Abstract
Although molecular characterization of the avocado germplasm started with the early development of molecular markers, the genetic relationships among the three botanical races are still uncertain. Here, we report the development of 47 new microsatellites in avocado (Persea americana Mill) and the results of various genetic studies carefully designed to address the unsolved questions. Forty high-quality, single-locus markers (25 simple sequence repeats (SSRs) and 15 expressed sequence tag–SSRs (EST-SSRs)) were evaluated in a selected group of 42 cultivated accessions, which represent the three described botanical races. A total of 455 alleles (11.4 alleles per locus) have been detected. The mean expected and observed heterozygosities averaged 0.831 and 0.674, respectively. All the analyzed genotypes could be unequivocally distinguished with an accumulated probability of identity value of 6.36 × 10−50. Seventy-five percent of the loci showed a significant departure from Hardy–Weinberg equilibrium, most likely due to the substructure of the accession set and kinship among some of the accessions. The genetic relationships among the accessions were explored using different methods. We demonstrate that the correct allocation of the avocado cultivars requires the complementary use of distance-based and model-based methods. All of the results agreed with the existence of three groups to which accessions were assigned based on their botanical race, with 25 % of the detected variation being partitioned among the groups. The diversity analysis within each group has allowed for the identification of unique alleles that are useful as race-specific markers. The effects of the different experimental parameters on the results are discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The cultivated avocado (Persea americana Mill.) is an important subtropical evergreen tree, native to the Mesoamerican area. There is evidence of human avocado consumption and domestication since approximately 8,000–7,000 bc and selection since 4,000–2,800 bc (Smith 1966). Currently, the world avocado production totals 3.8 million tons (Faostat 2012), although a few countries (Mexico, Chile, Indonesia, the USA, the Dominican Republic, Colombia, Brazil, Peru, and Guatemala) account for 70 % of the worldwide production, with Mexico being the principal worldwide producer (29 % of world production in 2010). The major exporters are Chile, Mexico, Spain, and Israel (Litz et al. 2007). Spain is a special case in avocado cultivation because it is the only European country with a significant commercial production (9,700 ha producing 72,000 t in 2009) (www.marm.es). Avocadoes were introduced to inland Spain in the late sixteenth to the early seventeenth century but were not commercially established until the 1970s. Between 1970 and 1975, commercial varieties were fixed, and the major growth of production began in 1976, mainly in the provinces of Malaga and Granada. In 2009, the productive area in this region was approximately 8,800 ha and produced approximately 72,000 t (www.marm.es). The second most productive region is the Canary Islands, where the variety of genotypes present are a result of exchanges and migratory movements established with America since the fifteenth century, but more significantly from Cuba in the nineteenth century (L. Gallo, personal communication). At the present time, the cultivated avocado covers a surface of approximately 800 ha in Canary Islands and produces approximately 8,000 t (www.marm.es).
P. americana belongs to the Lauraceae family, which is one of the oldest flowering plant families, and is a member of the clade that contains most of the primitive angiosperm, the Magnoliids. The family comprises approximately 50 genera and 2,500–3,000 species of trees, rare shrubs, and one genus of parasitic vines, the Cassytha (Chanderballi et al. 2008). P. americana is an extremely variable species with controversial taxonomy and remains ill-defined. Based on their presumed center of origin (Popenhoe 1941), a general consensus was reached for three different botanical races: Mexican [P. americana var. drymifolia (Schlecht. and Cham)], Guatemalan (P. americana var. guatemalensis L. Wms.), and West Indian (P. americana var. americana Mill.), which are originally from highland Mexico, highland Guatemala, and lowland (coastal) Guatemala to Costa Rica, respectively (Galindo-Tovar et al. 2008). The differences in fruit characteristics and environmental needs (temperature and humidity) among these three races are often used to circumscribe subgroups, with the Mexican and Guatemalan races adapted to cooler (Mediterranean and subtropical) climates, while the West Indian race requires warmer (tropical) conditions for optimal development. Other differential criteria are maturity season, anise scent, colored lenticels on young flesh, skin thickness and surface, and seed size (Bergh and Lahav 1996). Avocado has a synchronous protogynous dichogamous breeding system that promotes out-crossing, although self-fertilization does occur in orchards composed of a single genotype (Davenport 1986). Sterility barriers do not exist between the three races, leading to interracial hybridizations, whether natural or induced by man. In fact, commercial avocado production is currently based not only on selections within these three races, but also on interracial hybrids, most commonly Mexican and Guatemalan, with different degrees of hybridization (for example, “Hass,” “Fuerte,” and “Bacon” after Litz et al. 2007) which makes their identification in terms of racial origin difficult.
Traditionally, genotype allocation into a certain botanical race is based on morphological characters and, if known, pedigree information. However, the evaluation of morphological characters is inaccurate due to the influence of environmental factors, subjectivity, and the limited numbers of discriminating traits (Alcaraz and Hormaza 2007). The exact pedigree of a certain cultivar is rarely known due to the particular avocado flowering habit, which makes controlled pollination almost impracticable (Ashworth and Clegg 2003). When the pedigree is unknown, the allocation based on phenotype can result in errors because of segregation of phenotypic characters (Ashworth and Clegg 2003). For this reason, during the past decade, taxonomic inferences have been strengthened by analyses of genetic markers including isozymes, DNA fingerprinting (DFP) (Mhameed et al. 1997), random amplified polymorphic DNA (RAPD) (Fiedler et al. 1998), restriction fragment length polymorphism (RFLP) (Davis et al. 1998), single sequence repeat (SSRs) (Schnell et al. 2003; Ashworth and Clegg 2003; Alcaraz and Hormaza 2007), and single nucleotide polymorphism (SNP) (Chen et al. 2008, 2009). Some of those results suggest that the validity of previous race and species definitions within Persea should be treated with caution (Mhameed et al. 1997) and that the races are not as well-differentiated as assumed (Ashworth and Clegg 2003).
The lack of consistency in the genetic relationships among the three races, as deduced from different morphological and marker studies, corroborate the uncertainty of these classifications. Bergh and Lahav (1996) focused on morphological characters and revealed that the three races are equally distinct from each other. That point was supported by molecular studies implemented with isozymes (Goldring et al. 1985) or with RFLP (Furnier et al. 1990) or DFP markers (Lavi et al. 1991). Additionally, Fiedler et al. (1998), after analyzing 16 accessions with RAPDs, classified the ecological races of avocado into three equal ranking subspecies of Persea. The same conclusion arose in the extended work of Schnell et al. (2003), carried out with 14 SSRs over 224 accessions. On the other hand, Kopp (1966), using morphological parameters, and Bufler and Ben-Ya’acov (1992) and Davis et al. (1998), working with DNA markers, grouped the Guatemalan and West Indian races into one botanical variety distinct from the Mexican race. Williams (1977) separated the Guatemalan race from the other two and placed it as Persea nubigena L. Wms. var. guatemalensis L. Wms (subgenus Persea), while the Mexican race (var. drymifolia (Schlecht. and Cham.) Blake) and West Indian race (var. americana Mill.) remained varieties of P. americana. Williams was of the opinion that the West Indian race was selected from the Mexican race. In addition, mismatching data between phenotypic and molecular records are common in molecular avocado germplasm studies (Schnell et al. 2003; Ashworth and Clegg 2003; Alcaraz and Hormaza 2007), thus underlying the need for a better understanding of the relationship between these botanical races and their molecular traits. With this general aim, we undertook a new marker analysis of the three avocado races, using SSRs as the markers of choice.
Microsatellites, or SSRs, are particularly suitable for studying the population structure and culture history of domesticated species because of the high level of allelic diversity (Garris et al. 2005). In avocado, several types of microsatellite markers have previously been reported, including 50 from a non-enriched library (Sharon et al. 1997), 25 from dinucleotide- and trinucleotide-enriched libraries (Ashworth et al. 2004), and 70 SSRs developed from avocado expressed sequence tags (EST-SSRs) (Borrone et al. 2007). These markers have been used for linkage map construction (Sharon et al. 1997; Borrone et al. 2009), molecular characterization of avocado germplasm collections (Alcaraz and Hormaza 2007), and diversity analysis (Schnell et al. 2003); however, not all of these microsatellite markers have been shown to amplify consistently in all avocado varieties (Ashworth and Clegg 2003; Ashworth et al. 2004; Borrone et al. 2007). A high proportion of amplification failure (35 out of 75 microsatellites) has been observed by our group (data not shown) and also reported by other authors (Ashworth and Clegg 2003; Ashworth et al. 2004; Borrone et al. 2007) which promoted the development of new markers, mainly EST-SSRs (Borrone et al. 2007). However, the existing EST-SSRs have shown a high rate of various loci amplification in avocado (Borrone et al. 2009), hindering accurate genetic analysis (diversity studies, parental analysis, linkage studies, etc.). To increase the number of quality markers available for advanced genetic studies, we have developed new microsatellite markers using two approaches: the isolation of genomic SSRs from an enriched library and EST screening.
Material and methods
Plant material and genomic DNA extraction
A total of 42 P. americana accessions, including rootstocks, commercial varieties, and Spanish local selections, were sampled from one of the three avocado collections maintained in Spain: at the Instituto de Hortofruticultura Subtropical y Mediterranea La Mayora UMA-CSIC (Algarrobo-costa, Málaga), the Instituto de Investigación y Formación Agraria y Pesquera (IFAPA) Centro de Churriana (Churriana, Málaga), and the Instituto Canario de Investigaciones Agrarias (ICIA) (Tenerife, Canary Islands).
Table 1 lists the origins and information on these 42 genotypes. The initial classification of the accession by racial (Mexican, Guatemalan, or West Indian) or hybrid group was based on the previous records provided by the Variety Database of the University of California at Riverside (http:/www.ucavo.ucr.edu) and other avocado articles (Kadman and Ben-Ya’acov 1980; Lahav et al. 1989, 1992a; Young 1992; Lavi et al. 1997; López-López et al. 2001; Newett et al. 2002; Schnell et al. 2003; Ashworth and Clegg 2003; Gallo et al. 2003a; Ben-Ya’acov et al. 2003; Sanchez-Colín et al. 1998). In total, 14 Mexican, 14 West Indian, 1 Guatemalan, 10 interracial hybrids, and 3 unknown race accessions were studied. In six cultivars (Bacon, “Edranol”, Fuerte, Hass, “Negra de la Cruz,” and “Plowman”), the racial information available was questionable or contradictory. Genomic DNA was extracted from fresh leaves following the methods described by Viruel et al. (1995).
Construction and screening of a microsatellite-enriched library
A small-insert library was developed from the DNA of the avocado cultivar, Hass, digested with the restriction enzyme HaeIII (New England Biolabs, Beverly, MA, USA) and enriched with (CT) n sequences following the procedure described by Viruel and Hormaza (2004). Recombinant clones were PCR-amplified and transferred to a nylon membrane that was hybridized with a (CT)15 probe labeled with digoxigenin (DIG oligonucleotide 3′-end labeling kit, Roche Applied Science, Indianapolis, IN). Positive clones were sequenced to identify the flanking regions, which were then used to design primers with the program Primer3 (Whitehead Institute for Biochemical Research, Cambridge, MA). The primer pair name is preceded with the code LMAV (LM: La Mayora; AV: avocado) followed by a consecutive number.
Development and nomenclature of EST-SSR
Avocado ESTs were obtained from the National Center for Biotechnology Information database Entrez (http://www.ncbi.nlm.nih.gov/Entrez). Microsatellites within the genomic DNA sequences were identified with the program Sputnik (Abajian 1994), which employs a recursive algorithm to search for repeated patterns of nucleotides between 2 and 5 base pairs (bp). Sputnik does not search against a library of known microsatellites but, instead, applies simple scoring rules to identify the simple repeats for which the maximum score is above a certain threshold. Parameters were set as defaults. Imperfect repeats and those with insertions, deletions, and mismatches are allowed by Sputnik, although such imperfections reduce the score of the microsatellite. Redundant sequences and those with a score lower than 20 were excluded. Primers of the flanking regions were also designed using Primer3 software. Each primer pair was named with the code ESTAV (EST: expressed sequence tag; AV: avocado) followed by the repeated motif and a consecutive number.
Microsatellite analysis
The EST-SSR and the genomic SSR primers that were obtained were initially used to study a reduced group of four avocado genotypes by PCR amplification in a total volume of 25 μL, which contained 16 mM of (NH4)2SO4, 67 mM of Tris–HCl (pH 8.8), 0.01 % of Tween 20, 0.4 μM of each primer, 0.1 mM of each dNTP, 2 mM of MgCl2, and 0.5 U of Biotaq™ DNA Polymerase (Bioline, London, UK). Reactions were carried out on an I-Cycler (Bio-Rad Laboratories, Hercules, CA, USA) thermocycler using the following temperature profile: an initial step of 1 min at 94 °C; 35 cycles of 30 s at 94 °C; 30 s at 55 °C; 1 min at 72 °C; and a final step of 5 min at 72 °C. Amplification products were resolved by 3 % high-resolution agarose gel electrophoresis (Metaphor; FMC Bioproducts). Primers showing clear and scorable fragments were selected for further simple sequence repeat analysis using a CEQ 8000 capillary DNA analysis system (Beckman Coulter, Fullerton, California). PCR reactions were performed as previously described in a final volume of 15 μL, with an additional label of WellRED fluorescent dyes D2, D3, and D4 on the forward primers of each primer pair (Sigma-Aldrich, St Louis, MO). The annealing temperature for all of the primer pairs was 55 °C. Analyses were repeated at least two times to ensure the reproducibility of the results.
Statistical analysis
The allelic composition of each EST-SSR and genomic SSR marker was determined for each accession, and putative alleles were indicated by the estimated size in bp. The genetic information was assessed for only the 40 single-locus microsatellites using the following parameters: number of alleles per locus (A); observed heterozygosity (Ho); expected heterozygosity [He(1 − ∑p i 2) where p i is the frequency of the ith allele; Nei 1987]; effective number of alleles [Ne(1 / 1 − He)]; Wright's fixation index [F(1 − Ho / He) Wright 1951]; and the probability of identity \( \left[ {\mathrm{PI}\left( {1-\sum {p_i^4+\sum {\sum {{{{\left( {2{p_i}{p_j}} \right)}}^2}} } } } \right)} \right] \), where p i and p j are the frequency of the ith and jth alleles, respectively, which measures the probability that two randomly drawn diploid genotypes will be identical assuming observed allele frequencies and random assortment (Paetkau et al. 1995). The program CERVUS 3.0 (Kalinowski et al. 2007) was used to calculate A, Ho, and He. The software IDENTITY 1.0 (Centre for Applied Genetics, University of Agricultural Sciences, Vienna) was used to calculate probability of identity (PI). Deviations from Hardy–Weinberg equilibrium at individual loci were tested using a Markov chain method and calculated with the program Arlequin 3.11 (Excoffier et al. 2005).
A similarity matrix was obtained based on the Lynch shared band similarity index (Sxy = 2Nxy / (Nx + Ny)), where Nxy is the number of bands in common produced by microsatellite amplification and Nx and Ny are the numbers of these bands in the two individuals being compared (Lynch 1990). The similarity data matrix was then used to establish the relationships between the different avocado accessions through an unweighted pair group method average (UPGMA) dendogram and a principal coordinate analysis (PCoA) diagram to depict a non-hierarchical relationship among the accessions. The cophenetic correlation coefficient was calculated to measure the goodness of fit between the original similarity matrix and the dendrogram matrix. All analyses were computed with the program NTSYSpc 2.11 (Exeter Software, Stauket, NY).
The marker data were also analyzed with a model-based method using the STRUCTURE v2.3.1 software (Prichard et al. 2000) to infer any putative population structure present in the group of 42 accessions reported. The number of subpopulations (K) was estimated using the admixture model with correlated allele frequencies between populations, as this configuration was considered the best in cases of subtle population structure (Falush et al. 2003). The degree of admixture alpha was inferred from the data, and Lambda, the parameter of the distribution of allele frequencies, was set to 1, as recommended by the STRUCTURE authors. We used a predefined number of subpopulations (K) ranging from 1 to 10. Five runs were performed for each K with 105 iterations, following a burning period of 105 iterations to find the optimal number of subpopulations. The ad hoc statistic (ΔK), based on the change in the log probability of data between successive K values, was used to estimate the number of subpopulations (Evanno et al. 2005).
Finally, an analysis of molecular variance (AMOVA), based on the number of different alleles and 10,000 permutations, was performed using Arlequin 3.11 software (Excoffier et al. 2005) to determine the distribution of genetic diversity both among and within the subpopulations identified in previous analyses. The differentiation index among subpopulations (F ST) and the fixation index within the whole population (F IT) and subpopulations (F IS) were calculated. Pairwise F ST values were used to measure the genetic differentiation between populations and its significance (1,000 bootstrap replicates).
Results
Genomic SSR marker development
An enriched genomic library for GA/CT repeats was constructed from the cultivar Hass. A total of 96 randomly chosen clones were screened by hybridization with the probe (CT)15, of which 92 (96 %) gave a positive signal. Fifty-one of these clones were sequenced, producing a total of 44 (86 %) readable sequences. After discarding no-microsatellite, redundant, and incomplete sequences, 36 putative SSR primers were designed. Finally, 29 new SSR markers (Table 2) showing consistent and clear amplification patterns were used to amplify the whole avocado collections. The microsatellite sequences have been deposited in Genbank (accession numbers from JX130406 to JX130434). The efficiency of the library was estimated to be 55 %.
Based on the structure of the microsatellite motif, SSRs were classified as simple or compound, and each class was further characterized as perfect or imperfect (Varshney et al. 2000). Most of the motifs were simple, perfect (59 %), or imperfect (27 %). Only four SSRs (14 %) were compound, three of them perfect and only one imperfect (Table 2). The number of repeats varied between 9 and 23, with most microsatellites displaying between 9 and 16 repeats (Fig. 1).
EST-SSR development
From a total of 8,897 avocado EST sequences, 368 sequences (4 %) contained the SSR motif. Tri- and dinucleotide repeats were the most abundant (47 and 33 %, respectively), followed by pentanucleotide (11 %) and tetranucleotide repeats (9 %). One hundred and twenty-one sequences (33 %) were selected on the basis of a Sputnik value ≥20. After discarding 24 redundant sequences, the 30 sequences with the highest Sputnik value were selected for primer design. On the final amplification test, 18 primer pairs (60 %) were selected to amplify the whole cultivar collection (Table 3). Seventeen EST-SSRs contained the AG/TC motif and one the AT/TA motif. The number of repeats varied between 13 and 32 (Fig. 1), and all were classified as simple and perfect. Of these markers, seven were derived from clones with homology to proteins of known function.
Polymorphism markers and genetic diversity
The polymorphism level of the 47 newly designed primer pairs was investigated in the 42 avocado accessions. Two markers (LMAV.28 and ESTAVTC.17) produced a monomorphic pattern, and more than two amplification products were found in five markers (LMAV.01, LMAV.11, LMAV.20, ESTAVTC.15, and ESTAVTC.18). Those markers were discarded for the diversity analysis calculations, for the remaining markers, diversity values are presented in Table 4.
The 40 markers that were analyzed (25 SSRs and 15 EST-SSRs) detected a total of 455 alleles ranging from 5 (LMAV.27, LMAV.34, and ESTAVGA.01) to 18 (LMAV.07, LMAV.31, and ESTAVTC.03) alleles, with an average of 11.4 alleles per locus. The allele frequencies ranged from 0.012 to 0.513, with a mean of 0.088. No fixed alleles (frequency >0.9) were present, and 222 of the alleles (49 %) can be considered rare (frequency <0.05). One hundred and nine of these rare alleles (24 % out of total) obtained with 35 of the 40 markers were detected in only one accession. Twenty-eight genotypes showed the presence of at least one of these accession-specific alleles. They are local accessions from inland Spain (“La Piscina” and “Cónsula9”), the Canary Islands (BG25, BG28, BG31, BG32, CA2, CA3, CA9, and CA23), or other countries (“Tochimilco 3” and “Guayabamba”) as well as traditional and modern breeding cultivars from different origins (“Mexicola”, Bacon, Edranol, Negra de la Cruz, “Scott,” “Topa Topa,” “Gordon,” Plowman, “Adi,” “Anaheim,” “Eden,” “Iriet,” “Maoz,” “Mckee,” “Merensky 1,” and “Toro Canyon”). Most of the genotypes (10) contained two specific alleles, although the number of specific alleles varied from 1 in Maoz and “CA9” to 10 in Negra de la Cruz. A significant departure over Hardy–Weinberg equilibrium (p ≤ 0.05) was observed for 30 (75 %) out of the 40 loci studied. The accumulated PI for the 40 loci was 6.36 × 10−50. In general, the diversity parameters associated with genomic SSR and EST-SSR were very similar
Genetic relationships and population structure of the analyzed avocado accessions
Band profiles obtained with the 40 new markers unambiguously identify the 42 analyzed accessions. Pairwise comparison of accessions profiles, through the Lynch band similarity index, resulted in a similarity matrix (not shown) with index ranging from 0 (Maoz and “G6”), to 0.97 (Gordon and Guayabamba) and an average value of 0.29. UPGMA cluster analysis resulted in a single dendogram (Fig. 2a), with a cophenetic correlation coefficient of 0.82. Three main groups could be observed: group I comprises 14 West Indian race accessions; group II comprises 10 accessions, of which 8 are Guatemalan × Mexican hybrids and 2 (Merensky 1 and Tochimilco 3) are Mexican accessions; and group III comprises 18 accessions, mostly Mexican except Plowman (Guatemalan hybrid), Fuerte (Mexican/Mexican × Guatemalan hybrid), and Anaheim (Guatemalan/Mexican × Guatemalan hybrid). The three unknown race accessions, Gordon, Mckee, and Cónsula9, also clustered in this last group. PCoA (Fig. 3) and the model-based analysis carried out with STRUCTURE software (Fig. 2b) produced similar draws.
a Dendogram of the 42 avocado accessions studied based on the UPGMA analysis using the similarity matrix generated by the Lynch shared band similarity index after amplification with 40 pairs of SSR and EST-SSR primers. b Bar plot of the 42 avocado accessions based on the STRUCTURE results (K = 3). Each bar represents a single individual and its inferred admixture proportion of every putative subpopulation: green, Mexican; blue, Guatemalan × Mexican hybrid; and red, West Indian
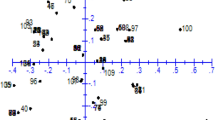
Principal coordinate analysis of the 42 avocado accessions based on the 40 SSR and EST-SSR patterns. I West Indian group, II Guatemalan × Mexican group, III Mexican group, with subgroups a and b. Dotted lines mark putative transition regions between groups. Every accession is represented with a figure depending on its assigned race (see the legend)
In PCoA, the three main axes explained approximately 32 % of the total variation, with 15.4, 10.2, and 6.4 % individually. Some subgroups, connecting closer accessions, could also be recognized in the Mexican group: a cluster with most of the rootstock selected by the University of California (“Barr Duke,” “Duke6,” “Duke9,” and G6) and a group of traditional rootstocks from local selections (Topa Topa, Scott, “Thomas”, Plowman, and the Spanish local selection La Piscina).
In the model-based analysis, both the log likelihood [LnP(D)] and the Evanno's ΔK linked to each K value agreed with the presence of three subpopulations in our set of avocado cultivars (Fig. 4), showed as green, Mexican; blue, Guatemalan × Mexican hybrid; and red, West Indian in Fig. 2b. STRUCTURE software, also, allows the contribution from any of the three putative ancestral subpopulations in the genome of every individual to be expressed as a percentage (Fig. 2b). A genotype was allocated in a population when more than three quarters (75 %) of the genetic background belonged to this population; otherwise, it was treated as an ambiguous (or hybrid) class member. Following this criterion, 71 % of the accessions (30 out of 42) were classified unambiguously into one of the three distinct subpopulations. A model with K = 2 grouped populations II and III into one unique Mexican–Guatemalan population. With the model K = 4, the putative Mexican population was split into two. The new population included some selections from the University of California (Duke6, Duke9, Barr Duke, and G6) and part of the genomes of Gordon and Guayabamba.
a Average of the estimated log likelihood (LnP(D)) for each value of K. b ΔK as calculated as described by Evanno et al. (2005) for different K values
Race-specific markers
A detailed analysis of the diversity parameters for each subpopulation was achieved with the aim of detecting race-specific markers. The accessions were grouped into the four subpopulations identified in our previous analysis: the Mexican, composed of 14 accessions, and the Guatemalan × Mexican and the West Indian, with eight accessions each. Eleven genotypes allocated between them were grouped into one hybrid subpopulation. Values of A, Ne, He, Ho, and F for each subpopulation are presented in Table 5. The hybrid subpopulation showed maximum values in all the diversity parameters calculated and an F value close to 0 (F = 0.030). With regards to the remaining subpopulations, the Mexican one was the most diverse (A, Ne, and He), while the West Indian one showed the lowest genetic diversity (A, Ne, and He). The Guatemalan × Mexican subpopulation presented intermediate diversity values, but showed the highest observed heterozygosity (Ho = 0.780) and was unique in showing a negative value of F, which indicates the presence of an excess of heterozygous genotypes.
A high proportion of observed alleles were not shared among the different subpopulations, either because they were accession-unique or they were present in more than one accession but belonging to one unique subpopulation (Table 5). Forty-five percent of the alleles present in the Mexican group, 34 % in the Guatemalan × Mexican group, and 33 % in the West Indian group were absent in the other two groups. As average, every Mexican genotype contained three accession-unique alleles, 2.1 every Guatemalan × Mexican genotype, and 1.5 every West Indian genotype.
From a practical viewpoint, alleles were considered useful as race markers when they were present at a high frequency (superior to 0.40) of the race group and absent in the other two, and, in addition, they were found in most of the individuals genotyped (Table 6). Alleles from seven markers were specific to the Guatemalan × Mexican subpopulation. The specific alleles from ESTAVCT.07 and LMAV.33 were present in every genotype of this subpopulation and the ones from LMAV.18 and LMAV.03 in every scored accession. Specific alleles from LMAV.02, LMAV.06, and ESTAVTC.20 were less informative: LMAV.02 because it was absent in two accessions, which contained genotype-unique alleles, and LMAV.06 and ESTAVTC.20 because two alleles were needed to mark all the accessions. Six microsatellites were useful for distinguishing the West Indian race. The most informative, LMAV.27, was fixed in this accession set. ESTAVTC.04 and LMAV.33 needed two alleles to mark all the accessions, with the exception of Maoz, which showed a low-frequency (0.12), race-specific allele (249 bp) in LMAV.33. The LMAV.03 and LMAV.33 markers exhibited race-specific alleles matching both the Guatemalan × Mexican and the West Indian subpopulations. High frequency alleles were absent in the Mexican subpopulation, but the combination of two alleles from two markers (LMAV.21 and ESTAVGA.11) marked most of the Mexican accessions. Exceptions were Scott, which has two unique alleles, and Mexicola, which contains a different (165 bp) low-frequency (0.18), race-specific allele from ESTAVGA.11. The hybrid accessions contain a mixture of those alleles, in accordance with their genomic composition.
Distribution of genetic diversity
The distribution of genetic diversity among the subpopulations identified was estimated through AMOVA. The partitioning of the genetic variance of the total population shows that the majority of the allelic diversity revealed with these 40 new markers occurred within populations and that the subpopulations considered were significantly different (F ST = 0.25, p < 0.001) (Table 7).
When only the three subpopulations (Mexican, Guatemalan × Mexican, and West Indian) were considered, 25 % of the variation occurred among subpopulations; however, this percentage dropped to 16 % when the hybrid subpopulation was included in the analysis (data not shown), as well as the variation within subpopulations that dropped from 2.68 to 1.93 %.
The three pairwise comparisons of F ST were significant (p < 0.001). The closest subpopulations (with the lowest number of different alleles) were Mexican and Guatemalan × Mexican (F ST = 0.17); the West Indian subpopulation was equally distant from the Mexican and Guatemalan × Mexican subpopulations (F ST = 0.30). The inbreeding coefficient in the total population was low (F IS = 0.04) and was significant within only the West Indian subpopulation (F IS = 0.24, p < 0.001).
Discussion
In general, molecular studies in tropical and subtropical fruit trees have been neglected; however, avocado is an exception to this rule (Lavi et al. 1994), and molecular markers have been developed and used both for genetic mapping and germplasm analysis (Sharon et al. 1997; Mhameed et al. 1997; Fiedler et al. 1998; Davis et al. 1998). In recent years, the available molecular tools have increased in both quantity and quality with the development of markers like SSRs (Schnell et al. 2003; Ashworth et al. 2004), EST-SSRs (Borrone et al. 2007), and SNPs (Chen et al. 2008). These markers have made more precise genetic studies possible, greatly enhancing avocado germplasm classification (Schnell et al. 2003) and developing new insight into the relationships of the different avocado races (Ashworth et al. 2004; Chen et al. 2009). Broadly speaking, molecular studies of cultivated accessions agree with the horticultural race classifications based on phenotypic criteria, although results at odds with the previous botanical assignment are always detected (Schnell et al. 2003; Ashworth and Clegg 2003; Alcaraz and Hormaza 2007). Additionally, despite the statement that each race is distinct from each other, until now, no useful race-specific markers have been identified (Schnell et al. 2003). Recently, Ashworth and Clegg (2003) suggested that interracial hybridization events occurred more recently than previously thought, such that there are fewer differences among the botanical races than are expected.
We planned this work to overcome the limits of previous similar studies, introducing the adjustments suggested by their authors, which includes finding a good balance between the number of entries (42) and markers (40), defining a convenient choice for every individual accession and marker, and performing a deep analysis of the molecular results through a survey of different statistical methods.
Microsatellite marker development and experimental approach
Forty good quality microsatellite markers (25 SSRs and 15 EST-SSRs) were newly developed. Comparing the effectiveness between the two developing methods is difficult because of the different factors involved, such as the initial number and quality of sequences/clones examined, the percentage of redundancies, the primer design condition, the number of primer pairs tested, etc. As a general rule, high-quality markers conflict with high developmental yield because they are correlated with the strength of the restrictive conditions imposed in every step. Our priority was to obtain high-quality markers with clear profiles that each detect a single locus, but even so, the efficiency of the library (55 %) and EST-SSR development (4 % of microsatellite sequences) was quite high. Although the SSRs and EST-SSRs had a different origin and structural characteristics (number, sequence, and organization of the repetitive motif), they have provided similar levels of genetic information, as is illustrated by the diversity parameter values obtained.
The choice of the number and type of accessions is as noteworthy as the choice of markers. In some previous studies, the number of genotypes included was limited (16 genotypes in Fiedler et al. (1998) and 12 genotypes in Borrone et al. (2007)), which makes it difficult to extrapolate the conclusions to a larger sample. In others, the number of markers used was low (14 SSRs in Schnell et al. (2003), 25 SSRs in Ashworth and Clegg (2003), and 16 SSRs in Alcaraz and Hormaza (2007)), and these markers failed to define sufficient differences among the different avocado genomes. In most cases, only the two Mediterranean races (Mexican and Guatemalan) are represented (Fiedler et al. 1998; Ashworth and Clegg 2003; Alcaraz and Hormaza 2007; Chen et al. 2008, 2009).We intended to include the “pure” accessions from the three avocado races, i.e., Mexican, Guatemalan, and West Indian, but a large enough sample was not available in the Guatemalan group. This under-representation corresponds to the low number of Guatemalan genotypes that are maintained in the collections used as a tree source (CSIC La Mayora, IFAPA Churriana, ICIA Tenerife) and is a reflection of the limited use of pure Guatemalan genotypes in Spanish avocado-growing areas. Therefore, we considered the interracial hybrid Guatemalan × Mexican as representative of the Guatemalan gene pool.
The statistical approaches used in a germplasm analysis may also affect the final results, thus, we used a broad range of different methods, two similarity-based methods (the UPGMA cluster method and the PCoA ordination method) and the model-based method (also called assignation method) conducted by the program STRUCTURE. The three analyses reached similar and complementary results, confirming that, contrary to previous statements (Chen et al. 2009), the conclusions inferred from diversity molecular data depend more on the information provided by the markers than on the statistical methods employed.
Genetic relationships, population structure, and race assignation of accessions
The analysis carried out with this new set of 40 SSR markers has unequivocally distinguished our 42 genotypes (accumulated PI = 6.36 × 10−50) and have solved the issues related to their race assignation, mainly (a) the elucidation of ambiguously assigned accessions, (b) the new assignation of unknown race accessions, and (c) the identification of wrongly assigned accessions.
The results of the three analyses performed (UPGMA cluster, PCoA, and model-based method) agree with the existence of three groups or subpopulations that gather all accessions on the basis of their botanical race: Mexican, interracial Guatemalan × Mexican, and West Indian. The STRUCTURE model-based inferences were essential in understanding the genomic relationships among the different avocado accessions. The main criteria used to catalog a cultivar in terms of its botanical origins are its pedigree and its fruit and cultural characteristics. In most of the cultivars, the pedigree is unknown or based only on the mother tree, and allocation based on phenotypic assessment can be tentative (Ashworth and Clegg 2003) because of character segregation and, very often, because modern and local cultivars are complex mixtures of different horticultural races (Davis et al. 1998; Chen et al. 2009; Scora et al. 2002; Ashworth and Clegg 2003; Schnell et al. 2003). The marker allele frequency differences considered in the model-based analysis provide new data about cultivar genome ancestries, and every accession can be described based on the composition of the putative ancestral subpopulation. This additional outlook is crucial to uncovering the genetic background and racial allocation of the accessions and provides evidence that some of the cultivars regarded as a “pure race” are not. In our accession set, Tochimilco 3, Guayabamba, and Toro Canyon were considered pure Mexican genotypes but were found to have Guatemalan ancestors as well. Residual West Indian genome components have been found in some Mexican accessions, such as Anaheim, Consula9, and Merensky 1. Similarly, some West Indian accessions (BG27, BG29, BG31, BG32, CA2, CA3, and CA24) contain a substantial proportion of the Guatemalan × Mexican genome, and they most likely are complex hybrids containing genomic material from all three races. The position of the last group of accessions in the PCoA diagram is at the transition area between the West Indian and the Guatemalan × Mexican groups, but they cluster with the West Indian accessions in the UPGMA dendogram, showing that the dendogram is not the optimal method to display genetic relationships in avocado germplasm.
The six cultivars with ambiguous assignation are a mixture of different degrees of the ancestral Mexican and Guatemalan × Mexican subpopulations. Bacon and Negra de la Cruz are composed of 50 % of each subpopulation. That fact, combined with their position in the PCoA diagram being closer to the Mexican group, seems to indicate that they contain a higher proportion of the Mexican subpopulation. Edranol, Fuerte, Hass, and “Pinkerton” were assigned to the Guatemalan × Mexican subpopulation and grouped there in the PCoA diagram. Included in the Mexican group are the newly assigned cultivars Gordon, which seems to be a combination of Mexican and Guatemalan gene pools; Cónsula9, with some of the West Indian genome; and Mckee, which is pure Mexican. Further analysis using pure Guatemalan accessions will help to identify the exact percentage of Mexican and Guatemalan gene pools present in the Guatemalan × Mexican hybrids.
The lack of pure Guatemalan genotypes in our accession set hinders us from stating definitive conclusions about the Guatemalan race. Guatemalan × Mexican subpopulation is the only subpopulation that shows an excess of heterozygotes than is expected under Hardy–Weinberg equilibrium (F = −0.114), pointing to its hybrid nature. Nevertheless, the results suggest that the Guatemalan gene pool is represented in that subpopulation, as the pairwise F ST calculation and the grouping analysis clearly separate the Guatemalan × Mexican subpopulation from the Mexican subpopulation. The 65 alleles (34 % from total) that are present in the Guatemalan × Mexican subpopulation and absent in the Mexican subpopulation likely belong to the Guatemalan gene pool. In addition, the total average gene diversity (He = 0.83) found in the present study is similar to the one obtained by Schnell et al. (2003) in a larger sample with accessions representing all racial groups and interracial hybrids.
Some accessions were assigned to different races than previously defined. Anaheim was reported as Guatemalan, and Plowman was reported as a Guatemalan hybrid; however, both of these accessions were allocated to the Mexican group in our study. Meresnsky 1 and Tochimilco 3 were previously defined as Mexican but were included in the Guatemalan × Mexican group using our parameters. The STRUCTURE results confirmed and quantified this new genome composition. It was estimated that Anaheim has 80 % and Plowman has 99 % of its genome in common with the ancestral Mexican subpopulation, and approximately 93 % of the Meresnsky 1 genome is related to the Guatemalan × Mexican subpopulation. The Mexican and Guatemalan × Mexican ancestors equally formed the Tochimilco 3 genome; in this case, incorrect race assignation could be explained by character segregation. Other incorrect clustering may be due to genotype miscalling. Schnell et al. (2003) estimated an error ratio of 7 % associated with propagation of their avocado collection. Mistakes during the process of material exchange and conservation are common and cannot be discarded as the origin of these discrepancies. Future revisions that compare the band pattern of those trees with the ones conserved in their country of origin will clarify whether we are dealing with the wrong genotype or wrong race assignation.
Race relationships: diversity distribution and race-specific markers
The different diversity and phylogenetic studies have found controversial relationships between the three avocado races (Williams 1977; Davis et al. 1998; Fiedler et al. 1998). The evaluation of population and race structure has important implications for traditional considerations in genetic resources management and for the use of advanced genetics approaches, like linkage disequilibrium and association mapping. Chen et al. (2009) showed that admixture among avocado races is a confounding factor that induces substantial interlocus linkage disequilibrium, and accurate characterization of the genetic structure within samples is needed before undertaking an association mapping approach. However, when the establishment of genetic relationship of avocado races is based on cultivated genotypes, the final picture is conditioned by several factors.
Hybrid nature of accessions
An accurate control of the hybrid nature of cultivated genotypes is needed to determine the true genetic and phylogenetic distances between races, as the hybrid character alters the diversity distribution. In our work, subpopulations representing the three common races (Mexican, Guatemalan × Mexican, and West Indian) were different from each other, as Schnell et al. (2003) and Fiedler et al. (1998) previously noticed. However, the inclusion of the hybrid subpopulation in the variation partitioning analysis decreases the diversity among subpopulations (from 25 to 16 %) and, therefore, the ability of markers to distinguish them. Our hybrid subpopulation is formed by 11 interracial hybrids, but only two of them (Bacon and Negra de la Cruz) were previously defined as such. The inclusion of interracial accessions, often described as pure races, could be one explanation for the slight differences among botanical races found in previous studies (Ashworth and Clegg 2003).
Race representation in the accession set studied
Our results indicate that the Mexican and Guatemalan races are closer to each other than either is to the West Indian race. This result agrees with the remarks by Chen et al. (2009) that genotypes from South Mexico are similar to the Guatemalan race, but contrasts with the findings of Schnell et al. (2003), which state that equal distance exists between the three avocado races. This disagreement could be due to the different composition of the accession sets studied. Our use of the hybrid Guatemalan × Mexican subpopulation to represent the Guatemalan gene pool could be overestimating the genetic similarity between the Mexican and Guatemalan races. However, a small distance between the Mexican and Guatemalan genomes fits with the high number of Mexican × Guatemalan interracial hybrids, either artificially or naturally obtained, and their common ecological niche in Central America highlands (Ashworth and Clegg 2003). On the other hand, our West Indian group is represented mainly by accessions growing in the Canary Islands, both seedlings from traditional local trees or offspring from Cuban trees. Avocado was known in the Canary Islands since shortly after the Spaniards discovered the New World in 1492, but most of the West Indian trees arrived from Cuba in the nineteenth century (L. Gallo, personal communication). During the last several years, the West Indian genome has been introduced into the rootstock selection programs as an alternative to overcome various soil problems (Gallo et al. 2003a), meaning that our accessions represent only a subset of the diversity present in the West Indian race and are different from the subset selected in Florida and studied by Schnell et al. (2003). In accordance with that, the West Indian subpopulation contains the lower diversity values (He = 0.562), is farther from Hardy–Weinberg equilibrium (F = 0.238), and has a significant inbreeding coefficient (F IS = 0.24, p < 0.001).
Size and kinship of accessions in race subpopulations
The small size of the subpopulations obtained can distort the values of inbreeding coefficients, but global results indicate that a behavior as a non-random mating group occurred only within the West Indian subpopulation. However, Wright's fixation index points that some degree of kinship within every race group cannot be discarded, with the exception of the hybrid subpopulation, the only population that represents a mixture of the three races.
Accession origin
Additionally, some bias can be due to the selection carried on during the breeding and culture history of cultivated genotypes. We have tried to represent the three races in rootstocks and cultivars, including both traditional and contemporary genotypes. Traditional cultivars were introduced early in the culture times and are well-adapted to the original environment conditions. Modern cultivars have been selected more recently from locally grown seedlings that were produced by open pollination or, rarely, by seedlings produced from a controlled cross (Bergh 1975). The rootstock breeding has been more fruitful than the cultivar breeding. The first rootstocks that were planted were seedlings of Mexican local avocados (ex. Mexicola and Topa Topa). Since 1977, when the avocado propagation technique was optimized, clonal avocado rootstocks resulting from rootstock program selections (ex. Barr Duke, Duke6, Duke9, Toro Canyon, Merensky 1, and Maoz) started to spread worldwide (Ben-Ya’acov and Michelson 1995), combining with local traditional rootstocks (ex. Scott and Thomas). However, the cultivar market is mainly composed of traditional cultivars (Hass, Fuerte, Bacon, etc.), and only some are the result of breeding programs (Iriet, Adi, and Eden) (Lahav et al. 1989, 1992a, b; Lavi et al. 1997). In our accession set, the Mexican group rootstock, selected from the University of California, is grouped apart from other Mexican rootstocks and local selections. All the West Indian representatives are rootstocks selected for high tolerance to salinity and soil diseases (Gallo et al. 2003b), and most of the cultivars are interracial Guatemalan and Mexican hybrids. Thus, the genetic relationship among the three botanical races is most likely influenced by the non-uniformity of seedling rootstocks, the selection for ease of propagation in clonal rootstocks, and the market and grower preferences in clonal cultivars and rootstocks.
A more accurate portrayal of the racial organization should be undertaken based on the study of wild avocado populations coming from every center of origin. Chen et al. (2009) delineated the racial boundaries de novo using haplotype data of four nuclear genes from a sample of 21 wild avocado accessions. Despite the small number of accessions and markers involved, a more complex structure of the Central Mexico populations was revealed, where two subpopulations were distinguished based on elevation and latitude. The difficulties and contradictions found when trying to describe a genealogical relationship based only on a small sample of the cultivated avocado, or a small sample of the total genome, could be explained by a greater complexity inside each botanical race. In our accession set, the extraordinary proportion of rare (49 %) and accession-unique alleles (25 %) indicates the existence of a high degree of variability that is hardly represented in our collection (Barkley et al. 2006). Within every subpopulation, the Mexican group accumulates the highest proportion of group and accession-specific alleles. A larger sample size and diversity of our Mexican group could explain it, but the fact that the Mexican race contains higher diversity was also noted in previous works (Schnell et al. 2003; Ashworth and Clegg 2003) and was again illustrated here with the broad dispersion in the PCoA diagram and the STRUCTURE results under the of K = 4 model. The hybrid subpopulation contained only four specific alleles (1 %), fitting with its interracial character and the mixture of gene pools represented in the other subpopulations.
All the factors mentioned above that shift the diversity distribution among races (hybrid nature, sampling variability within races, kinship, local, and breeding selection) and the increase in diversity within races (heterogeneity of seedlings of cultivars/rootstocks and germplasm management mistakes) hamper the establishment of clear boundaries among botanical races and the identification of race-specific markers. Because of higher diversity, it has been exceptionally difficult to identify Mexican-specific markers. In fact, only two markers are able to tag the Mexican group and only when two alleles of each are considered. In contrast, seven and six different markers are able to label Guatemalan × Mexican and West Indian accessions, respectively. An analysis of the alleles detected by these 13 markers in a different and wider group of accessions will help to validate their utility as universal race-specific markers.
Conclusions
The application of single-copy, easy-to-use, and highly reproducible markers, such as microsatellites, in addition to a range of complementary analytical methods, have been proven to be sufficient and necessary to better understand the complex relationships in the avocado germplasm. A high number of good quality markers, accessions representing the three avocado races, and analyses with both ordination and model-based methods are needed to reach consistent conclusions concerning genetic relationships and to establish the race composition of each individual.
The inferences of avocado race relationships based on cultivated genotypes present a range of factors that alter the depiction of diversity distribution and distort their genetic relationships. From this study, it is important to note that many genotypes considered to be “pure races” contain two or more gene pools to different degrees. Additionally, there is a bias in diversity depiction due to several features of the genotype set analyzed, namely, the hybrid nature of the genotypes, the kinship among the accessions, the agronomical and breeding selection of accessions, and an insufficient representation of the total racial diversity. In that sense, a real picture of the avocado germplasm requires the study of the wild genome or the choice of a broad range of local and breeding cultivars from different ecological areas and accurate control of all factors. Understanding the complex structure of the avocado germplasm therefore requires a higher investment (high number of informative markers and high number of accessions) than that required for simpler gene pools.
The practical importance of this study lies in the ability to unravel the complex hybrid origins of various cultivars and the first identification of race-specific markers. The evaluation of race structure and the hybrid nature of the accessions should lead to better management of avocado genetic resources for breeding programs and germplasm conservation. These results also have important implications in the increasingly exploited association mapping approach where an accurate characterization of the structure within samples is needed before it (Chen et al. 2009). In this work, we have presented some tools and tips to achieve this first step.
References
Abajian C (1994) Sputnik program http://rast.abajian.com/sputnik. Accessed October 2008
Alcaraz ML, Hormaza JI (2007) Molecular characterization and genetic diversity in an avocado collection of cultivars and local Spanish genotypes using SSRs. Hereditas 144:244–253
Ashworth VETM, Clegg MT (2003) Microsatellite markers in avocado (Persea americana Mill.). Genealogical relationships among cultivated avocado genotypes. J Hered 94:407–415
Ashworth VETM, Kobayashi MC, De la Cruz M, Clegg MT (2004) Microsatellite markers in avocado (Persea americana Mill.): development of dinucleotide and trinucleotide markers. Sci Hortic 101:255–267
Barkley N, Roose M, Krueger R, Federici C (2006) Assesing genetic diversity and population structure in a citrus germplasm collection utilizing simple sequence repeat markers (SSRs). Theor Appl Genet 112:1519–1531
Ben-Ya’acov A, Michelson E (1995) Avocado rootstocks. In: Janick J (ed) Horticultural reviews, volume 17. Wiley, New York, pp 381–429, Posted with permission of J. Janick and the International Society for Horticultural Science
Ben-Ya’acov A, Zilberstaine M, Goren M (2003) The Israeli avocado germplasm bank: where and why the items had been collected. Proc V World Avocado Congr :13–19
Bergh BO (1975) Avocados. In: Janick J, Moore JN (eds) Advances and fruit breeding. Purdue University Press, Lafayette, pp 541–567
Bergh BO, Lahav E (1996) Avocados. In: Janick J, Moore JN (eds) Fruit breeding, vol 1. Tree and tropical fruits. Wiley, New York, pp 113–166
Borrone JW, Schnell RJ, Violi HA, Ploetz RC (2007) Seventy microsatellite markers from Persea americana Miller (avocado) expressed sequence tags. Mol Ecol Notes 7:439–444
Borrone JW, Brown JS, Tondo CL, Mauro-Herrera M, Kuhn DN, Violi HA, Sautter RT, Schnell RJ (2009) An EST-SSR-based linkage map for Persea americana Mill. (avocado). Tree Genet Genomes 5:553–560
Bufler G, Ben-Ya’acov A (1992) A study of the avocado genetic resources. III. Ribosomal DNA repeat unit polymorphism in avocado. In: Proc, 2nd World Avocado Congress, Riverside, CA 2:545–555
Chanderballi AS, Albert VA, Ashworth VTEM, Clegg MT, Litz RE, Soltis DE, Soltis PS (2008) Persea americana (avocado): bringing ancient flowers to fruit in the genomics era. Bioassays 30:386–396
Chen H, Morrel PL, Ashwoth VETM, De la Cruz M, Clegg MT (2008) Nucloetide diversity and linkage disequilibrium in wild avocado (Persea americana Mill.). J Hered 99(4):382–389
Chen H, Morrel PL, Ashwoth VETM, De la Cruz M, Clegg MT (2009) Tracing the geographic origins of mayor avocado cultivars. J Hered 100:56–65
Davenport TL (1986) Avocado flowering. Hortic Rev 8:257–289
Davis J, Henderson D, Kobayashi M, Clegg MT (1998) Genealogical relationships among cultivated avocado as revealed through RFLP analyses. J Hered 89(4):319–323
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evolutionary. Bioinformatics 2005(1):47–50
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Faostat (2012) Agriculture data. http://faostat.fao.org/. Accessed January 2012
Fiedler J, Bufler G, Bangerth F (1998) Genetic relationships of avocado (Persea americana Mill.) using RAPD markers. Euphytica 101:249–255
Furnier GR, Cummings MP, Clegg MT (1990) Evolution of the avocados as revealed by DNA restriction fragment variation. J Hered 81:183–188
Galindo-Tovar ME, Ogata-Aguilar N, Arzate-Fernandez AM (2008) Some aspects of avocado (Persea americana Mill.) diversity and domestication in Mesoamerica. Genet Resour Crop Evol 55:441–450
Gallo L, Rodríguez A, Silverio de la Rosa F, Díaz S, Domínguez P (2003a) Uso potencial de la raza Antillana como fuente de resistencia a la podredumbre radicular del aguacate. V Congreso Mundial del Aguacate, Málaga, España. Actas: Vol I. p: 61–67
Gallo L, Siverio de la Rosa F, Rodríguez Pérez A, Domínguez Correa P, Pérez Zárate S, Díaz Hernández S (2003b) Evaluación en campo de patrones clonales de aguacate de raza mexicana y antillana tolerante-resistentes a Phytophtora cinnamomi rands. Proceedings V World Avocado Congress. 573–578
Garris AJ, Tai TH, Coburn J, Kresovich S, McCouch S (2005) Genetic structure and diversity in Oryza sativa L. Genetics 169:1631–1638
Goldring A, Zamir D, Degani C (1985) Duplicated phosphoglucose isomerase genes in avocado. Theor Appl Genet 71:491–494
Kadman A, Ben-Ya’acov A (1980) G.A.-13 avocado rootstock selection. HortSci 15:206
Kalinowski ST, Taper ML, Marshall TC (2007) Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol Ecol 16:1099–1106
Kopp L (1966) A taxonomic of the genus Persea in the western hemisphery (Persea-Lauracea) New York Bot. Garden, pp 15–20
Lahav E, Lavi U, Zamet D, Degani C, Gazit S (1989) Iriet—a new avocado cultivar. HortSci 24(5):865–866
Lahav E, Lavi U, Degani C, Zamet D, Gazit S (1992a) ‘Adi’, a new avocado cultivar. HortSci 27(11):1237
Lahav E, Steinhardt R, Kalmar D (1992b). Water requirements and the effect of salinity in an avocado orchard on clay soil. In Lovatt, C.J. (ed.) Proc. Sec. World Avocado Cong, pp 323–330
Lavi U, Lahav E, Genizi A, Degani C, Gazit S, Hillel J (1991) Quantitative genetic analysis of traits in avocado cultivars. Plant Breed 106:149–160
Lavi U, Akkaya M, Bhagwat A, Lahav E, Cregan PB (1994) Methodology of generation and characteristics of simple sequence repeat DNA markers in avocado (Persea Americana M.). Euphytica 80:171–177
Lavi U, Sharon D, Kaufman D, Saada D, Chapnik A, Zamet D, Degani C, Lahav E (1997) ‘Eden’—a new avocado cultivar. HortSci 32(1):151
Litz RE, Raharjo SHT, Gomez Lim MA (2007) Avocado. In: Davey M (ed) Biotechnology in agriculture and forestry, vol. 60. Transgenic crops V. EC Pua. Springer, Berlin
López-López L, Barrientos-Priego A, Ben-Ya’acov A (2001) Study of avocado genetic resources and related kinds species at the Fundacion Salvador Sanchez Colin Cictamex S.C. Memoria Fundación Salvador Sánchez Colin CICTAMEX S.C. Coatepec Harinas, México 1998–2001:188–201
Lynch M (1990) The similarity index and DNA fingerprinting. Mol Biol Evol 7:478–484
Mhameed S, Sharon D, Kaufman D, Lahav E, Hillel J, Degani C, Lavi U (1997) Genetic relationships within avocado (Persea americana Mill) cultivars and between Persea species. Theor Appl Genet 94:279–286
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Newett SDE, Crane JH, Balerdi CF (2002) Cultivars and rootstocks. In: Whiley AW, Schaffer B, Wolstenholme BN (eds) Avocado: botany, production and uses. CAB International, Wallingford
Paetkau D, Calvert W, Stirling I, Strobeck C (1995) Microsatellite analysis of population structure in Canadian polar bears. Mol Ecol 4:347–354
Popenhoe W (1941) The avocado—a horticultural problem. Trop Agric 18:3–7
Prichard JK, Stephens M, Donnely R (2000) Inference of population structure using multilocus genotype date. Genetics 155:945–959
Sanchez-Colín S, Mijares-Oviedo P, Lopez-Lopez L, Barrientos-Priego AF (1998) Historia del Aguacate en Mexico. Memoria Fundación Salvador Sánchez Colin 1998–2001. Coatepec Harinas (México): CICTAMEX p 171–187
Schnell RJ, Brown JS, Olano CT, Power EJ, Krol CA, Kuhn DN, Motamayor JC (2003) Evaluation of avocado germplasm using microsatellite markers. J Amer Soc Hort Sci 128:881–889
Scora RW, Wolstenholme BN, Lavi U (2002) Taxonomy and botany. In: Whiley AW, Schaffer B, Wolstenholme BN (eds) The avocado: botany, production and uses. CABI, Wallingford, pp 15–38
Sharon D, Cregan PB, Mhameed S, Kuharska M, Hillel I, Lahav E, Lavi U (1997) An integrated genetic linkage map of avocado. Theor Appl Genet 95:911–921
Smith CE (1966) Archaeological evidence for selection in avocado. Econ Bot 20:169–175
Varshney RK, Kumar A, Balyan HS, Roy JK, Prasad M, Gupta PK (2000) Charachterization of microsatellites and development of chromosome specific STMs markers in bread wheat. Plant Mol Biol Rep 18:5–16
Viruel MA, Hormaza JI (2004) Development, characterization and variability analysis of microsatellites in lychee (Litchi chinensis Sonn., Sapindaceae). Theor Appl Genet 108:896–902
Viruel MA, Messeguer R, de Vicente MC, García-Mas J, Puigdomènech P, Vargas F, Arús P (1995) A linkage map with RFLP and isozyme markers for almond. Theor Appl Genet 91:964–971
Williams LO (1977) The avocados, a sinopsis of the genus Persea, subg. Persea. Econ Bot 31:315–320
Wright S (1951) The genetical structure of populations. Ann Eugen 15:323–354
www.marm.es. Ministerio de Agricultura, Alimentación y Medio Ambiente (2012) Accessed January 2012
Young PJ (1992) Inherent influence of rootstock race on avocado fruit maturity. Proc. of Second World Avocado Congress: 149–154
Acknowledgments
The financial support for this work was provided by the Spanish Ministry of Education and Science through the Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria project grant RTA2006-00027-00-00 and a predoctoral grant given to E.G.G. We gratefully acknowledge A. Barcelo and L Gallo for providing plant material and information on the avocado germplasm.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Dandekar
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(XLSX 29 kb)
Rights and permissions
About this article
Cite this article
Gross-German, E., Viruel, M.A. Molecular characterization of avocado germplasm with a new set of SSR and EST-SSR markers: genetic diversity, population structure, and identification of race-specific markers in a group of cultivated genotypes. Tree Genetics & Genomes 9, 539–555 (2013). https://doi.org/10.1007/s11295-012-0577-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-012-0577-5