Abstract
Retrotransposons are an ubiquitous component of plant genomes, especially abundant in species with large genomes. Populus trichocarpa has a relatively small genome, which was entirely sequenced; however, studies focused on poplar retrotransposons dynamics are rare. With the aim to study the retrotransposon component of the poplar genome, we have scanned the complete genome sequence searching full-length long-terminal repeat (LTR) retrotransposons, i.e., characterised by two long terminal repeats at the 5′ and 3′ ends. A computational approach based on detection of conserved structural features, on building multiple alignments, and on similarity searches was used to identify 1,479 putative full-length LTR retrotransposons. Ty1-copia elements were more numerous than Ty3-gypsy. However, many LTR retroelements were not assigned to any superfamily because lacking of diagnostic features and non-autonomous. LTR retrotransposon remnants were by far more numerous than full-length elements, indicating that during the evolution of poplar, large amplification of these elements was followed by DNA loss. Within superfamilies, Ty3-gypsy families are made of more members than Ty1-copia ones. Retrotransposition occurred with increasing frequency following the separation of Populus sections, with different waves of retrotransposition activity between Ty3-gypsy and Ty1-copia elements. Recently inserted elements appear more frequently expressed than older ones. Finally, different levels of activity of retrotransposons were observed according to their position and their density in the linkage groups. On the whole, the results support the view of retrotransposons as a community of different organisms in the genome, whose activity (both retrotransposition and DNA loss) has heavily impacted and probably continues to impact poplar genome structure and size.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Class I transposons or retrotransposons (REs) represent the majority of the repetitive component of eukaryotic genomes. REs propagate via a “copy and paste” mechanism in which, after RE transcription, enzymes encoded by the RE synthesize double-stranded DNA copies that are integrated back in the host genome. This mechanism resembles the replication cycle of retroviruses (Wicker et al. 2007).
REs can be separated into LTR and non-LTR retrotransposons, depending on the presence of long terminal repeats (LTRs) flanking the coding portion at both 5′ and 3′ ends. Such repeats are identical at the time of insertion of the new element in the chromosome. They range from a few hundred to several thousand base pairs in length. LTR retrotransposon (LTR-RE) transcription starts in the 5′-LTR, where the TATA box usually occurs; within LTR, cis-regulatory motifs can be found that regulate RE transcription (Sugimoto et al. 2000). An LTR is typically delimited by two dinucleotides TG…CA, has terminal inverted repeats (TIRs) of 6 bp, and is flanked by target site duplications (TSDs) of 4–6 bp. Both TIR and TSD may, however, be imperfect as result of mutations subsequent to LTR-RE insertion.
Internal to the 5′ and 3′ LTRs, respectively, are present the primer binding site (PBS) and the polypurine tract (PPT). They provide the signals for reverse transcription of RE transcripts into the complementary DNA (cDNA) that will be integrated in the genome. The PBS is complementary to a portion of a host encoded transfer RNA (tRNA), which can act as a primer for retrotranscription (Wicker et al. 2007).
The two LTRs flank an internal portion that typically contains one or more open reading frames encoding the enzymes for retrotransposition (Boeke and Corces 1989; Kumar and Bennetzen 1999): gag (encoding a capsid protein) and pol (encoding aspartic proteinase, integrase, reverse transcriptase and RNaseH).
LTR-REs are subdivided into autonomous and non-autonomous elements, depending on the presence, in the internal region flanked by LTRs, of genes encoding the retrotransposition machinery. Among autonomous LTR-REs, superfamilies Ty1-copia and Ty3-gypsy differ in the enzyme order within pol (Wicker et al. 2007). Both superfamilies are ubiquitous throughout the eukaryotes and have been present since the divergence of plants, animals, and fungi.
Non-autonomous LTR-REs have the PBS, PPTs, and LTRs needed for transcription, replication, and integration as cDNA (Sabot and Schulman 2006), but they do not carry genes for retrotransposition and are mobilized in trans using enzymes produced by autonomous LTR-REs. Among non-autonomous LTR-REs, two main groups have been described: terminal-repeat retrotransposons in miniature (TRIMs) and large retrotransposon derivatives (LARDs) (Witte et al. 2001; Kalendar et al. 2004).
Because of the error-prone nature of transcription and reverse transcription, the replicative mechanism of LTR-REs has generated different families. LTR-RE sequence heterogeneity is found in the coding, transcribed portion, and especially in the LTRs (Beguiristain et al. 2001).
The replicative activity of retrotransposons has determined the structure of eukaryotic genomes. Genome expansion by insertion of REs occurred frequently during evolution; on the other hand, retrotransposons have been the object of sequence removal—and, in part, they also have favoured DNA loss—mediated by unequal homologous recombination or by illegitimate recombination (Devos et al. 2002; Ma et al. 2004; Grover et al. 2008). The rates of both genome expansion and genome contraction processes appear to vary between species (Bennetzen et al. 2005; Vitte and Bennetzen 2006), allowing some genomes to shrink while others expand. Within a genome, for example in rice, the occurrence of illegitimate and unequal homologous recombination can be related to the gene density, being higher in coding sequences rich regions (Tian et al. 2009). Rearrangements and illegitimate and unequal homologous recombination are the processes driving DNA removal in plants by multiple mechanisms, including repair of double-strand breaks (nonhomologous end-joining) and slipstrand mispairing (Kalendar et al. 2000; Ma and Bennetzen 2004; Neumann et al. 2006; Ammiraju et al. 2007; Hawkins et al. 2008; Morse et al. 2009).
A survey of the dynamics of different RE superfamilies in eukaryotic genomes is facilitated by the availability of whole genome sequence or, at least, sequence of large portions of the genome, as bacterial artificial chromosome (BAC) clones. In plants, LTR-REs have been largely surveyed in species whose genome has been entirely sequenced and in species for which the sequence of large portions of the genome are available. Gypsy and Copia superfamilies are differently represented in the genome, depending on the species, with respective ratios of 5:1 in papaya (Ming et al. 2008), 4:1 in Sorghum (Paterson et al. 2009), 3:1 in rice (The International Rice Genome Sequencing Project 2005), and 1:2 in grapevine (The French-Italian Public Consortium for Grape Genome Characterization 2007). Maize shows a similar abundance of the two classes (Meyers et al. 2001), with Gypsy elements especially concentrated in gene-poor regions and Copia REs overrepresented in gene-rich ones (Schnable et al. 2009; Baucom et al. 2009a). Similar data are reported for other cereal species with large genomes such as wheat and barley (Vicient et al. 2005; Paux et al. 2006). Species of the Gossypium genus show a variable proportion of Gypsy versus Copia elements with Gypsy elements prevailing in species with larger genome sizes (Hawkins et al. 2006).
Recent reports have shown that retrotransposon sequences can have an impact on the expression of nearby genes (Kashkush et al. 2003) by their presence or absence in the cis-regulatory sequences of genes of the host species. Therefore, the identification and characterisation of LTR-REs are a priority in analyzing the genome of crop species.
Among species whose genome has been sequenced, poplar (Populus trichocarpa), grapevine, and papaya are the only perennial plants, and it is plausible that perennial habit affects genome dynamics in a different way from annually sexually propagated species.
In their report on poplar genome sequencing, Tuskan et al. (2006) reported that class I elements (Ty1-copia-like, Ty3-gypsy-like, LINEs, and unidentified retroelements) are the most abundant (over 5000 copies). Poplar genome is relatively small (550 Mbp) and retroelements cover approximately 176 Mbp (42% of the genome). A prevalence of Gypsy over Copia RE sequences was reported (Tuskan et al. 2006); however, unidentified elements account for 120 Mbp.
Recently, a database of repetitive elements (RepPop) has been released (Zhou and Xu 2009). However, a comprehensive analysis of LTR retrotransposon dynamics in the poplar genome is still not available (Klevebring et al. 2009). With the aim of studying the dynamics of LTR retrotransposons in the poplar genome, we identified putative full-length retrotransposons based on the occurrence of both LTRs and established phylogenetic relationships among them according to LTR sequence similarity.
Materials and methods
LTR-REs identification
Putative LTR-REs were identified in the sequenced genome of P. trichocarpa (Tuskan et al. 2006) deposited at EMBL (accession number AARH00000000.1) using LTR-FINDER software (Xu and Wang 2007). LTR-FINDER uses a suffix-array-based algorithm to construct all exact matching pairs, which are extended to long highly similar pairs. Alignment boundaries are obtained adjusting the ends of LTR pair candidates using the Smith–Waterman algorithm. These boundaries are re-adjusted, based on the occurrence of typical LTR-RE features such as: (1) being flanked by the dinucleotides TG and CA, at 5′ and 3′ ends, respectively; (2) the presence of a TSD of 4–6 bp; (3) the presence of a putative PBS, complementary to a tRNA at the end of putative 5′-LTR; and (4) the occurrence of a putative polypurine tract just upstream of the 5′ end of the 3′ LTR. The following parameters were used: LTR sequence length from 80 to 5,000 bp and maximum distance between LTRs of 20,000 bp. The sequences between two putative LTRs were subsequently analysed by BLASTX and BLASTN searches (E value threshold, 10−5) against public non-redundant databases at GenBank and against REPBASE (Jurka et al. 2005). Sequences are available at the Department of Crop Biology of Pisa University repository website (http://www.agr.unipi.it/Sequence-Repository.358.0.html).
All sequences were masked against RepPop database (Zhou and Xu 2009) using RepeatMasker (developed by A.F.A. Smit, R. Hubley, and P. Green; http://www.repeatmasker.org/).
LTR-REs were annotated using both structure- and homology-based methods. Relationships between LTR-REs were established according to sequence similarity between LTRs. All putative LTRs were clustered using CAP3 software (Huang and Madan 1999) using an overlap length cut-off of 80% and an overlap identity cut off of 80%, following the guidelines for transposable element annotation proposed by Wicker et al. (2007).
Mutation rate estimation
Based on the estimation that separation between tacamahaca and Populus sections (to which P. trichocarpa and Populus alba belong, respectively) occurred in the Miocene between 18 and 23.3 MYA (Eckenwalder 1996), a synonymous substitution rate was calculated comparing protein coding sequences of P. alba (Maestrini et al. 2009) to orthologous sequences in the P. trichocarpa genome. Thirty-one sequences (longer than 320 bp) out of 150 available P. alba sequences [aligned at high similarity (>e −80) with only one sequence in the P. trichocarpa genome] were selected for analysis. Rates of synonymous and nonsynonymous nucleotide substitution for each gene were calculated by the method of Nei and Gojobori (1986) with the Jukes–Cantor correction as implemented in the DnaSP program (Rozas and Rozas 1999). The average synonymous substitution number for 31 genes was estimated.
LTR-REs insertion time estimation
Retrotransposon insertion age was estimated comparing the 5′- and 3′-LTRs of each putative full-length retrotransposon. The two LTRs of a single retrotransposon are identical at the time of insertion because they are mostly copied from the same template. The two LTRs were aligned with ClustalX software (Thompson et al. 1994), indels were eliminated, and the number of nucleotide substitutions was counted using the DnaSP program (Rozas and Rozas 1999). The insertion times of retrotransposons with both LTRs were dated using the Kimura two parameter method (K2P, Kimura 1980), calculated using DnaSP, and a synonymous substitution rate that is twofold the one calculated for genes, according to SanMiguel et al. (1998) and to Ma and Bennetzen (2004).
LTRs copy number estimation
To estimate the number of LTR-RE remnants in the genome, we have measured the number of hits obtained by BLASTN searches against P. trichocarpa genome at Genbank (http://blast.ncbi.nlm.nih.gov/Blast.cgi) using the LTR sequences of each putative full-length LTR-RE as queries. The occurrence of sequences with at least 80% similarity to putative LTRs in EST databases of P. trichocarpa was scored by BLASTN search against such databases at the same NCBI site (E value threshold, 10−5).
Other sequence and statistical analyses
In other analyses, we used the TandemRepeat Finder program (Benson 1999) in conjunction with BLAST analysis against poplar genome at NCBI, to search putative centromeric repeats.
Statistical analyses were carried out using GraphPad Prism Software.
Results
Identification and classification of REs with complete LTRs
An intact LTR retrotransposon was defined as one that contains two relatively intact LTRs and identified PPT and PBS sites and is also flanked by TSDs (Ma et al. 2004), irrespective of encoding or not enzymes for retrotransposition. Using this definition, we started our analyses searching for every sequence flanked by two highly similar sequences longer than 80 bp and with the above specified typical features.
We mined putative LTR-REs of poplar from the entire P. trichocarpa genome using LTR-FINDER software (Xu and Wang 2007). False positives were eliminated by careful checking each sequence separately. To estimate the frequency of false negatives, we masked the sequence of chromosome I with all identified poplar LTR-REs using RepeatMasker. Then, the masked and unmasked sequences of chromosome I were analyzed by tBLASTn using two poplar sequences, a Copia retrotranscriptase and a Gypsy integrase. The unmasked chromosome I showed 172 hits for the Copia sequence and 88 hits for the Gypsy sequence; the masked chromosome I showed only one Gypsy sequence that revealed a retrotransposon fragment. Hence, we estimated that the number of false negatives was negligible.
On the whole, we collected 325 intact elements. Moreover, putative LTR REs with two or one of the above described three typical LTR-RE features (PPT, PBS, and TSD) were identified (1,150 and four elements, respectively). Hereafter, the complete set of 1,479 putative LTR-REs is referred as full-length LTR-REs. Their sequences are available at the Department of Crop Biology of Pisa University repository website (http://www.agr.unipi.it/Sequence-Repository.358.0.html, see also Supplemental file 1).
The collected elements were masked against repetitive sequences present in the RepPop database (Zhou and Xu 2009) using RepeatMasker. Beside the overlaps, there are significant portions unique to both sets. Forty-three percent of bases of our dataset were unmasked. Moreover, 132 out of 1,479 LTR-REs were masked only for 0–15% of their sequence; hence, it can be considered as specific to our dataset.
Nearly all elements found using this approach are isolated, i.e., apparently adjacent to sequences of the host genome. In only 31 loci were we able to recognise nested elements, i.e., an element within another one. We cannot exclude the possibility that more complex nested structures are present in the poplar genome, as observed for example in maize (SanMiguel et al. 1996). However, we decided to limit our search to full-length and linear elements to analyze a homogeneous RE sample.
The recorded putative LTRs had a mean length of 566 bp, but large length variability was observed (up to 4,848 bp, standard deviation = 631.82 bp). As for full-length retrotransposons, the mean length was 7,225 bp, again with a large standard deviation (5,436 bp).
The full-length LTR-REs were compared with the GenBank nr database by BLAST analysis (E value threshold, 10−5) to explore whether sequences encoding RE enzymes were present. Of 1,479 putative LTR-REs, only 595 (40.2%) were found to contain at least one of the coding domains needed for retrotransposition.
LTR-REs were first classified as belonging to Ty3-gypsy, Ty1-copia, or unknown superfamilies according to BLAST analysis of their internal portion (i.e., between LTRs) in comparisons with GenBank and REPBASE databases.
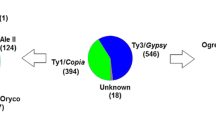
Table 1 reports the number of full-length Ty1-copia-like, Ty3-gypsy-like, and unknown LTR-REs identified in the poplar genome. Unknown putative elements are the most represented in our sample, followed by Ty1-copia- and Ty3-gypsy-like ones.
Concerning unknown full-length elements (855 LTR-REs), in some cases, BLAST analysis showed the presence of coding sequences with similarity to non-LTR retrotransposons (34 elements), to DNA transposons (44 elements), or to helitrons (6 elements) between the putative LTRs. These elements possibly originated by insertion of such sequences in previously existing LTR-REs. In 41 cases, BLAST analysis showed the occurrence of pol or gag encoding sequences, but the attribution to a superfamily was not allowed. The internal domain of other unknown LTR-REs (730 elements) lacked strong homology to any known LTR-RE proteins.
According to Wicker et al. (2007), all elements lacking typical LTR-RE protein encoding sequences can be classified as TRIMs when they had a length <4 kbp and as LARDs when longer than 4 kbp. On the whole, elements not showing any RE enzyme coding portion or elements containing sequences with similarity to DNA transposons or non-LTR-REs and not sharing their LTR sequence with any Copia or Gypsy superfamily were classified as unknown (Wicker et al. 2007).
Chromosome distribution of LTR-REs
Table 1 reports the number of full-length LTR-REs in the 19 linkage groups (LGs) of P. trichocarpa. The putative full-length REs identified in our analysis represent 3.47% of the poplar genome, i.e., a mean of one full-length retroelement every 208,141 bp. The distribution in the 19 LGs is somewhat different, from 6.29% in the LG XIX to 2.00% in the LG VI. Copia LTR-REs are especially frequent in the LG I. Gypsy LTR-REs are more frequent than Copia in five out of the 19 LGs.
In Fig. 1 and Supplemental file 2, the distribution of the 1,479 LTR-REs on the 19 linkage groups of P. trichocarpa is reported. REs are mostly dispersed throughout the chromosomes. Unfortunately, the current Populus genome sequence does not annotate the centromeric regions (Klevebring et al. 2009). Moreover, a complete cytogenetic map of the poplar, based on linkage groups as determined by whole genome sequencing, is still to be established (see Islam-Faridi et al. 2009). The fact that, in some cases, Gypsy-like and unknown LTR-REs are especially clustered in one chromosome position might suggest that this is the centromere position, where Gypsy REs are usually very frequent (Santini et al. 2002 and references therein).
To determine if clustered LTR-REs are actually centromeric, we searched for putative centromeric satellites in the poplar genome using the TandemRepeat Finder software. We identified two types of putative centromeric repeats. The first type, whose consensus sequence is 107 bp long, should allow the identification of the centromere position in chromosomes IV, V, VIII, X, XI, XII, XIII, XIV, and XV. The second, a consensus sequence 142 bp long, should identify the centromere of chromosomes I, III, IX, XVI, XVIII, and XIX (Giordani and Cossu, unpublished data, see Supplemental file 3). No putative centromeric repeats were found in chromosomes II, VI, VII, and XVII, probably because of underrepresentation of repetitive sequences in the currently available poplar genome sequence (Klevebring et al. 2009). It is to be noted that the 142-bp long sequence shows high similarity to a 145-bp tandem repeat sequence isolated by Rajagopal et al. (1999) in Populus deltoides and Populus ciliata, which was described as putatively centromeric.
We overlapped a map track of putative centromeric repeats for each chromosome with the distribution of Copia, Gypsy, and unknown LTR-REs along chromosomes (Fig. 1 and Supplemental files 2a, b). In all chromosomes in which the centromere position seemed to be identified, there was a significant overlap between the putative centromeric position and the accumulation of full-length Gypsy LTR-REs, suggesting the association between centromeric repeats and Gypsy LTR-REs. It is, however, to be recalled that the definition of the centromere position requires biochemical and cytological validation, for example by BAC in situ hybridization (Islam-Faridi et al. 2009).
Family distribution and frequency of LTR-REs in the poplar genome
Usually, structural and sequence similarities are used for the classification of non-autonomous LTR retrotransposons into families; such a classification is used, for example, in Repbase, a database of eukaryotic repetitive and transposable elements (Jurka et al. 2005). Wicker et al. (2007) established application rules to a hierarchical transposable element classification similar to that used in Repbase and defined a family of retrotransposons as a group of REs that have high DNA sequence similarity in their coding region (if present) or internal domain, or in their LTR. Specifically, they proposed that two REs are assumed to belong to the same family if at least 80% of the aligned sequence (LTRs, or internal portion, or both) show 80% or more similarity, analyzing segments longer than 80 bp.
We classified the full-length LTR-REs into families based on their LTR sequence similarity. We used LTR sequences to classify families rather than more commonly used retrotranscriptase (RT) coding domain sequences because many nonautonomous LTR-REs lack an intact RT domain.
The set of 1,479 LTR pairs (longer than 80 bp) were compared using CAP3 algorithm, setting 80% identity of 80% LTR length, with reference to the so called 80–80–80 rule, according to Wicker et al. 2007). A schematic representation of LTR alignments of the four most redundant Gypsy families are reported in Fig. 2 as an example. In the case of the G126 family, all 12 LTRs overlap. In the other cases in Fig. 2, overlapping is not complete; some LTRs do not share their sequence with other LTRs that have been attributed to the same family. Such attribution is justified because if members A and B fulfill the 80–80–80 rule, then they should belong to the same family, and if members B and C also fulfill that rule, then also members A and C should belong to the same family because they should share a common ancestor. Such transitivity might induce errors in classification, as reported by Seberg and Petersen (2009). However, no alternatives have been proposed at present.
Based on this classification, in some cases, autonomous, defective, and non-autonomous elements could be attributed to one and the same family, even in the absence of the coding portion. In such cases, we assumed that non-autonomous and defective elements originated from autonomous elements with which they share LTR sequence.
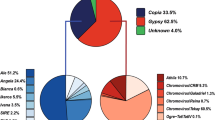
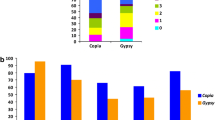
One-hundred twenty-six LTR-RE families were established by this method. Nine hundred eighty-one elements did not cluster and remained single. The mean number of full-length elements per family was 3.94. The distribution of LTR-RE families in relation to the number of components is reported in Fig. 3. The vast majority of families comprise two to three components, and only ten families had more than eight components. Copia and Gypsy families were also analysed separately and Gypsy families resulted more redundant than Copia ones (Fig. 3). The majority of Copia and Gypsy families were specific to poplar. Analysis using RepBase showed, in four cases, similarity to Tto1 Copia elements of Nicotiana tabacum. Some Gypsy families were similar to Diaspora elements of Asparagus officinalis.
In another analysis, the LTR sequence of each full-length RE was compared to the whole poplar genome to measure the frequency of LTR-RE remnants containing that LTR, hence belonging to the same LTR-RE family. The LTR-RE remnants include solo-LTR and isolated LTR fragments and REs with only one complete or fragmented LTR. The frequency of RE remnants was calculated for each LTR-RE family (126 entries) and for single LTR-REs (981 entries), keeping Copia, Gypsy, and unknown elements separate (Table 2).
A correlation occurs between the number of full-length LTR-REs and the number of LTR-RE remnants (not shown); accordingly, the most numerous family (G011) showed the highest number of LTR-RE remnants in the genome. The mean number of LTR-RE remnants per family or single LTR-RE is by far higher for Gypsy than for Copia elements.
The above-described correlation is especially true for Gypsy elements, being not significant for Copia REs (not shown). This should indicate that retrotransposition activity and DNA loss (by rearrangements and by homologous and illegitimate recombination) of Gypsy elements is more ancient than that of Copia elements and/or that mechanisms of DNA loss in Gypsy elements are more efficient (possibly because they are longer than Copia).
Putative insertion dates of LTR-REs
The availability of both complete LTRs allows the insertion time of a LTR-RE to be estimated. Insertion time estimates are based on the occurrence of nucleotide substitutions in the LTRs, which are supposed to be identical at the retroelement insertion time, using a nucleotide substitution rate suitable for such elements (SanMiguel et al. 1998; Ma and Bennetzen 2004). It should be noted that the calculation of insertion date by the number of mutations in sister LTRs is subjected to error because it assumes the same mutation rate in all LTR-RE sequences and all chromosome positions. However, this method appears as the most suitable to study LTR-RE dynamics.
We estimated the synonymous substitution rate by comparing orthologous cDNA sequences of P. alba and P. trichocarpa, i.e., 31 coding sequences for a total of 18,344 bp. The mean number of synonymous substitutions per site (K s) was 0.0483 (Table 3).
Based on the dating of fossil leaves in the second part of the Miocene, the separation between the sections tacamahaca and Populus (to which P. trichocarpa and P. alba belong, respectively) is estimated as 18–23 MYA, i.e., a common ancestor should have existed in the early Miocene (Eckenwalder 1996, and references therein). Recent data based on dating polyploidization events in different Populus species indicates that genus speciation occurred 8–13 MYA (Sterck et al. 2005; Tuskan et al. 2006). The difference in dating Populus speciation was attributed to the use of substitution rates calculated in herbaceous monocots and dicots (Sterck et al. 2005), considering that the generation time of a species is known to affect its nucleotide-substitution rate (Gaut 1998) and that poplar has a much longer generation time than herbaceous species.
Assuming an average of 20.5 MY as insertion date and a K s of 0.0483, the resulting synonymous substitution rate was 2.36 × 10−9 substitutions per years. It has been suggested that mutation rates for LTR retrotransposons may be approximately twofold higher than silent site mutation rates for protein coding genes (Xu and Wang 2007). Consequently, a substitution rate per year of 4.72 × 10−9 was used in our calculations of LTR-RE insertion dates.
LTR pairs were compared in their sequence, excluding deletions from comparisons, and the putative insertion date was calculated for each full-length LTR-RE based on the number of substituted nucleotides per site. When the whole set of usable retrotransposons was taken into account, the nucleotide distance (K) between sister LTRs showed large variation between retroelements (0 to 0.602, Kimura two-parameter method), representing a time span of at most 124 MYA. The putative mean age of analysed LTR-REs is 10.4 MY, with great variability (standard deviation = 8.9 MY). The distribution of full-length LTR-REs according to their putative insertion date is reported in Fig. 4. As expected, since the most ancient LTR-REs should have accumulated the largest variations in their sequences (being not recognised by LTR-FINDER), the frequency of LTR-REs with older insertion date reduces progressively. Analysis of the insertion date profiles provides evidence for overlapping among retrotransposition waves of Gypsy, Copia, and unknown full-length LTR-REs (Fig. 4). When taking into consideration the last 20 MY (i.e., after the separation of poplar sections), peaks of retrotransposition by Gypsy and Copia elements alternate. However, it is to be considered that most full-length LTR-REs were not assigned to any family. If Gypsy- and Copia-related unknown elements in this class were not distributed with nearly 1:1 ratio, different profiles would be observed.
The mean insertion dates of the most numerous Gypsy (6) and Copia (4) families (with number of full-length LTR-REs ≥ 9) show that different families underwent amplification in different time spans (Fig. 5), as indicated also by one-way ANOVA (Table 4).
The profiles of LTR-RE insertion age along the 19 linkage groups are reported in Fig. 6 and Supplemental file 4. Comparisons between the profiles and the mean insertion age of each LG or of the entire genome suggest that retrotransposition occurred at different times in the different chromosomes and chromosome positions (see, for example, LGX) or that mutation rate changes according to chromosome positions. Actually, the concentration of older elements in pericentromeric regions might reflect the suppressed recombination in these areas (Tian et al. 2009).
Distributions of estimated insertion ages (MYA) of full-length LTR-REs along the poplar linkage groups IV and X. For each linkage group two horizontal lines are reported, representing the mean of all full-length LTR-REs in the genome (dotted line) and in each linkage group (dashed line). The box represents the putative position of the centromere as indicated by the occurrence in that position of centromeric repeats
Transcriptional activity of LTR-REs
The transcriptional activity of LTR-REs of our sample was computationally evaluated by BLASTN searches of putative LTR sequences against the available EST databases of P. trichocarpa. Such evaluation represents just a qualitative indication of RE activity, and it should be confirmed by RT-PCR experiments. The available EST collection includes 139,007 sequences from terminal vegetative buds (two libraries), young and mature leaves, along with green shoot tips (one library) phloem and cambium (one library), outer xylem (three libraries) (Ralph et al. 2006), and 17,727 sequences from male catkins, female catkins, and floral buds (Sterky et al. 2004). We are conscious that RE-related EST might result from DNA contamination of the EST library, mostly because of the repetitiveness of RE sequences in the genomes. Moreover, finding ESTs with similarity to LTR sequences could be also related to the expression of small interfering (siRNA): it has been shown that, in young leaves of poplar, the majority of 24 nt short RNA correspond to LTR elements (Klevebring et al. 2009). However, as we found numerous EST matches to LTR-RE sequences, this should be a strong indication that those elements are expressed. We established a threshold of five EST matches to consider a LTR-RE as transcriptionally active. The distribution of full-length Copia, Gypsy, and unknown LTR-REs according to their expression and insertion date is reported in Table 5. Actually, for the vast majority (1188/1479) of LTR-REs, no match to EST sequences was found. The percentages of active full-length LTR-REs (with number of EST matches > 5) range from 3.91 (for Copia REs) to 11.65 (for Gypsy REs). Gypsy REs are apparently more active than Copia ones. Although variations are not significant, there is a tendency for completely inactive full-length LTR-REs (showing no EST matches) to be older than the mean of their superfamily, indicating that transcriptional activity is maintained mostly by young LTR-REs and ancient elements are repressed.
We also related RE transcriptional activity to the frequency of RE remnants for each family. Low copy number families are generally more expressed than highly redundant ones (not shown). This result confirms data in the literature that low copy number REs are the most active (Meyers et al. 2001; Yamazaki et al. 2001).
Relationship between RE density and activity
To study the effect of LTR-RE density on LTR-RE activity, we established two subsets of full-length LTR-REs; the first subset, called clustered LTR-REs, contained the elements found in 400,000-bp long regions in which at least ten full-length LTR-REs are present; the second subset, called dispersed elements, contained the elements found in 1 million bp long regions, in which only one full-length element is present. A descriptive statistics of these two subsets compared to the entire sample of poplar LTR-REs is reported in Table 6. It is to be noted that the two subgroups are placed in opposition to the data of the entire set; LTRs of dispersed elements are less represented in the genome. These elements show lower transcriptional activity and are putatively younger than the entire full-length LTR-RE population. On the contrary, LTR of clustered elements are more common in the genome and these elements are more transcribed and older than the mean of the whole full-length LTR-RE population. The observed different transcriptional activities between the two subsets might suggest that silencing is more efficient when a LTR-RE is dispersed. Concerning the putative insertion age, dispersed elements show more similar sister LTRs; therefore, they should be younger than clustered ones.
Discussion
We have analysed poplar LTR-retrotransposons based on sister LTRs identification. By this approach, only putative full-length retroelements, i.e., with two very similar LTRs, are scored. On the whole, we have isolated 1,479 full-length LTR-REs, of which 132 were identified for the first time, being absent in the existing database of poplar repeated sequences, RepPop (Zhou and Xu 2009), and so adding new retroelements to those already available.
Our data show that Copia full-length retroelements are more common than Gypsy ones (Table 1). However, Gypsy RE remnants were much more common in the genome than Copia ones (Table 2).
Our analysis also showed that the majority of full-length LTR-REs of poplar are of unknown nature, without any apparent coding sequence. Some unknown elements are to be classified as LARDs or TRIMs. To account for the origin of LARDS, it has been proposed that they are the product of transduction of a genomic sequence from the host genome, flanked by two solo LTRs. Alternatively, LARDs may have originated from the virus-like particle by co-encapsulation of a messenger RNA (mRNA) of the autonomous element with a mRNA of any host gene, followed by strand exchange between the two during the reverse transcription step (Jiang et al. 2002). LARDS and TRIMS could also have originated by rearrangements, deletions, and/or illegitimate recombination of old functional elements, both Gypsy and Copia. Some of the LARDs identified in our analyses have probably maintained the capacity to retrotranspose, as indicated by the presence of families with genetically uniform LTRs (Table 2), by the putative very recent insertion dates of some of them (Table 5) and by the occurrence of such sequences in EST libraries (Table 5). Examples of recently inserted nonautonomous LTR-REs are known in other plant species, such as Glycine max (Wawrzynski et al. 2008).
The occurrence of retrotransposon families in poplar was established according to sequence similarity of their LTRs (Wicker et al. 2007). The number of full-length LTR-REs per family is generally low. Gypsy families contain more members than Copia ones (Fig. 3). No family is made of a large number of elements; only ten families show more than eight LTR-REs. Prevalence of small LTR-RE families has been observed also in medium- to large-sized genome angiosperms as maize (Schnable et al. 2009) and sunflower (Cavallini et al. 2010).
Our data show a direct relationship between the number of full-length LTR-REs of a family and the number of LTR-RE remnants of that family in the genome. For instance, the LTR sequence of the largest family, G011, made of 37 full-length elements, shows high similarity with 3,754 sequences in the genome, indicating that this family has been active in ancient times and the vast majority of components of this family are now LTR-RE remnants. This aspect is generally true for poplar full-length LTR-retrotransposons (Table 2). The equilibrium between enlargement of the genome by retrotransposition and RE DNA loss affects the genome size of a species (Devos et al. 2002; Ma et al. 2004; Grover et al. 2008). Our data suggest that, in poplar, a small-sized genome species, the equilibrium between retrotransposition activity and loss of DNA is biased towards DNA loss and that, probably, many REs have been active also in ancient times.
Analysis of sister LTR similarity indicates that, in poplar, both Gypsy and Copia REs have been active in the same period. Nearly all the identified full-length elements appear to be mobilised in a time span of 40 MY (Fig. 4). It is conceivable that more ancient REs are no more recognizable because of accumulation of variability between sister LTRs.
The mean insertion date of poplar Copia full-length REs is lower than that of Gypsy ones (9.301 vs. 10.259 MY, Table 5). The insertion date profiles indicate that, after separation of poplar sections, Copia and Gypsy REs have both been active, but with different time courses. It can also be observed that different Copia and Gypsy families show different mean insertion times (Fig. 5, Table 4). Similar results have been reported in other species, in which retrotransposon superfamilies are subjected to different amplification histories during the evolution of the host; for instance, in wheat, Copia and Gypsy superfamilies are differently represented in the A and B genomes (Charles et al. 2008). Another example of different amplification histories among LTR-RE families was reported for Copia elements of Vitis vinifera (Moisy et al. 2008).
Concerning LTR-RE activity, a search for LTR sequences in EST databases of P. trichocarpa showed that only a small number of families appear to be transcriptionally active, often composed of one or at most two full-length elements. Generally, ancient full-length LTR-REs are inactive or less active than young ones, probably because of the accumulation of mutations determining premature stop codons in the coding portion of the LTR-RE, as observed in rice (Baucom et al. 2009b). Moreover, there is also a strong control of retrotransposon activity by the host species; it has been established that retrotransposons are especially silenced by siRNA (Lisch 2009). It is plausible that the large number of LTR-RE fragments spread throughout the poplar genome can produce siRNAs that silence related retroelements. Many 24-nt small RNAs associated to LTRs have been recently discovered in the poplar (Klevebring et al. 2009).
LTR-REs are present in poplar chromosomes at different densities. No loci are found with more than 16 full-length REs inserted therein. Nonsignificant variations are observed for mean insertion age between chromosomes, though such values range from 7.9 to 12.2 MY (Table 1). Within chromosomes, large regions are found in which the mean insertion age of full-length retrotransposons are either higher or lower than the mean insertion age of LTR-REs in the whole chromosome (Fig. 6). Not only have LTR-REs inserted in different positions at different ages, but their retrotransposition activity appears to be somehow specific to their position in the chromosome (Table 6). In fact, LTR-REs inserted in regions with high full-length elements density belong to families whose LTR is largely represented in the genome (the number of LTR-RE remnants containing single LTRs or LTR fragments related to those elements is higher than the general mean), a feature related to the past activity of a LTR-RE family. On the other hand, dispersed full-length LTR-REs belong to families with lower numbers of related remnants than the general mean, i.e., with low past activity. In addition, a parameter indicating present activity (LTR-RE transcription) shows a difference between clustered (higher than the general mean) and dispersed elements (lower than the general mean).
Dispersed elements seem also younger than clustered ones because of a higher similarity of sister LTRs. This result could, however, be explained hypothesising that the mutation rate of LTR-REs is higher in clustered than in dispersed elements. In fact, clustered elements are found in regions with a low number of predicted genes, on the contrary, dispersed elements lie in gene-rich regions, that are probably preserved from retrotransposition and, in general, from mutations; in this sense, the higher identity shown by sister LTRs of dispersed elements should depend more on the region in which the element is found and less on the insertion age of the retrotransposon. Such a conclusion should support the hypothesis of the existence of different mutation rates in different kinds of transposon sequences or in different chromosome positions (Zuccolo et al. 2010) and would also indicate that insertion ages measured on sequence dissimilarity between LTR pairs are to be taken with caution.
Our analyses show the relationships between sequence characteristics, estimated age of LTR retrotransposons and their transcriptional activity in poplar LTR-REs. They are similar to those observed in other plant species and support the theory of a “life-history” common to all LTR-REs, which includes birth through transposition, followed by silencing and then death by both random mutation and possibly deletion from the genome (Baucom et al. 2009b). However, we observed that different superfamilies and families are subjected to transposition in different time spans and show different transcription levels, suggesting that if dynamics are similar, the factors inducing such dynamics might be different in different families and possibly related to the “ecosystem” in which the REs interact and compete, as proposed by Le Rouzic et al. (2007). In this sense, according to Venner et al. (2009), we suggest that poplar REs are a community of different organisms in the genome, with RE superfamilies, which can be described as species, and with “subspecies” characterised by different LTR sequence, activity, and evolution history.
Abbreviations
- RE:
-
Retrotransposon
- LTR RE:
-
LTR retrotransposon
- LTR:
-
Long terminal repeat
- MY:
-
Million of years
- MYA:
-
Million years ago
References
Ammiraju JS, Zuccolo A, Yu Y, Song X, Piegu P, Chevalier F, Walling JG, Ma J, Talag J, Brar DS, SanMiguel PJ, Jiang N, Jackson SA, Panaud O, Wing RA (2007) Evolutionary dynamics of an ancient retrotransposon family provides insights into evolution of genome size in the genus Oryza. Plant J 52:342–351
Baucom RS, Estill JC, Chaparro C, Upshaw N, Jogi A, Deragon JM, Westerman RP, SanMiguel PJ, Bennetzen JL (2009a) Exceptional diversity, non-random distribution, and rapid evolution of retroelements in the B73 maize genome. PLoS Genet 5:e1000732. doi:10.1371/journal.pgen.1000732
Baucom RS, Estill JC, Leebens-Mack J, Bennetzen JL (2009b) Natural selection on gene function drives the evolution of LTR retrotransposon families in the rice genome. Genome Res 19:243–254
Beguiristain T, Grandbastien MA, Puigdomenech P, Casacuberta JM (2001) Three Tnt1 subfamilies show different stress-associated patterns of expression in tobacco. Consequences for retrotransposon control and evolution in plants. Plant Physiol 127:212–221
Bennetzen JL, Ma J, Devos KM (2005) Mechanisms of recent genome size variation in flowering plants. Ann Bot 95:127–132
Benson G (1999) Tandem Repeat Finder: a program to analyze DNA sequences. Nucl Acids Res 27:573–580
Boeke JD, Corces VG (1989) Transcription and reverse transcription of retrotransposons. Ann Rev Microbiol 43:403–434
Cavallini A, Natali L, Zuccolo A, Giordani T, Jurman I, Ferrillo V, Vitacolonna N, Sarri V, Cattonaro F, Ceccarelli M, Cionini PG, Morgante M (2010) Analysis of transposons and repeat composition of the sunflower (Helianthus annuus L.) genome. Theor Appl Genet 120:491–508
Charles M, Belcram H, Just J, Huneau C, Viollet A, Couloux A, Segurens B, Carter M, Huteau V, Coriton O, Appels R, Samain S, Chalhoub B (2008) Dynamics and differential proliferation of transposable elements during the evolution of the B and A genomes of wheat. Genetics 180:1071–1086
Devos KM, Brown JKM, Bennetzen JL (2002) Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res 12:1075–1079
Eckenwalder JE (1996) Systematics and evolution of Populus. In: Stettler RF, Bradshaw HD, Heilman PE, Hinckley TM (eds) Biology of Populus and its implications for management and conservation. NRC Research Press, National Research Council of Canada, Ottawa, pp 7–32
Gaut BS (1998) Molecular clocks and nucleotide substitution rates in higher plants. In: Hecht MK, Macintyre RJ, Clegg MT (eds) Evolutionary Biology, vol 30. Plenum, New York, pp 93–120
Grover C, Hawkins J, Wendel J (2008) Phylogenetic insights into the paceand pattern of plant genome size evolution. In: Volff JN (ed) Plant genomes. Genome dynamics, vol 4. Karger, Basel, pp 57–68
Hawkins JS, Kim HR, Nason JD, Wing RA, Wendel JF (2006) Differential lineage-specific amplification of transposable elements is responsible for genome size variation in Gossypium. Genome Res 16:1252–1261
Hawkins JS, Hu G, Rapp RA, Grafenberg JL, Wendel JF (2008) Phylogenetic determination of the pace of transposable element proliferation in plants: Copia and LINE-like elements in Gossypium. Genome 51:11–18
Huang X, Madan A (1999) CAP3: A DNA sequence assembly program. Genome Res 9:868–877
Islam-Faridi MN, Nelson CD, DiFazio SP, Gunter LE, Tuskan GA (2009) Cytogenetic analysis of Populus trichocarpa—ribosomal DNA, telomere repeat sequence, and marker-selected BACs. Cytogenet Genome Res 125:74–80
Jiang N, Jordan IK, Wessler SR (2002) Dasheng and RIRE2. A nonautonomous long terminal repeat element and its putative autonomous partner in the rice genome. Plant Physiol 130:1697–1705
Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J (2005) Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res 110:462–467
Kalendar R, Tanskanen J, Immonen S, Nevo E, Schulman AH (2000) Genome evolution of wild barley (Hordeum spontaneum) by BARE-1 retrotransposon dynamics in response to sharp microclimatic divergence. Proc Natl Acad Sci USA 97:6603–6607
Kalendar R, Vicient CM, Peleg O, Anamthawat-Jonsson K, Bolshoyb A, Schulman AH (2004) Large retrotransposon derivatives: abundant, conserved but nonautonomous retroelements of barley and related genomes. Genetics 166:1437–1450
Kashkush K, Feldman M, Levy AA (2003) Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat Genet 33:102–106
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Klevebring D, Street NR, Fahlgren N, Kasschau KD, Carrington JC, Lundeberg J, Jansson S (2009) Genome-wide profiling of Populus small RNAs. BMC Genomics 10:620
Kumar A, Bennetzen J (1999) Plant retrotransposons. Annu Rev Genet 33:479–532
Le Rouzic A, Dupas S, Capy P (2007) Genome ecosystem and transposable elements species. Gene 390:214–220
Lisch D (2009) Epigenetic regulation of transposable elements in plants. Annu Rev Plant Biol 60:43–66
Ma J, Bennetzen JL (2004) Rapid recent growth and divergence of rice nuclear genomes. Proc Natl Acad Sci USA 101:12404–12410
Ma J, Devos KM, Bennetzen JL (2004) Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res 14:860–869
Maestrini P, Cavallini A, Rizzo M, Giordani T, Bernardi R, Durante M, Natali L (2009) Isolation and expression analysis of low temperature-induced genes in white poplar (Populus alba). J Plant Physiol 166:1544–1556
Meyers BC, Tingey SV, Morgante M (2001) Abundance, distribution, and transcriptional activity of repetitive elements in the maize genome. Genome Res 11:1660–1676
Ming R, Hou S, Feng Y, Yu Q, Dionne-Laporte A, Albert H, Suzuki JY, Tripathi S, Moore PH, Gonsalves D (2008) The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus). Nature 452:991–997
Moisy C, Garrison KE, Meredith CP, Pelsy F (2008) Characterization of ten novel Ty1/Copia-like retrotransposon families of the grapevine genome. BMC Genomics 9:469
Morse AM, Peterson DG, Islam-Faridi MN, Smith KE, Magbanua Z, Garcia SA, Kubisiak TL, Amerson HV, Carlson JE, Nelson CD, Davis JM (2009) Evolution of genome size and complexity in Pinus. PLoS One 4:e4332
Nei M, Gojobori T (1986) Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol 3:418–426
Neumann P, Koblizkova A, Navratilova A, Macas J (2006) Significant expansion of Vicia pannonica genome size mediated by amplification of a single type of giant retroelement. Genetics 173:1047–1056
Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, Schmutz J, Spannagl M, Tang H, Wang X, Wicker T, Bharti AK, Chapman J, Feltus FA, Gowik U, Grigoriev I, Lyons E, Maher CA, Martis M, Narechania A, Otillar RP, Penning BW, Salamov AA, Wang Y, Zhang L, Carpita NC, Freeling M, Gingle AR, Hash CT, Keller B, Klein P, Kresovich S, McCann MC, Ming R, Peterson DG, Rahman M, Ware D, Westhoff P, Mayer KFX, Messing M, Rokhsar DS (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457:551–556
Paux E, Roger D, Badaeva E, Gay G, Bernard M, Sourdille P, Feuillet C (2006) Characterizing the composition and evolution of homoeologous genomes in hexaploid wheat through BAC-end sequencing on chromosome 3B. Plant J 48:463–474
Rajagopal J, Das S, Khurana DK, Srivastava PS, Lakshmikumaran M (1999) Molecular characterization and distribution of a 145-bp tandem repeat family in the genus Populus. Genome 42:909–918
Ralph S, Oddy C, Cooper D, Yueh H, Jancsik S, Kolosova N, Philippe RN, Aeschliman D, White R, Huber D, Ritland CE, Benoit F, Rigby T, Nantel A, Butterfield YSN, Kirkpatrick R, Chun E, Liu J, Palmquist D, Wynhoven B, Stott J, Yang G, Barber S, Holt RA, Siddiqui A, Jones SJM, Marra MA, Ellis BE, Douglas CJ, Ritland K, Bohlmann J (2006) Genomics of hybrid poplar (Populus trichocarpa × deltoides) interacting with forest tent caterpillars (Malacosoma disstria): normalized and full-length cDNA libraries, expressed sequence tags, and a cDNA microarray for the study of insect-induced defences in poplar. Mol Ecol 15:1275–1297
Rozas J, Rozas R (1999) DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15:174–175
Sabot F, Schulman AH (2006) Parasitism and the retrotransposon life cycle in plants: a hitchhiker’s guide to the genome. Heredity 97:381–388
SanMiguel P, Tikhonov A, Springer PS, Edwards KJ, Zakharov D, Melake-Berhan A, Springer PS, Edwards KJ, Lee M, Avramova Z, Bennetzen JL (1996) Nested retrotransposons in the intergenic regions of the maize genome. Science 274:765–768
SanMiguel P, Gaut BS, Tikhonov A, Nakajima Y, Bennetzen JL (1998) The paleontology of intergene retrotransposons of maize. Nature Genet 20:43–45
Santini S, Cavallini A, Natali L, Minelli S, Maggini F, Cionini PG (2002) Ty1/Copia- and Ty3/Gypsy-like DNA sequences in Helianthus species. Chromosoma 111:192–200
Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, Minx P, Reily AD, Courtney L, Kruchowski SS, Tomlinson C, Strong C, Delehaunty K, Fronick C, Courtney B, Rock SM, Belter E, Du F, Kim K, Abbott RM, Cotton M, Levy A, Marchetto P, Ochoa K, Jackson SM, Gillam B, Chen W, Yan L, Higginbotham J, Cardenas M, Waligorski J, Applebaum E, Phelps L, Falcone J, Kanchi K, Thane T, Scimone A, Thane N, Henke J, Wang T, Ruppert J, Shah N, Rotter K, Hodges J, Ingenthron E, Cordes M et al (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326:1112–1115. doi:1126/science.1178534
Seberg O, Petersen G (2009) A unified classification system for eukaryotic transposable elements should reflect their phylogeny. Nature Rev Genet 10:276
Sterck L, Rombauts S, Jansson S, Sterky F, Rouzé P, Van de Peer Y (2005) EST data suggest that poplar is an ancient polyploid. New Phytol 167:165–170
Sterky F, Bhalerao RR, Unneberg P, Segerman B, Nilsson P, Brunner AM, Charbonnel-Campaa L, Lindvall JJ, Tandre K, Strauss SH, Sundberg B, Gustafsson P, Uhlen M, Bhalerao RP, Nilsson O, Sandberg G, Karlsson J, Lundeberg J, Jansson S (2004) A Populus EST resource for plant functional genomics. Proc Natl Acad Sci USA 101:13951–13956
Sugimoto K, Takeda S, Hirochika H (2000) MYB-related transcription factor NtMYB2 induced by wounding and elicitors is a regulator of the tobacco retrotransposon tto1 and defense-related genes. Plant Cell 12:2511–2528
The French-Italian Public Consortium for Grape Genome Characterization (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449:463–467
The International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436:793–800
Thompson JD, Desmond G, Gibson H, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res 22:4673–4680
Tian Z, Rizzon C, Du JC, Zhu L, Bennetzen JL, Jackson SA, Gaut BS, Ma J (2009) Do genetic recombination and gene density shape the pattern of DNA elimination in rice long terminal repeat retrotransposons? Genome Res 19:2221–2230
Tuskan GA, DiFazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, Schein J, Sterck L, Aerts A, Bhalerao RR, Bhalerao RP, Blaudez D, Boerjan W, Brun A, Brunner A, Busov V, Campbell M, Carlson J, Chalot M, Chapman J, Chen GL, Cooper D, Coutinho PM, Couturier J, Covert S, Cronk Q, Cunningham R, Davis J, Degroeve S, Déjardin A, dePamphilis C, Detter J, Dirks B, Dubchak I, Duplessis S, Ehlting J, Ellis B, Gendler K, Goodstein D, Gribskov M, Grimwood J, Groover A, Gunter L, Hamberger B, Heinze B, Helariutta Y et al (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313:1596–1604
Venner S, Feschotte C, Biemont C (2009) Dynamics of transposable elements: towards a community ecology of the genome. Trends Genet 25:317–323
Vicient CM, Kalendar R, Schulman AH (2005) Variability, recombination, and mosaic evolution of the barley BARE-1 retrotransposon. J Mol Evol 61:275–291
Vitte C, Bennetzen JL (2006) Analysis of retrotransposon structural diversity uncovers properties and propensities in angiosperm genome evolution. Proc Natl Acad Sci USA 103:17638–17643
Wawrzynski A, Ashfield T, Chen NWG, Mammadov J, Nguyen A, Podicheti R, Cannon SB, Thareau V, Ameline-Torregrosa C, Cannon E, Chacko B, Couloux A, Dalwani A, Denny R, Deshpande S, Egan AN, Glover N, Howell S, Ilut D, Lai H, Martin del Campo S, Metcalf M, O’Bleness M, Pfeil BE, Ratnaparkhe MB, Samain S, Sanders I, Ségurens B, Sévignac M, Sherman-Broyles S, Tucker DM, Yi J, Doyle JJ, Geffroy V, Roe BA, Saghai Maroof MA, Young NA, Innes RW (2008) Replication of nonautonomous retroelements in soybean appears to be both recent and common. Plant Physiol 148:1760–1771
Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, Paux E, SanMiguel P, Schulman AH (2007) A unified classification system for eukaryotic transposable elements. Nature Rev Genet 8:973–982
Witte CP, Le QH, Bureau T, Kumar A (2001) Terminal-repeat retrotransposons in miniature (TRIM) are involved in restructuring plant genomes. Proc Natl Acad Sci USA 98:13778–13783
Xu Z, Wang H (2007) LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucl Acids Res 35:W265–W268
Yamazaki M, Tsugawa H, Miyao A, Yano M, Wu J, Yamamoto S, Matsumoto T, Sasaki T, Hirochika H (2001) The rice retrotransposon Tos17 prefers low-copy-number sequences as integration targets. Mol Genet Genomics 265:336–344
Zhou F, Xu Y (2009) RepPop: a database for repetitive elements in Populus trichocarpa. BMC Genomics 10:14
Zuccolo A, Sebastian A, Yu Y, Jackson S, Rounsley S, Billheimer D, Wing RA (2010) Assessing the extent of substitution rate variation of retrotransposon long terminal repeat sequences in Oryza sativa and Oryza glaberrima. Rice 3:242–250
Acknowledgments
This research work supported by PRIN-MIUR, Italy, Project “Verso la delucidazione delle basi molecolari dell’eterosi nelle piante coltivate: variazione cis-regolatoria ed espressione genica in ibridi di pioppo.” Thanks are due to Dr. Andrea Zuccolo (Arizona Genomics Institute, USA) and Dr. John A. Walsh (Warwick University, UK) for their critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J. Wegrzyn
Rights and permissions
About this article
Cite this article
Cossu, R.M., Buti, M., Giordani, T. et al. A computational study of the dynamics of LTR retrotransposons in the Populus trichocarpa genome. Tree Genetics & Genomes 8, 61–75 (2012). https://doi.org/10.1007/s11295-011-0421-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-011-0421-3