DNA-based approaches provides a way of quantifying similarities among populations so that these differences in preference would be associated with more accurate population classifications. Combining molecular-based phylogenies with populations selected as genetically distinct may provide a means for targeting populations with specific adaptation profiles for use in landscape genomic studies (Sork et al. 2013).
While it is possible to make comparisons among all population preferences simultaneously, care must be taken to constrain inferences to make it clear that they are estimates from populations that were evaluated within a specific climatic space. For example, Fig. 4 provides comparisons among populations across the two ecotypes even though these populations are experimentally disconnected; they were not tested in common trial and each ecotype was targeted to environments that were more similar to where they were sourced from (Supplementary Figures 1, 2, 3, 4 and 5). Defining the climate envelope that each population experienced in the field trials provides the environmental boundaries that must be used to constrain inferences on preference. Estimates of preference across a climatic gradient should be paired with a description of the trial environments as these environments define the climates that the population has experienced. In a similar manner, inferences on rotation-age adaptability are extrapolations and should refer to the early development of P. tecunumanii plantations given that data comes from 5-year-old assessment. Given the high age-age correlations and moderately high site-site correlations for pine growth traits, extrapolating temporally is likely to be less problematic than extrapolating environmentally. Survival is a trait that in under genetic control and clearly reflects adaptability that this was not accounted for in this study. One option would be to use total volume in the row plots to provide a better indication of overall adaptability, with mean annual increment calculated from later-age block plots in replicated trials providing the most robust estimates of genetic change associated with to adaptability (Brawner et al. 1999; Stanger et al. 2010). In any case, this specific analysis is interpreted with selection of 5-year-old individual tree volume as the phenotype of interest.
As well as displaying similarities or differences among populations and environmental variables, the location of populations with respect to the environmental variables provides an indication of how they are inter-related. Further information can be gained by inspecting the distance of populations or climatic variables from the origin with relatively responsive populations positioned far from the origin and relatively unresponsive populations such as San Esteban (16), San Vincente (21), San Jeronimo (18) and San Lorenzo (19) located near the origin. Interestingly, this similarity in stability is reflected by geographical proximity with the San Vincente, San Jeronimo and San Lorenzo populations located along the same mountain ridge and the first two populations separated by just 10 km (Fig. 1). Climate variables located nearer the origin, such as precipitation in wettest quarter and precipitation in the wettest month, are those that produce the least change among the populations.
Application in the conservation and use of forest genetic resources
Understanding populations’ responsiveness to climatic variables may be used to prioritise conservation efforts in view of a changing climate (Hamann and Aitken 2013). The preference of populations may be used to target resources to conserve germplasm from populations that are close to their environmental limits (Benito-Garzón et al. 2013; Razgour et al. 2013). For example, the low-elevation populations of La Esperanza (10) and San Francisco (17) are oriented away from environmental variables associated with higher temperatures, indicating these populations prefer cooler temperatures more than the other populations. The negative responses to increased temperature may be avoided if the populations were to migrate to higher elevations. However, stands of individuals within these populations that are already located near the tops of hills or ridges may require assisted migration sooner than populations that are insensitive to temperature increases to ensure rare alleles contained within these stands may be conserved (Ponce-Reyes et al. 2013). Mitigating the risks of population losses or reductions in genetic diversity from lowland attrition or range-shift gaps from upward range shifts may be possible if a populations’ climatic preferences are understood (Kreyling et al. 2010).
A real-world application of this method may be found in the development of P. tecunumanii planted forests in an area where the species has never been grown, using seed from selections made from field trials established in South Africa. Planted forests are a recent development on the Lichinga plateau in Mozambique, an area characterised by a long and hot dry season marked by a high seasonality in precipitation. A natural first step in the domestication process for this new environment is to take seed from the populations that have been shown to be highly productive in South Africa, such as Montebello (13) and San Francisco (17). Referring to the biplot in Fig. 4, it is clear that neither Montebello nor San Francisco prefer higher seasonality of precipitation, as the vectors for both populations are nearly at 90° angles from the seasonality of precipitation climate variable. However, Montebello responds positively to higher temperatures, and San Francisco responds positively to higher precipitation in the driest quarter. While both populations are of a similar genetic merit, this evidence would suggest that Montebello may provide a more robust choice. On the other hand, El Carrizal (5) and Mountain Pine Ridge (14) are two populations with estimates of genetic merit close to the species mean and may therefore be passed over using breeding-value analyses. Inspection of Fig. 4 would indicate that these populations are relatively more productive when the seasonality of precipitation and seasonality of temperatures are high, which suggests that selections from within these populations could be used to increase the resilience of planted forests established where large fluctuations of precipitation and temperature are typical. Combining information from standard genetic analyses and the biplot approach also identifies Yucul (23) as a population of interest since it combines a high across-sites genetic merit and moderate increases in relative productivity when planted in environments where the seasonality of precipitation is high.
Climatic preferences are not predictable using climate data from the native habitat
This study could not find significant evidence to support the concept that populations that are more or less reactive to environmental change may be identified using the climate of their native habitat. This could support the hypothesis that many tree species have native ranges that reflect genetic changes during the Pleistocene or some other climatic period (Crisp et al. 2004) rather than the present climate. While this may initially be counter-intuitive, three points may be considered to support this finding: the first relates to the representativeness of current climate data compared to the climate experienced over time period required to develop adaptive differences, the second is associated with the precision and level of sampling of genetic and environmental variables and the third relates to the pine mating system. Firstly, when one considers the time scales that are required for pine populations to migrate, it is evident that the climatic trajectory that was followed during the process of producing genetically distinct populations within P. tecunumanii is likely to have had a stronger influence than the 10 to 50 years represented by the Bioclim variables (Franklin 2010). Secondly, while populations of P. tecunumanii differ significantly in their productivity, there is also genetic variation within populations. Downscaling coarse climatic data should provide a more precise classification of environments (Storlie et al. 2013) for investigating relationships between climatic preference and current climate and may be required to provide further support for the concept of local environment shaping environmental preferences (Gillingham et al. 2012; Blanquart et al. 2013). Thirdly, knowledge of the highly outcrossing mating system and extremely long distance pollen transfer that is possible in P. tecunumanii would facilitate a wide mixing of alleles among and within populations with wide-crossing effectively reducing the fixation of adaptive alleles that would lead to distinct adaptation strategies developing divergent populations.
Conclusions
The preference-based biplot presented herein provides an alternative view of the adaptive features of populations with respect to the environmental variables that characterise experimental locations. The main strength of presenting GxE interactions as they are displayed in Fig. 4 is the abstraction away from location-specific data to the climatic variables that drive changes in performance. While useful, this methodology requires further information for breeding climate-resilient crops, which is readily retrieved from standard genetic analyses. Nevertheless, these combined analyses would be useful for the development of different breeds with adaptive patterns tailored to distinct environments.
For the populations investigated in this study, variables related to precipitation were able to explain a large proportion of the variation in climatic preferences compared to variables related to temperature. Predictable changes in population productivity across climatic gradients or genotype by environmental variable interactions are effectively presented in a concise manner using this preference analysis approach. Providing a direct link between changes in productivity among populations evaluated in field trial networks and the native population climatic variables associated with these changes may be used to identify populations that merit conservation efforts, to direct the deployment of germplasm for existing replanting programs and to identify germplasm suitable for future and alternative climatic conditions.
Using population classifications derived from genomic data rather than locality of seed collection provides an alternative means of evaluating the manner in which environment shapes genetic variation. Testing the significance of associations between molecular-based phylogenies or haplotypes and changes in relative productivity across environmental gradients provides a means for identifying the genomic regions involved in regulating the environmental responses of germplasm. This may lead to a more refined understanding of how changes in climatic variables will differentially impact the genetic composition of our forests.
References
Almeida AC, Siggins A, Batista TR, Beadle C, Fonseca S, Loos R (2010) Mapping the effect of spatial and temporal variation in climate and soils on Eucalyptus plantation production with 3-PG, a process-based growth model. For Ecol Manag 259:1730–1740
Baltunis BS, Brawner JT (2010) Clonal stability in Pinus radiata across New Zealand and Australia. I. Growth and form traits. New For 40:305–322
Basford KE, Williams ER, Cullis BR, Gilmour A (1996) Experimental design and analysis of variety trials. In: Adaptation P, Improvement C (eds) Cooper M & Hammer GL). CAB International Wallingford, Oxon, pp 125–138
Benito-Garzón M, Ruiz-Benito P, Zavala MA (2013) Interspecific differences in tree growth and mortality responses to environmental drivers determine potential species distributional limits in Iberian forests. Glob Ecol Biogeogr 22:1141–1151
Bivand R., Keitt T. & Rowlingson B. (2013). rgdal: Bindings for the Geospatial Data Abstraction Library. R package, version 0.8-9
Blanquart F, Kaltz O, Nuismer SL, Gandon S (2013) A practical guide to measuring local adaptation. Ecol Lett 16:1195–1205
Brawner JT, Carter DR, Huber DA, White TL (1999) Projected gains in rotation-age volume and value from fusiform rust resistant slash and loblolly pines. Can J For Res 29:737–742
Brawner JT, Meder R, Dieters M, Lee DJ (2012) Selection of Corymbia citriodora for pulp productivity. South For 74:121–131
Brawner JT, Lee DJ, Meder R, Almeida AC, Dieters MJ (2013) Classifying genotype by environment interactions for targeted germplasm deployment with a focus on Eucalyptus. Euphytica 191:403–414
Carroll JD (1972) Individual differences and multidimensional scaling. Seminar Press, New York
Costa e Silva J, Potts BM, Dutkowski GW (2006) Genotype by environment interaction for growth of Eucalyptus globulus in Australia. Tree Genet Genomes 2:61–75
Crisp MD, Cook LG, Steane DA (2004) Radiation of the Australian flora: what can comparisons of molecular phylogenies across multiple taxa tell us about the evolution of diversity in present-day communities? Phil Trans R Soc Lond B 359:1551–1571
Cullis BR, Smith AB, Beeck CP, Cowling WA (2010) Analysis of yield and oil from a series of canola breeding trials. Part II. Exploring variety by environment interaction using factor analysis. Genome 53:1002–1016
DeLacy IH, Basford KE, Cooper M, Bull JK, McLaren CG (1996) Analysis of multi-environment trials - an historical perspective. In: Adaptation P, Improvement C (eds) Cooper M & Hammer GL). CAB International Wallingford, Oxon, pp 39–124
Development Core Team R (2013) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Dutkowski GW, Potts BM (2012) Genetic variation in the susceptibility of Eucalyptus globulus to drought damage. Tree Genet Genomes 8:757–773
Dvorak WS (1986) Provenance/progeny testing of Pinus tecunumanii. Proc IUFRO Breeding theory, progeny testing and seed orchard management, Williamsburg, pp 299–309
Dvorak WS, Raymond RH (1991) The taxonomic status of closely related closed cone pines in Mexico and Central America. New For 4:291–307
Dvorak W, Hodge G, Gutiérrez E, Osorio L, Malan F, Stanger T (2000) Pinus tecunumanii. Conservation and testing of tropical and subtropical forest tree species by the CAMCORE Cooperative. College of Natural Resources, North Carolina State University, Raleigh
Dvorak WS, Jordan AP, Romero JL, Hodge GR, Furman BJ (2001) Quantifying the geographic range of Pinus patula var. longipenduculata in southern Mexico using morphological and RAPD marker data. South Afr For J 192:19–30
Eisen EJ, Saxton AM (1983) Genotype by environment interactions and genetic correlations involving two environmental factors. Theor Appl Genet 67:75–86
Estes LD, Bradley BA, Beukes H, Hole DG, Lau M, Oppenheimer MG, Schulze R, Tadross MA, Turner WR (2013) Comparing mechanistic and empirical model projections of crop suitability and productivity: implications for ecological forecasting. Glob Ecol Biogeogr 22:1007–1018
Franklin J (2010) Moving beyond static species distribution models in support of conservation biogeography. Divers Distrib 16:321–330
Gabriel KR (1971) Biplot graphic display of matrices with application to principal component analysis. Biometrika 58:453–467
Gillingham PK, Huntley B, Kunin WE, Thomas CD (2012) The effect of spatial resolution on projected responses to climate warming. Divers Distrib 18:990–1000
Hamann A, Aitken SN (2013) Conservation planning under climate change: accounting for adaptive potential and migration capacity in species distribution models. Divers Distrib 19:268–280
Hardner CM, Dieters M, Dale G, DeLacy I, Basford KE (2010) Patterns of genotype-by-environment interaction in diameter at breast height at age 3 for eucalypt hybrid clones grown for reafforestation of lands affected by salinity. Tree Genet Genomes 6:833–851
Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A (2005) Very high resolution interpolated climate surfaces for global land areas. Int J Climatol 25:1965–1978
Hodge GR, Dvorak WS (1999) Genetic parameters and provenance variation of Pinus tecunumanii in 78 international trials. For Genet 6:157–180
Hodge GR, Dvorak WS (2004) The CAMCORE international provenance/progeny trials of Gmelina arborea: genetic parameters and potential. New For 28:147–166
Hodge GR, Dvorak WS (2012) Growth potential and genetic parameters of four Mesoamerican pines planted in the Southern Hemisphere. South For 74:27–49
Kreyling J, Wana D, Beierkuhnlein C (2010) Potential consequences of climate warming for tropical plant species in high mountains of southern Ethiopia. Divers Distrib 16:593–605
Kroonenberg PM (2008) Applied multi-way data analysis. Wiley, Hoboken
Leibing C, Zonneveld M, Jarvis A, Dvorak W (2009) Adaptation of tropical and subtropical pine plantation forestry to climate change: Realignment of Pinus patula and Pinus tecunumanii genotypes to 2020 planting site climates. Scand J For Res 24:483–493
Perry JP (1991) The pines of Mexico and Central America. Timber Press, Portland
Ponce-Reyes R, Nicholson E, Baxter PWJ, Fuller RA, Possingham H (2013) Extinction risk in cloud forest fragments under climate change and habitat loss. Divers Distrib 19:518–529
Razgour O, Juste J, Ibáñez C, Kiefer A, Rebelo H, Puechmaille SJ, Arlettaz R, Burke T, Dawson DA, Beaumont M, Jones G (2013) The shaping of genetic variation in edge-of-range populations under past and future climate change. Ecol Lett 16:1258–1266
Sork VL, Aitken SN, Dyer RJ, Eckert AJ, Legendre P, Neale DB (2013) Putting the landscape into the genomics of trees: approaches for understanding local adaptation and population responses to changing climate. Tree Genet Genomes 9:901–911
Stanger TK, Galloway GM, Retief ECL (2010) Final results from a trial to test the effect of plot size on Eucalyptus hybrid clonal ranking in coastal Zululand, South Africa. South For 73:131–135
Storlie CJ, Phillips BL, VanDerWal JJ, Williams SE (2013) Improved spatial estimates of climate predict patchier species distributions. Divers Distrib 19:1106–1113
Yan W. & Hunt L.A. (2002). Biplot anlaysis of multi-environment trail data. In: Quantitative Genetics, Genomics and Plant Breeding, CAB International (ed. Kang MS), pp. 289-303
Acknowledgments
We would like to thank the CSIRO Climate Adaptation Flagship’s Adaptive Primary Industries, Enterprises and Communities research theme for supporting the development of this work. The insightful comments from the associate editor and Tree Genetics and Genomics editorial review process, as well as those from CSIRO Plant Industry colleagues David Bush and Scott Chapman greatly improved this manuscript. We are also grateful for the support of all Camcore members who have managed the program over the past 35 years, particularly in those organisations that actively manage the field trials and conservation parks of Pinus tecunumanii.
Data Archiving Statement
Data required to reproduce any estimates or figures provided in this manuscript is provided in the Supplementary Materials attachment to this manuscript. The main figures in the manuscript (3 and 4) describing the clustering and ordination are readily reproduced with a call to the R functions using the options identified in the text based on the matrix provided in Supplementary Table 1. This table contains the correlation coefficients estimated from the population within trial estimates meeting trial and population minimum specifications. Specifications required for inclusion in the meta-analysis are provided in the methods. Detailed results from populations (Table 1) involved in the trial network are provided in Southern Forests—Growth potential and genetic parameters of four Mesoamerican pines planted in the Southern Hemisphere 2012, GR Hodge and WS Dvorak doi: 10.2989/20702620.2012.686192 #.Uomk1CcdPK0. All climatic data associated with native range or trial locations used for this study is stored on the Bioclim website http://www.worldclim.org/bioclim. Table 1 describes the location of the native range populations (Fig. 1) evaluated in the trial network. The weather variables used to compare the environmental coverage of each population across the trial network (Fig. 2) are summarised in Supplementary Figures 1, 2, 3, 4 and 5.
No genomic data was used in this study.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by R. Burdon
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 1614 kb)
Rights and permissions
About this article
Cite this article
Brawner, J.T., Hodge, G.R., Meder, R. et al. Visualising the environmental preferences of Pinus tecunumanii populations. Tree Genetics & Genomes 10, 1123–1133 (2014). https://doi.org/10.1007/s11295-014-0747-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-014-0747-8