Abstract
Genetics of control mechanisms that underlies sex differentiation in date palm is not known. Sex of the plants becomes known only at the time of first flowering, which takes around 5 years. In comparison, molecular diagnosis (if available/feasible) promises quick and reliable identification of sex types very early when plantlets are growing in seedbeds. To develop such an assay, genomic DNA from 45 individual plants (25 female and 20 male) belonging to different varieties of date palm was subjected to PCR amplification using 100 random amplified polymorphic DNA (RAPD) and 104 intersimple sequence repeat (ISSR) primers. Initially, two bulk genomic DNA samples (each made by pooling DNA from ten male and female plants, separately) were used. A primer showing sex-specific band in bulked samples was further used for amplification of the genomic DNA of the individual samples of that bulk. Only one RAPD primer, OPA-02, amplified a fragment of ~1.0 kb in all the individual samples of male genotypes, whereas this fragment was absent in all the female genotypes. This male-specific fragment was cloned and sequenced (GenBank accession no. JN123357), and a sequence-characterized amplified region (SCAR) primer pair was designed that amplified a 406-bp fragment in both female and male genotypes and a unique fragment of 354 bp in only male genotypes. The SCAR marker was further validated using 25 female and ten male date palm plants belonging to different varieties collected from different locations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Date palm (Phoenix dactylifera L.) is a long-living, dioecious evergreen fruit tree (2n = 36) cultivated mainly in North Africa, South Asia, the USA, and Australia. Date palm is an important plantation crop due to the high nutritional quality of its fruit. Like all other dioecious plants, female date palm plants produce fruits and male plants are used for pollination only. Since sexuality cannot be distinguished prior to floral initiation, identification of sex in date palm at an early stage is of utmost importance from the commercial point of view as only about 8–10 % of male plants are required for pollination. Excess males if weeded out at early stages, can avoid wastage of important plantation resources. In absence of information on chromosomal and genetic basis of sex determination in date palm, molecular marker-based identification of male and female genotypes is worth exploring, which promises fast and reliable approach to sex the plants regardless of reproductive age.
A number of sex-specific molecular markers have been identified in several dioecious plants like Pistacia vera (Hormaza et al. 1994), Actinidia chinensis (Gill et al. 1998), Salix viminalis (Gunter et al. 1998), Piper longum (Banerjee et al. 1999), Myristica fragrans (Shibu et al. 2001), Actinidia deliciosa (Shirkot et al. 2002), Cannabis sativa (Torjek et al. 2002), Mercurialis annua (Khadka et al. 2002), Calamus simplicifolius (Yang et al. 2005), Carica papaya (Parasnis et al. 2000; Deputy et al. 2002; Gangopadhyay et al. 2007), and Simmondsia chinensis (Agrawal et al. 2007; Agarwal et al. 2011; Ince et al. 2010). In date palm, some putative sex-linked random amplified polymorphic DNA (RAPD) and intersimple sequence repeat (ISSR) markers have earlier been reported (Singh et al. 2006; Younis et al. 2008), but these could not be verified on use of more number of genotypes. Recently, Mohsenzadeh and Pasalari (2010) have reported one RAPD marker, which produced a male plant-specific 520-bp fragment in “Zamardan,” an Iranian date palm cultivar.
The sequence-characterized amplified region (SCAR) markers, generally developed from RAPD and ISSR fragments, are locus-specific and more reliable and more reproducible for molecular identification (Paran and Michelmore 1993). SCAR markers linked to sex-specific genes have been developed in many dioecious plants like C. papaya (Bedoya and Nunez 2007), P. vera L. (Yakubou et al. 2005), and Cycas circinalis (Gangopadhyay et al. 2007), and Ginkgo biloba L. (Liao et al. 2009). In the present study, we have developed a sex-specific SCAR marker for reliable sexing of date palm plants.
Material and methods
Plant material
Identification and development of sex-specific marker/s
For identification and development of sex-specific marker(s), young leaf samples were collected from fully mature trees of a total of 45 genotypes of P. dactylifera L. (ESM_Table S1) that comprised 20 male genotypes of unknown origin and 25 female genotypes belonging to seven different commercially important varieties [“Hillawi” (H1–H8), “Zaidi” (ZA1–ZA4), “Khadrawy” (KH1 and KH2), “Zaglool” (ZG1 and ZG2), “Shamran” (SH1), “Egypt” (EG1), and “Medjool” (MD1)] as well as some female plants of unknown origin (UF1–UF6). All trees are growing at the Horticulture Farm Area, Chaudhary Charan Singh Haryana Agricultural University, Hisar, Haryana, and these represent some of the commerecial cultivars introduced from the USA, Pakistan, and Middle Eastern countries, which harbored considerable genetic diversity (Mitra et al. 2011).
Validation of sex-specific SCAR marker
Young leaf samples collected from 25 female plants and ten male plants belonging to different varieties growing at Date Palm Research Centre, Swami Keshwanand Rajasthan Agricultural University, Bikaner, Rajasthan, India (ESM_Table S2) were used for validation of sex-specific SCAR marker. In addition, young leaves were also collected from ten date palm seedlings (grown in pots in a net house at the Horticulture Farm Area, Chaudhary Charan Singh Haryana Agricultural University, Hisar, Haryana) for prediction/possible future use.
Genomic DNA isolation and PCR amplification
Genomic DNA was isolated from young leaf samples using cetyltrimethylammonium bromide (CTAB) extraction method of Murray and Thompson (1980), as modified by Saghai-Maroof et al. (1984) and by Xu et al. (1994). Isolated DNA samples were dissolved in TE buffer (10 mM tris–HCl and 1 mM ethylenedinitrilo-tetraacetic acid (EDTA), pH 8.0) and their quality/quantity was determined by visual assessment and band intensity comparisons with standard uncut lambda DNA (Fermentas, USA) on 0.8 % agarose gel made/run in 1× TBE buffer (89 mM Tris base, 89 mM Boric acid, 2 mM EDTA, pH 8.0). All DNA samples were diluted to a concentration of ~50 ng/μl using TE buffer and stored at −20 °C.
Since sex was the target trait, two bulked DNA samples were prepared by pooling an equal amount (50 ng) of genomic DNA from ten male and ten female plants, separately. A total of 100 RAPD primers (Operon Technologies Inc., USA) and 104 ISSR primers (University of British Columbia, USA; synthesized locally by Sigma-Aldrich Chemical Pvt. Ltd., New Delhi, India) were used to amplify the bulked DNA samples. On the basis of clear, scorable, and reproducible amplification banding patterns, 37 RAPD and 53 ISSR primers were selected for individual genotype analysis (ESM_Table S3). Polymerase chain reaction (PCR) amplifications were carried out using MyCycler Thermal Cycler (Bio-Rad Laboratories Ltd., UK).
Each reaction for RAPD-PCR (20 μl) consisted of 1× PCR buffer, 250 μM dNTPs (Fermentas, USA), 0.25 μM primer, 1.5 mM MgCl2, 1.5 U Taq DNA polymerase (Bangalore Genei Ltd., India), and 50 ng of template DNA. PCR amplifications were performed as follows: an initial denaturation step of 4 min at 94 °C, followed by 40 three-step amplification cycles, each comprising of 1 min at 94 °C, 1 min at the annealing temperature (36 °C), and 2 min extension at 72 °C. A final extension step at 72 °C for 8 min was included.
Each ISSR-PCR reaction (10 μl) contained 1× PCR buffer, 500 μM dNTPs (Fermentas, USA), 0.5 μM primer, 3.0 mM MgCl2, 1.5 U Taq DNA polymerase (Bangalore Genei Ltd., India), and 25 ng of template DNA. PCR amplification was carried out as follows: an initial denaturation step of 4 min at 92 °C, followed by 40 cycles of 1 min at 92 °C, annealing for 1 min at 36–66 °C (depending upon the guanine-cytosine (GC) content of the primer), and extension at 72 °C for 2 min, with a final extension step at 72 °C for 8 min.
All PCR-amplified DNA fragments were resolved by electrophoresis in submerged horizontal agarose gels (1.5 % w/v), stained with 0.5 μg ml−1 ethidium bromide, visualized, and photographed using the AlphaDigiDoc Pro™ documentation system (Alpha Innotech Corp., San Leonardo, CA, USA).
Data analysis
The RAPD and ISSR primer-generated banding profiles of the two bulked DNA samples were compared to identify presence of amplified fragment(s) that were apparently sex-specific. The RAPD/ISSR primers amplifying sex-specific bands (ESM_Table S4) were tested further on the individual samples that were used to prepare the male (M1–M10) and female (F1–F10) DNA bulks (ESM_Table S1) and, finally, validated by testing on male and female date palm plants (ESM_Table S2) that were not used earlier.
Cloning and sequencing of male-specific RAPD marker
The ~1,000-bp putative male-specific fragment amplified by RAPD marker, OPA-02, was eluted from 1.5 % agarose gel using QIAquick gel extraction kit (Qiagen Inc., USA) and cloned into pJET1.2 plasmid using CloneJET™ PCR cloning kit (Fermentas, USA). Cloned DNA fragment was amplified using universal primers, and the resulting amplicon was sequenced directly for both strands after ExoSAP-IT clean up kit (USB Corporation, USA) on the automated sequencer ABI-3730 (Applied Biosystems, USA), and the sequence was deposited in GenBank (accession no. JN123357).
SCAR primer designing and PCR amplification using the SCAR primers
SCAR primer pair (sequence shown in Table 1) was designed (patent filed; application no. 1513/DEL/2010 dated 29/6/10) based on the sequence of male-specific RAPD marker using the Primer-BLAST tool (http://www.ncbi.nlm.nih.gov/tools/primer-blast/). This primer pair was tested on DNA of male and female date palm genotypes using PCR amplification conditions as follows: 94 °C for 4 min, 35 cycles at 94 °C for 1 min, at the annealing temperature of 55 °C for 1 min and 72 °C for 2 min, and a final extension at 72 °C for 8 min. This thermal profile was standardized according to the specific T m of the primer pair. PCR reactions were carried out in 20 μl of the reaction mix containing 1× PCR buffer, 250 μM dNTP mix (Fermentas, USA), 0.125 μM each of forward/reverse primer, 1.5 mM MgCl2, 1.5 U Taq DNA polymerase (Bangalore Genei Ltd., India), and 50 ng of template DNA. Amplified DNA fragments were resolved on 1.5 % TBE agarose gel, stained with ethidium bromide, and visualized under ultra violet (UV) light.
Results
Identification of putative sex-specific marker
Six RAPD primers and ten ISSR primers showed differences in banding pattern of amplified fragments in the bulked DNA samples of male and female genotypes (ESM_Table S4). Subsequently, when tested further on individual male and female samples, only one RAPD primer, OPA-02, amplified a male-specific fragment of 1.0 kb consistently (Fig. 1), while all other primers amplified the putative sex-specific fragments in some of the genotypes but not exclusively in any one sex. Thus, only the OPA-02-derived male-specific fragment was selected for further studies, i.e., cloning/sequencing and conversion to SCAR marker.
RAPD profiles of male and female date palm genotypes generated using Operon primer OPA-02. The arrow indicates the male-specific putative fragment of ~1.0 kb. Lanes F1–F25 individual female genotypes, M1–M20 individual male genotypes, Bf bulked genomic DNA of female genotypes, Bm bulked genomic DNA of male genotypes, M 200-bp DNA ladder
Sequence analysis of OPA-02-derived putative male-specific fragment
The complete generated sequence of putative male-specific RAPD fragment was 1,031 bp in length (Fig. 2) having a GC content of 34.5 % (A = 338, C = 212, G = 144, T = 337) (Fig. 2). The sequence (GenBank accession number JN123357) didn't show homology to any database sequence on BLASTn search (http://www.ncbi.nlm.nih.gov/Blast.cgi). Similarly, the sequence was also searched for putative protein coding domains using open reading frame (ORF) finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html), which indicated relatively short four open reading frames of sizes 111 bp (1–111 bp), 117 bp (108–224 bp), 171 bp (556–726 bp), and 213 bp (818–1,030 bp). These ORFs, when compared with protein sequences available in the NCBI database using nBLASTp, revealed no homology for two of the ORFs (111 and 171 bp), while the other two ORFs showed partial similarity to database sequences, i.e., 39 % similarity of ORF of 117 bp (coding for 38 aa) with hypothetical protein ARALYDRAFT_908343 (Arabidopsis lyrata subsp. Lyrata) and 40 % similarity in the case of ORF of 213 bp (coding for 70 aa) with bacterial hypothetical proteins F7308_0042 (Francisella sp. TX077308), Fphi_0775 (Francisella philomiragia subsp. Philomiragia), and DESPIG_01975 (Desulfovibrio piger ATCC 29098).
Conversion of RAPD marker into SCAR marker
This primer pair designed from the sequenced RAPD fragment, when tested on individual samples, resulted in an amplicon of 406 bp in both female and male samples, but interestingly, also gave a unique amplicon of 354 bp only in male samples (Fig. 3), thus indicating it to be an informative male-specific SCAR marker. To further validate the results, the SCAR primer pair was also tested on another 25 female and ten male plant samples belonging to different varieties collected from Date Palm Research Centre, Swami Keshwanand Rajasthan Agricultural University, Bikaner. Significantly, the marker correctly differentiated all the male samples (characterized by the additional amplicon of ~300 bp) from the female samples (Fig. 4). We have also tried to predict the sex of ten young seedlings using the SCAR maker. The results suggest that three of the seedlings should be male as they carry male-specific amplicon, while the remaining seven seedlings should be females (Fig. 5).
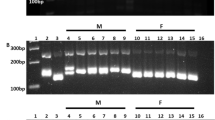
Profiles of male and female date palm genotypes showing amplicons generated using a SCAR primer pair (Table 1) developed in the study. Note that the 406-bp fragment is amplified in all samples, while the fragment of 354 bp is seen only in lanes of male genotypes. Lanes F1–F25 individual female genotypes, M1–M20 individual male genotypes, Bf bulked genomic DNA of female genotypes, Bm bulked genomic DNA of male genotypes, M 200-bp DNA ladder
Profiles of male and female date palm genotypes collected from Date Palm Research Centre, Bikaner showing amplicons generated using a SCAR primer pair developed in the study (Table 1). Note that the 406-bp fragment is amplified in all samples, while the fragment of 354 bp is seen only in lanes of male genotypes. Lanes F1–F25 individual female genotypes, M1–M10 individual male genotypes, M 200-bp DNA ladder
Profiles of young seedlings of date palm genotypes showing amplicons generated using a SCAR primer pair (Table 1) developed in the study. Note that lanes S4, S7, and S9 have the male-specific fragment of 354 bp in addition to the 506-bp fragment, which is shared by all samples. Lanes S1–S10 ten seedling samples, M 200-bp DNA ladder
Discussion
Currently, there is no reliable method to identify sex of the date palm seedlings at the early stages, though there has been significant progress in our understanding of sex-determining mechanisms in date palm using traditional means. There are no easily distinguishable sex chromosomes in date palm, despite some cytological evidence that they exist (Siljak-Yakovlev et al. 1996). As biochemical studies have yielded little insight into how to identify the gender of immature plants (Qacif et al. 2007), the identification of DNA sequences or sequence polymorphisms that are gender-specific offers a promising alternative to efficiently determine date palm gender. In the present study, we report, for the first time (to the best of our knowledge), a robust male-specific SCAR marker that can discriminate male and female date palm plants quite accurately. Although some putative sex-linked RAPD and ISSR markers in date palm have been reported earlier (Singh et al. 2006; Younis et al. 2008), the same could not be verified when tested on additional unrelated samples (our unpublished data). Knowledge on the gender of the seedlings prior to their transplantation in the field is useful to obtain a desired ratio of male and female plants and also to make best use of the available resources, especially land usage.
Obtaining a marker linked to a gene or genomic region through molecular marker analysis generally requires screening of a very large number of primers, which seemingly is a direct outcome of the genome size and the genome coverage by the tested markers, albeit with some degree of a chance success. In this study, we tested a total of 204 primers (100 RAPD and 104 ISSR primers) to amplify/identify one potential male-specific marker. Hormaja et al. (1994) screened 1,000 primers in P. vera and found only one female-associated marker. In S. viminalis, Gunter et al. (1998) screened 700 primers for identification of a sex-linked marker. Shibu et al. (2001) used 60 RAPD primers for identification of sex-specific DNA markers in the dioecious tree, nutmeg. Yang et al. (2005) obtained a 500-bp male-specific DNA fragment after screening a total of 1,041 RAPD primers in C. simplicifolius. The present study also reconfirms the utility of bulk segregant analysis (BSA), especially when one needs to test a large number of markers on a large number of samples. Bulked segregant analysis has earlier been used successfully to identify molecular markers associated with sex determination in several dioecious plant species (Mulcahy et al. 1992; Hormaza et al. 1994). The male-specific fragment was cloned and sequenced, and its GC content was comparable to similar male-specific sequences reported in other plants, e.g., 39.9 % for MADC1 and 40.4 % for MADC2 in C. sativa L. (Mandolino et al. 1998) as well as 45 % for OPA10-710 and 33 % for OPN11-1095 in Aucuba japonica Thunb. (Maki 2009). Further, the male-specific sequence identified in this study did not reveal significant similarity to any known sequence in the NCBI database and showed only short ORFs, with only one of them showing relatively poor similarity to a hypothetical protein of a bacterium. Similar observations were reported for some of the sex type-linked molecular markers that are reported in other species. Liao et al. (2009) identified two sex-specific fragments in G. biloba L. by using RAPD primers, which did not share significant similarity to any known sequence in the NCBI database and had no ORFs. Similarly, Li et al. (2010) observed no sequence homology for a 500-bp male-specific fragment in C. simplicifolius, which also had short ORFs with no homology to any known gene. These observations are expected, as molecular tags identified using anonymous genomic markers are usually from the genomic region(s) controlling the associated trait but not part of the gene(s).
The main significance of our study is that one new SCAR markers has been developed from RAPD marker for sex identification in date palm. Although this RAPD marker (OPA-02) could discriminate male plants from female plants efficiently but SCAR markers possess several advantages over RAPD marker, such as: (1) robust reproducible PCR amplification (Paran and Michelmore 1993) with minimum effect of reaction conditions and (2) locus specificity amenable to easy detection/scoring on agarose gel. The SCAR primer pair (SCARdpF and SCARdpR) designed in the study was found efficient and accurate in identification of sex of date palm plants when tested repeatedly (at least twice) and on multiple unrelated samples of different varieties (90 samples including 50 female plants, 30 male plants, and ten seedlings) collected in nature from different geographical regions (HAU, Hisar and RAU, Bikaner).
Conclusion
In this study, one new SCAR marker was developed for sex identification of date palm. This SCAR marker was validated to be a reliable indicator of the sexuality of date palm. As this diagnostic procedure to accurately predict the sex of date palm plants can be used at a very early stage, it would be of immense importance to growers while establishing date palm plantations.
References
Agarwal M, Shrivastava N, Padh H (2011) Development of sex linked AFLP markers in Simmondsia chinensis. Plant Breeding 130(1):114–116
Agrawal V, Sharma K, Gupta S, Kumar R, Prasad M (2007) Identification of sex in Simmondsia chinensis (jojoba) using RAPD markers. Plant Biotech Rep 1(4):207–210
Banerjee NS, Manoj P, Das MR (1999) Male-sex-associated RAPD markers in Piper longum L. Curr Sci 77(5):693–695
Bedoya GC, Nunez V (2007) A SCAR marker for the sex types determination in Colombian genotypes of Carica papaya. Euphytica 153:215–220
Deputy JC, Ming R, Ma H, Liu Z, Fitch MM, Wang M, Manshardt R, Stiles JI (2002) Molecular markers for sex determination in papaya (Carica papaya L.). Theor Appl Genet 106:107–111
Gangopadhyay G, Roy SK, Kaushik G, Poddar R, Bandyopadhyay T, Basu D, Mukherjee KK (2007) Sex detection of Carica papaya and Cycus circinalis in pre-flowering stage by ISSR and RAPD. Curr Sci 92:524–526
Gill GP, Harvey CF, Gardner RC, Fraser LG (1998) Development of sex-linked PCR markers for gender identification in Actinidia. Theor Appl Genet 97:439–445
Gunter LE, Roberts GT, Lee K, Larimer FW, Tuskan GA (1998) RAPD and SCAR markers linked to femaleness in Salix viminalis L. Plant and Animal Genome VI Conference P362
Hormaza JI, Dollo L, Polito VS (1994) Identification of a RAPD marker linked to sex determination in Pistacio vera using bulked segregant analysis. Theor Appl Genet 89:9–13
Ince AG, Karaca M, Onus AN (2010) A reliable gender diagnostic PCR assay for jojoba (Simmondsia chinensis (Link) Schneider). Genet Resour Crop Evol 57(5):773–779
Khadka DK, Nejidat A, Tal M, Goldhirsh GA (2002) DNA markers for sex: molecular evidence for gender dimorphism in dioecious Mercurialis annua. Mol Breed 9:251–257
Li M, Yang H, Li F, Yang F, Yin G, Gan S (2010) A male-specific SCAR marker in Calamus simplicifolius, a dioecious rattan species endemic to China. Mol Breed 25:549–551
Liao L, Liu J, Dai Y, Li Q, Xie M, Chen Q, Yin H, Qiu G, Liu X (2009) Development and application of SCAR markers for sex identification in the dioecious species Ginkgo biloba L. Euphytica 169:49–55
Maki M (2009) Development of SCAR markers for sex determination in the dioecious shrub Aucuba japonica (Cornaceae). Genome 52:231–237
Mandolino G, Carboni A, Forapani S, Faeti V, Ranalli P (1998) Identification of DNA markers linked to the male sex in dioecious hemp (Cannabis sativa L.). Theor Appl Genet 98:86–92. doi:10.1007/s001220051043
Mitra C, Kharb P, Uppal S, Jain S (2011) Genetic diversity analysis in date palm (Phoenix dactylifera L.): a comparative assessment of ISSR and RAPD marker assays. J Hort Sci & Biotech 86(4):398–402
Mohsenzadeh H, Pasalari H (2010) Identification of a male-specific RAPD marker in Phoenix dactylifera. J Hort Sci & Biotech 85(2):144–146
Mulcahy DL, Weeden NF, Kesseli R, Carroll SB (1992) DNA probes for the Y chromosome of Silene latifolia, a dioecious angiosperm. Sex Plant Reprod 5:86–88
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acid Res 8:4321–4326
Paran I, Michelmore RW (1993) Development of reliable PCR based markers linked to downy mildew resistance genes in lettuce. Theor Appl Genet 85(8):985–993
Parasnis AS, Gupta VS, Tamhankar SA, Ranjekar PK (2000) A highly reliable sex diagnostic PCR assay for mass screening of papaya seedlings. Mol Breed 6:337–344
Qacif N, Baaziz M, Bendiab K (2007) Biochemical investigations on peroxidase contents of male and female inflorescences of date palm (Phoenix dactylifera L.). Sci Hortic (Amsterdam) 114:298–301
Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allerd RW (1984) Ribosomal spacer length polymorphism in barley: Mendelian inheritance, chromosomal location and population dynamics. Proc Natl Acad Sci USA 81:8014–8018
Shibu MP, Ravishankar KV, Anand L, Ganeshaiah KN, Shaanker U (2001) Identification of sex-specific DNA markers in the dioecious tree, nutmeg (Myristica fragrans Houtt.). PGR Newsl 121:59–61
Shirkot P, Sharma DR, Mohapatra T (2002) Molecular identification of sex in Actinidia deliciosa var. deliciosa by RAPD markers. Sci Hortic 94:33–39
Siljak-Yakovlev S, Cerbah M, Sarr A, Benmalek S, Bounaga N, Coba de la Pena T, Brown SC (1996) Chromosomal sex determination and heterochromatin structure in date palm. Sex Plant Reprod 9(3):127–132
Singh P, Kharb P, Pandey N, Batra P, Khatak S, Dhillon S, Uppal S, Chowdhury VK (2006) RAPD analysis for genetic diversity and identification of sex-specific marker in date palm (Phoenix dactylifera L.). Haryana J of Hort Sci 35:232–234
Torjek O, Bucherna N, Kiss E, Homoki H, Finta-Korpelova Z, Bócsa I, Heszky LE (2002) Novel male-specific molecular markers (MADC5, MADC6) in hemp. Euphytica 127:209–218
Xu Y, Shimoro X, Hofstra H (1994) Plant DNA isolation protocol. Nucleic Acids Res 22:2399–2403
Yakubou B, Barazani O, Golan-Goldhirsh A (2005) Combination of SCAR primers and touchdown-PCR for sex identification in Pistacia vera L. Sci Hort 103:473–478
Yang H, Si-Ming G, Huang-Tian Y, Huang-Can X (2005) Identification of random amplified polymorphic DNA markers linked to sex determination in Calamus simplicifolius. J Integr Plant Biol 47:1249–1253
Younis RAA, Ismail OM, Soliman SS (2008) Identification of sex-specific DNA markers for date palm (Phoenix dactylifera L.). Res J Agric Biol Sci 4(4):278–284
Acknowledgments
We thankfully acknowledge the Horticulture Depratment, CCS-HAU, Hisar and Date Palm Research Centre, Swami Keshwanand Rajasthan Agricultural University, Bikaner, Rajasthan, India for helping with the plant samples used in the study. This research was supported with a grant from the Department of Science and Technology, Government of India (SR/SO/PS-58/2007) provided to Dr. Pushpa Kharb.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by W.-W. Guo
Supplementary information A patent has been filed in India for the developed marker vide application no. 1513/DEL/2010 dated 29/6/10.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 242 docx)
Rights and permissions
About this article
Cite this article
Dhawan, C., Kharb, P., Sharma, R. et al. Development of male-specific SCAR marker in date palm (Phoenix dactylifera L.). Tree Genetics & Genomes 9, 1143–1150 (2013). https://doi.org/10.1007/s11295-013-0617-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-013-0617-9