Abstract
Macaranga (Euphorbiaceae) has received much ecological and evolutionary research attention as a genus that includes some of the most conspicuous pioneer trees of Southeast Asian tropical rainforests and because of its manifold associations with ants, including about 30 species that are obligate ant-plants (myrmecophytes). We used sequence data from three chloroplast DNA loci (ccmp5, ccmp6, atpB-rbcL) to assess phylogeographical patterns in species of Macaranga, section Pruinosae, sampled from various regions of Borneo and the Malay Peninsula. Forty-nine chloroplast DNA haplotypes (HT) were identified among 768 specimens from five species, Macaranga gigantea (N = 329; 23 HT), Macaranga pearsonii (N = 347; 21 HT), Macaranga puberula (N = 24; 4 HT), Macaranga hosei (N = 48; 6 HT), and Macaranga pruinosa (N = 20; 5 HT). Forty-one haplotypes were species-specific, whereas eight haplotypes were shared by two, three, or four species and occupied internal positions in a parsimony network. Population genetic parameters based on haplotype frequencies proved to be in a similar range in the non-myrmecophytic M. gigantea and in the ant-associated M. pearsonii, which have overlapping distributions in northern and eastern Borneo. A comparison of GST and NST values revealed a strong phylogeographic structure in both species, whereas colonization pathways suggested by the network topology were different. Both species exhibited similar levels of haplotypic diversity and moderate to high levels of population differentiation. There were no obvious indications for an influence of the symbiotic ant partners on the population structure of their host plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The island of Borneo is part of Sundaland, which is one of the four large bioregions of Southeast Asia (Woodruff 2010) and one of the 25 worldwide biodiversity hotspots for conservation priorities suggested by Myers et al. (2000). Unfortunately, much of the biodiversity of Southeast Asia as a whole is currently in threat of extinction (Brooks et al. 2002; Sodhi et al. 2004). For Borneo, for example, annual deforestation averages 1.7%, and only 57% of the land surface remained forested in 2002 (Brooks et al. 2002; FAO 2006). Considering the rapid loss of habitat and the need for protection measures, surprisingly little is known about the distribution of botanical richness on Borneo (Slik et al. 2003; Cannon and Manos 2003; Raes et al. 2009).
As a consequence of logging, road building, and other human activities, pioneer trees have become dominant elements in many tropical landscapes, including the lowland dipterocarp forests of Borneo. Despite their ecological significance, the evolutionary history of tropical pioneer trees has been addressed in few studies yet. Most of these are dealing with the Neotropics (e.g., Alvarez-Buylla and Garay 1994; Caron et al. 2000; Dutech et al. 2000; Cavers et al. 2005; Dick and Heuertz 2008), whereas still little is known about genetic diversity, genetic structure, and underlying evolutionary processes in paleotropical pioneer trees (e.g., Muloko-Ntoutoume et al. 2000; Cannon and Manos 2003; Ng et al. 2004; Born et al. 2008). Given the important role that pioneer trees play in the regeneration dynamics of tropical forests in general, knowledge about these processes may be of crucial importance for future management considerations.
Some of the most important pioneer trees of lowland dipterocarp forests of the Malay Archipelago belong to the genus Macaranga Thouars (Euphorbiaceae). Macaranga includes about 280 dioecious species that are distributed from Africa through Southeast Asia to some remote Pacific islands (Whitmore 2008). While Macaranga pioneers are mostly restricted to forest gaps, stream banks, and forest margins in primary forests, they have become tremendously abundant following the anthropogenic destruction of rainforests and nowadays can form almost pure stands in degraded areas. At the same time, Macaranga populations have become increasingly isolated on a larger scale, due to the fragmentation of formerly continuous forests into small patches and the extension of oil palm plantations. The occurrence of certain Macaranga species was shown to be closely related with the level of disturbance in a forest, and these species can therefore serve as indicators of the general type of forest structure (Slik et al. 2003).
A remarkable feature of Macaranga is its manifold associations with ants (Fiala et al. 1989; 1999). Roughly 30 Macaranga species from three taxonomical sections (i.e., Pachystemon, Pruinosae, and Winklerianae) are so-called myrmecophytes or ant-plants, in which the plants provide food and nesting space for the ants in their hollow stems (Davies 2001; Fiala et al. 1989; Whitmore 2008). In exchange, the ants protect their host plants against herbivores and vines. Macaranga myrmecophytes are confined to the ever-wet rainforests of Sundaland and form a diversity center on Borneo (reviewed in Fiala et al. 1999; Blattner et al. 2001).
We are investigating the phylogeography of selected Macaranga species to contribute to a better understanding of biodiversity patterns of myrmecophytic and non-myrmecophytic pioneer trees in the Sundaic bioregion and Borneo in particular. In earlier studies, we successfully reconstructed phylogenetic trees of Macaranga myrmecophytes with the help of nuclear ribosomal ITS sequences (Blattner et al. 2001) and amplified fragment length polymorphisms (Bänfer et al. 2004). Our previous attempts at using chloroplast DNA (cpDNA) haplotypes for generating species-specific phylogeographies were however hampered by the frequent occurrence of shared haplotypes among closely related Macaranga species (Vogel et al. 2003). The problem was particularly pronounced in the bancana group of Macaranga section Pachystemon, where the distribution of cpDNA haplotypes apparently followed a geographical rather than a taxonomical pattern (Bänfer et al. 2006). These results indicated a considerable influence of hybridization and introgression and/or the persistence of ancient haplotypes in this group. In such a situation, phylogeographic analyses of chloroplast data for single species can be misleading (see also Jakob and Blattner 2006).
In the present study, we set up a cpDNA-based phylogeography for five species of Macaranga section Pruinosae with emphasis on comparing the non-myrmecophytic Macaranga gigantea with the ant-associated Macaranga pearsonii. Both species are pioneer trees in lowland dipterocarp rainforests and share an overlapping distribution area in northern and eastern Borneo.
The overall aims of our study were (1) to assess the extent of cpDNA haplotype sharing among Macaranga species of section Pruinosae, (2) to investigate levels of cytoplasmic genetic diversity and genetic structure of two typical pioneer tree species, M. gigantea and M. pearsonii, with a focus on northern and eastern Borneo, and (3) to examine and compare phylogeographical patterns in the two species. We discuss the obtained results in the light of biogeographical, historical, and ecological aspects and present a preliminary hypothesis on the evolution of M. gigantea and M. pearsonii on Borneo. Finally, we discuss whether the obligate association with ants may influence the population structure of myrmecophytic Macaranga pioneer trees.
Materials and methods
Study species
Members of Macaranga section Pruinosae in the Malay Archipelago have been shown to be monophyletic in all molecular studies so far (Blattner et al. 2001; Davies et al. 2001; Bänfer et al. 2004, 2006; Kulju et al. 2007). According to Whitmore (2008), the section comprises nine species, five of which are myrmecophytes. In molecular trees, the non-myrmecophyte M. gigantea is sister to all other species in the section (Bänfer et al. 2004). M. gigantea is locally very abundant and has a wide distribution range covering the Malay Peninsula, Sumatra, and Borneo. Another non-myrmecophyte in section Pruinosae is the rare Macaranga pentaloba which is endemic to the Kelabit Highlands on Borneo (Davies 2001). The likewise non-myrmecophytic Macaranga nicobarica and Macaranga siamensis do not occur in the region considered in this paper. Macaranga pruinosa has a somewhat peculiar status because it has never been found associated with ants on Borneo but is polymorphic for this trait in the rest of its range, i.e., Sumatra and the Malay Peninsula. It is absent from Sabah in the northeast of Borneo. The remaining four species of section Pruinosae are myrmecophytic across their whole distribution ranges. Macaranga hosei is found on the Malay Peninsula, Sumatra, and Borneo except for Sabah where it is replaced by M. pearsonii. Macaranga rufescens is endemic to Brunei and northern Sarawak, where it occurs sympatrically with M. hosei. Macaranga puberula is restricted to high altitudes and only occurs in mountainous habitats of northern Borneo.
All species of section Pruinosae are early successional, light-demanding pioneer trees. Their seeds, which are often produced in large quantities, are covered in a violet-colored aril and are preferentially taken by small birds and squirrels (Davies 2001). Occasionally, hornbills, monkeys, and orangutans have also been observed while feeding on large fruiting trees (B. Fiala and D. Guicking, personal observation). Pollinators are mainly thrips, but in M. gigantea and M. pruinosa, small heteropterans were also found as visitors in male and female flowers (Moog et al. 2002; Moog 2002).
Plant material
A total of 768 specimens from five species of Macaranga section Pruinosae were sampled from various regions of Borneo and the Malay Peninsula. A list of the sampling localities and sample sizes is provided in Table 1. Species identification was based on morphological characters (Davies 2001). Most of the samples were collected from the two main target species of the study, i.e., M. gigantea (N = 329) and M. pearsonii (N = 347). Additional specimens were included from M. puberula (N = 24), M. pruinosa (N = 20), and M. hosei (N = 48). In general, one or two young leaves per plant were collected, dried with silica gel, and stored at −80°C until DNA isolation. Samples were taken from larger seedlings, saplings, or adult trees to allow unambiguous species identification. Only in four cases was species identification questionable (see below). Almost all of the samples originated from roadsides, logging areas, or disturbed forests, whereas only few plants were found along trails or in gaps in the primary forest. In northern Borneo and on the Malay Peninsula, samples were collected from individuals with a minimum distance of 50 m to minimize the risk of sampling related individuals, whenever possible. Samples from East Kalimantan were collected in plots of 10 × 10 m which were spaced at 30-m intervals along straight transects of up to 900-m length. However, because information about the mode and distance of seed dispersal is lacking for the species, we cannot exclude that in a few cases specimens with the same genetic background might have been analyzed. In such cases, genetic diversity values for a population would have been underestimated. The different sampling sites, hereafter referred to as populations, were usually separated from each other by non-forested land (with the exception of those from the Danum Valley area). Due to the inaccessibility of some remote localities as well as natural variation in the abundance of some species, a certain sampling disparity could not be avoided.
DNA isolation and identification of chloroplast haplotypes
DNA was isolated from dried leaf material following a modified cetyl trimethylammonium (CTAB) protocol as described in Bänfer et al. (2006). Chloroplast DNA haplotypes were defined on the basis of the combined sequence information at three chloroplast loci which had previously been shown to be polymorphic within and between Macaranga species. These are the ccmp5 (microsatellite region 3′ to rps2, 65–73 bp) and ccmp6 (intron 2 of the Ycf3 gene, 70–79 bp) loci, each harboring a size-variable stretch of mononucleotides (chloroplast microsatellites; Weising and Gardner 1999) and an approximately 450–500-bp fragment of the atpB-rbcL intergenic region (Vogel et al. 2003; Bänfer et al. 2006). DNA polymorphisms at the target loci were identified by the polymerase chain reaction (PCR) followed by single-nucleotide sequencing (SNS) analysis of PCR products on an automated sequencer (LI-COR, model 4200 IR²), with details given in Guicking et al. (2008). As terminating dideoxynucleotides, ddCTP was chosen for the ccmp5 locus, ddATP for the ccmp6 locus, and ddTTP for the atpB-rbcL intergenic region. Base substitutions, the presence or absence of indels, and the numbers of bases at poly-(T) or poly-(A) repeats in each sample were inferred from the SNS patterns. This information was then converted into a set of unique haplotypes, each of which was defined by a specific string of character states (see Guicking et al. 2008). Base substitutions and indels were each assumed to represent single-mutation events. A stepwise mutation model was followed for coding microsatellite variation, i.e., each gain or loss of one repeat unit was counted as a single mutational event (Bänfer et al. 2006).
Network reconstruction
Parsimony networks were reconstructed to infer genetic relationships among haplotypes. To account for the presumably different mutation rates underlying base substitutions, indels, and microsatellites, we followed the two-step strategy described by Bänfer et al. (2006). In the first step, all length variation at chloroplast microsatellites was excluded from the analyses, and a backbone network of core haplotypes was reconstructed using the computer program TCS 1.21 (Clement et al. 2000). In the second step, variation at microsatellite loci was added manually at the appropriate network positions. Some closed loops could be resolved by assuming that connections between haplotypes from different species were less likely than those within a species. In one case, geographical origins of the samples were taken into account to resolve some unlikely loops (Crandall and Templeton 1993).
Population genetic analyses
Geographic populations were defined by pooling all sampled plants from a continuous forest stand that were less than 15 km apart. Parameters of genetic variation and population differentiation were estimated for the two main target species of our analysis, M. gigantea and M. pearsonii, following various approaches. Within-population diversities were calculated as h (haplotype diversity, defined as the probability that two randomly chosen haplotypes are different in the sample) and π (nucleotide diversity, the mean number of differences between all pairs of haplotypes in the sample) using the program ARLEQUIN 3.11 (Excoffier et al. 2005). Species-specific overall diversity measurements were estimated according to Pons and Petit (1995, 1996). The software PERMUT (available at http://www.pierroton.inra.fr/genetics/labo/Software/) was used to calculate the mean within-population gene diversity (h s), the total gene diversity (h T), and the gene differentiation over all populations (G ST, which might be viewed as a natural extension of Wright’s F ST; Pons and Petit 1995). We also estimated the equivalent parameters (ν S, ν T, N ST) that take into account the numbers of pairwise differences between haplotypes. To test for the presence of a phylogeographic structure, we checked whether N ST > G ST by comparing the directly measured N ST values with those obtained after 1,000 random permutations of haplotype identities (Burban et al. 1999).
A hierarchical analysis of molecular variance (AMOVA) was conducted to estimate the variance components at three levels: among regions, among populations within regions, and within populations. Five geographical regions were defined: (1) Malay Peninsula, (2) northwestern Borneo including Sarawak, Brunei, and part of Sabah, west of the Crocker Range, (3) northeastern Borneo (i.e., Sabah east of the Crocker Range), (4) northern East Kalimantan (Berau), and (5) southern East Kalimantan. Conventional F statistics were used as distance measures. All AMOVA analyses were conducted with ARLEQUIN 3.11 (Excoffier et al. 2005).
Populations from the lowlands of Sabah (bordered by mountain ranges to the west and to the south) were tested for isolation by distance. To do this, the relationships between genetic and geographic distances of populations were examined following Grivet and Petit (2002). Pairwise G ST and N ST values were calculated using the program DISTON (available at http://www.pierroton.inra.fr/genetics/labo/Software/) and were plotted as a function of geographic distance. In each distance class considered, mean pairwise diversities among populations were computed first, and the coefficients of differentiation G ST and N ST were then derived by taking the ratios of the means.
Results
Species specificity of chloroplast haplotypes in Macaranga, section Pruinosae
A total of 49 different chloroplast haplotypes were identified among 768 plants from five species (Tables S1 and S2). Of these, 23 haplotypes were found in M. gigantea (N = 329), 21 in M. pearsonii (N = 347), four in M. puberula (N = 24), six in M. hosei (N = 48), and five in M. pruinosa (N = 20) (Table 1, Fig. 1).
Statistical parsimony network based on cpDNA haplotype data in Macaranga section Pruinosae. Each rectangle represents a unique haplotype (indicated by numbers). Lines connecting the haplotypes represent mutational steps (thin lines: microsatellite mutations; bold lines: other indels and base substitutions). Small circles represent unsampled or extinct haplotypes. Hatched colors indicate that the respective haplotypes were found in more than one species. Haplotype 13 was found also in one sample of M. gigantea, probably representing a case of introgression (see text)
Forty-one haplotypes were species-specific, six were shared by two species, one was shared by three species, and one was shared by four species. Most noteworthy was haplotype 15 (HT 15), which was present in all studied species except for M. hosei. Three of the four haplotypes found in M. puberula also occurred in M. pearsonii, and one of these (HT 13) also occurred in M. gigantea. The dominant haplotype in M. hosei, HT 5, was also found once in M. pearsonii. HT 17 was present in M. pearsonii and M. gigantea, and HT 43 occurred in M. gigantea and M. hosei on the Malay Peninsula. Haplotypes HT 22 and HT 24 were frequently found in M. gigantea and also occurred in M. pruinosa. We may conclude that chloroplast haplotypes are mostly species-specific in Macaranga section Pruinosae (see “Discussion”). In the analyses described below, we therefore treat haplotype distributions by species.
Haplotype diversity within populations
The observed levels of within-population haplotypic diversity differed markedly among populations (Table 1). The majority of populations exhibited multiple haplotypes, with haplotype diversities up to h > 0.7. However, a subset of populations from all species were fixed for one particular haplotype (i.e., haplotype diversity h = 0), even if sample numbers were high. Nucleotide diversity values ranged from π = 0 to π > 0.01 (Table 1).
Region specificity of chloroplast haplotypes
Whereas most haplotypes were found in more than one population, haplotypes were almost always restricted to one particular region (e.g., Malay Peninsula, East Kalimantan, Sabah; Table S1, Fig. 2a, b). Among the few exceptions are HT 5 of M. hosei, which occurred in Brunei as well as in Berau (northern East Kalimantan), HT 15 of M. gigantea, which occurred both on the Malay Peninsula and in Berau, and HT 24 of M. gigantea, which was found on the Malay Peninsula, in Sarawak, and in East Kalimantan.
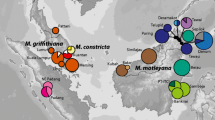
Geographic distribution (right panel) and genetic relationships (left panel) of cpDNA haplotypes in a M. gigantea and b M. pearsonii. Circle sizes are proportional to population sample sizes. Individual haplotypes are defined by numbers (see Fig. 1 and Table S1) as well as by different colors. The known localities of the two species according to Davies (2001) and own observations are denoted in the inset maps
Geographical distribution of haplotypes and genetic diversity in M. gigantea and M. pearsonii
The geographical distribution of chloroplast haplotypes and population genetic parameters based on haplotype diversities were analyzed in more detail for M. gigantea and M. pearsonii (Fig. 2a, b). In M. pearsonii, we were able to sample the distribution range quite extensively, except for populations in northern East Kalimantan (Fig. 2b). M. gigantea with its much larger distribution range could not be sampled over the whole area (Fig. 2a). Diversity parameters were calculated on the basis of 22 populations in M. gigantea and 16 in M. pearsonii (all populations with N < 4 excluded). In both species, haplotypic diversity proved to be generally higher in Sabah as compared with East Kalimantan (Fig. 2a, b, Table 1). Coefficients of population differentiation were generally moderate to high (Table 2). Whereas the N ST values were in a similar range for the two species, G ST values were higher in M. gigantea than in M. pearsonii (G ST 0.691 vs. 0.585, N ST 0.855 vs. 0.877; Table 2). The permutation test showed that N ST was significantly higher than G ST in both species, indicating a clear phylogeographic structure for M. gigantea and M. pearsonii chloroplast haplotypes.
The results of the AMOVA also suggested a considerable extent of population structure (Table 3). In both species, a considerable percentage of the total variation occurred among regions and among populations within regions (M. gigantea 43.7% and 29.9%, M. pearsonii 28.3% and 36.1%). M. gigantea showed higher differentiation among groups than M. pearsonii, irrespective of whether all sampled geographic locations or only those locations that were sampled for both species were included in the analysis.
Within Sabah, genetic differentiation among M. pearsonii populations clearly increased with geographic distance (Fig. 3), with N ST values mostly exceeding G ST values. In M. gigantea, no such pronounced effect of isolation by distance was found in Sabah (Fig. 3).
Discussion
Species specificity of chloroplast haplotypes in Macaranga section Pruinosae
The correct interpretation of intraspecific phylogeographic data might crucially rely on the knowledge of cpDNA haplotypic diversity in species closely related to the taxon under study, especially for species that frequently hybridize or show incomplete sorting of ancient polymorphisms (Petit et al. 2002; Cannon and Manos 2003; Jakob and Blattner 2006). Extensive sharing of cpDNA haplotypes among species and a geographical rather than taxonomic arrangement of haplotypes had been observed in the bancana group of Macaranga section Pachystemon (Bänfer et al. 2006). This is apparently not the case in section Pruinosae, where 41 out of 49 haplotypes were confined to a single species. Furthermore, the genealogy of the haplotypes (Fig. 1) also showed a clear differentiation of the five species included. These results may indicate an earlier radiation of section Pruinosae, as compared with the bancana group of section Pachystemon.
The few haplotypes that were shared by two or more species were mainly restricted to the central part of the network where the most ancient alleles are supposed to occur. These central haplotypes differed solely by different lengths of mononucleotide repeats, i.e., cpDNA microsatellites (Fig. 1). There is good evidence that recurrent and independent mutations frequently occur at chloroplast microsatellite loci (Doyle et al. 1998; Bänfer et al. 2006); hence, a high probability of homoplasy exists at these loci for old haplotypes. HT 17 might represent such a case of homoplasy. This haplotype was found in M. gigantea and M. pearsonii, and in both species it was separated from the neighboring haplotype by a single mutational step at a chloroplast microsatellite locus (Fig. 1). However, as all individuals bearing HT 17 were collected in the same region, we cannot exclude the possibility of introgression either.
Only a single individual was identified that clearly represents a recent hybrid. This refers to a M. gigantea specimen bearing HT 13 that is otherwise specific to M. pearsonii. Although morphologically unambiguously M. gigantea, this plant also harbored nuclear microsatellite alleles specific for M. gigantea as well as for M. pearsonii (unpublished data). Furthermore, it was found at a location (Danum Valley) where HT 13 frequently occurred in syntopic M. pearsonii, while all other M. gigantea specimens had different haplotypes. A second case of possible introgression refers to a few specimens that were morphologically identified as M. pruinosa but exhibited M. gigantea haplotypes (HT 22 and HT 24). In his revision, Davies (2001) noted that hybridization between M. gigantea and M. pruinosa is not unlikely. However, as the two species look very similar in the seedling and sapling stages which were analyzed in these particular cases, we cannot exclude the possibility that the specimens were simply misidentified.
Haplotype sharing among species can also be attributed to incomplete sorting of ancient lineages. The generally low numbers of missing intermediate haplotypes suggest that extinction rates of chloroplast lineages in section Pruinosae are rather low, which results in a low rate of lineage sorting. Furthermore, shared chloroplast types were found mostly in intermediate positions of the network (i.e., at the base of species-specific genealogical lineages, see Fig. 1) and were also shared between species with non-overlapping distribution areas, suggesting that these haplotypes represent ancestral alleles. The most striking case of haplotype sharing was the occurrence of HT 15 among four of the five species under study, M. hosei being the only exception. As sampling was limited in M. hosei and HT 15 represents an intermediate between two M. hosei haplotypes (HT 43 and HT 46), we may nevertheless assume that HT 15 has also survived in this species but was not yet found. The central position and the wide taxonomic and geographic distribution of this haplotype suggest that HT 15 might in fact represent the most basal haplotype in section Pruinosae. Incomplete lineage sorting of common ancient haplotypes is also likely to explain the shared occurrence of HT 13 and HT 14 in M. pearsonii and M. puberula. M. puberula is most closely related to M. pearsonii (Davies 2001) and probably evolved as an ecotype of higher elevations.
We conclude that persistent ancient polymorphisms provide the best explanation for most of the cases of shared chloroplast haplotypes in Macaranga section Pruinosae. Hybridization between species seems to be comparatively rare. As shared alleles proved to be restricted to a few ancient polymorphisms, species-specific population genetic and phylogeographic analyses of the chloroplast haplotypes therefore seem justified.
Biogeographic implications
Any discussion of the evolutionary history of a species must take into account the historical dynamics of the studied landscape (Woodruff 2010). The Southeast Asian continent has one of the most complex geological histories in the world, and its paleogeographic history has been greatly influenced by changing sea levels and the intermittent formation and destruction of land bridges across the Sunda shelf during the Pleistocene (Morley 2000; Hall 2009; Woodruff 2010). As was postulated for Southeast Asian tropical rain forests in general (Cannon et al. 2009), we may assume that Macaranga species, and pioneer species in particular, expanded their distribution ranges over the exposed Sunda shelf during glacial periods. We found support for formerly more widespread distribution ranges of individual haplotypes not only in section Pachystemon (Bänfer et al. 2006) but also in M. gigantea, where identical or closely related haplotypes occurred in East Kalimantan and on the Malay Peninsula (Table S1, Fig. 2a). This pattern most likely originated from migration across land connections between Borneo and the mainland during Pleistocene cold cycles.
The biogeography of plants and animals on Borneo is strongly influenced by the occurrence of a central mountain range that divides the island into a northwestern and a southeastern part and rises up to 4,010 m at Mount Kinabalu in Sabah. There is evidence from previous studies that this range forms an effective dispersal barrier for the lowland flora and fauna (e.g., Cannon and Manos 2003; Slik et al. 2003; Moyle et al. 2005; Bänfer et al. 2006; Raes et al. 2009). The distribution areas of some lowland Macaranga species are also bordered by this mountain range, e.g., those of Macaranga bancana, Macaranga trachyphylla, Macaranga indistincta, and Macaranga glandibracteolata (Davies 2001).
In the present study, we found clear evidence that the range also forms an intraspecific barrier both in M. gigantea and M. pearsonii (Fig. 2a, b). M. gigantea is distributed on both sides of the range, but eastern and western haplotypes were clearly differentiated from each other, suggesting a long-term independent evolution. A few eastern and western haplotypes were nevertheless also found on the “wrong” side (Fig. 2a), indicating that limited seed flow across the range may occur. M. pearsonii is mainly distributed east of the range; however, we also found some individuals on the western slopes of the range. This “western” M. pearsonii population was fixed for a single haplotype plus one related local type (Fig. 2b) and thus might have originated from a single founder event.
Haplotypes of M. pearsonii from Sabah and East Kalimantan were separated from each other by several mutational steps (Fig. 2b). Such a strong genetic differentiation can either be explained by the occurrence of geographic barriers and/or by the large geographical distance. Although the mountain ranges that presently separate Sabah and East Kalimantan are not very high, one has to take into account that vegetation zones shifted to lower altitudes during glacial periods of the Pleistocene (Cannon et al. 2009). Hence, mountain barriers may have had a stronger barrier effect on lowland vegetation in the past than they have today. Alternatively, isolation by distance may explain the observed differentiation of populations in Sabah and East Kalimantan. We found some indications for isolation by distance in M. gigantea, as haplotypes from Berau in northern East Kalimantan (HT 15 and HT 30, green in Fig. 2a) were in an intermediate network position between haplotypes from Sabah and southern East Kalimantan (Fig. 2a). Unfortunately, we were not able to get hold of material of M. pearsonii from northern East Kalimantan.
A preliminary hypothesis on the evolution of M. pearsonii and M. gigantea in Borneo
M. pearsonii is endemic to northern and eastern Borneo. Despite a certain sampling disparity, we may conclude that much higher levels of genetic diversity are present in Sabah as compared with southern East Kalimantan (Table 1). Assuming that high measures of genetic diversity are characteristic of regions with large and stable populations, whereas low measures of genetic diversity are indicative of a recent colonization (Petit et al. 2002; Lowe et al. 2004), we propose that M. pearsonii has a longer continuous presence in the state of Sabah than at the sampled locations in East Kalimantan. The small number of missing intermediate (ancestral) haplotypes in the network further argues for a continuous presence of M. pearsonii in the eastern lowlands of Sabah, consistent with the assumption of persistent ever-wet rainforests in northern Borneo (Gathorne-Hardy et al. 2002; Cannon et al. 2009). The considerable extent of genetic differentiation within Sabah (Fig. 2b) may then be related to the rugged topography of the region, which may enhance allopatric differentiation. In contrast, southern East Kalimantan might have experienced periods of more arid conditions, which could have given rise to founder events during subsequent re-colonization of savannah-like areas by rainforest plants (Morley 2000; Gathorne-Hardy et al. 2002). The relatively low diversity of Macaranga-associated Crematogaster ants in southern Kalimantan was also interpreted as an indicator for recent colonization of this region (Quek et al. 2007).
The situation is more complicated in the widespread M. gigantea. As in other Macaranga species, we found higher levels of genetic diversity in Sabah as compared with East Kalimantan and the Malay Peninsula. Assuming that interior positions in the network represent ancestral haplotypes and tip positions represent derived haplotypes, most of the M. gigantea haplotypes in Sabah are derived. Ancestral types, including the putative root of the section (HT 15), predominate in East Kalimantan as well as on the Malay Peninsula, suggesting either a mainland Malaysian or a southern Bornean origin of the species. Furthermore, in Sabah, haplotypes from different branches of the network co-exist, showing affinities to either haplotypes from western Borneo or from East Kalimantan (Fig. 2a). This pattern seems compatible with the assumption of at least two independent colonizations into Sabah, one from the west crossing the central mountain range and one from the south. High measures of genetic diversity in Sibu, Sarawak, and Luasong, Sabah, then most likely represent secondary contact zones between formerly differentiated lineages.
Genetic diversity of tropical pioneer trees
Due to their restriction to forest gaps and edges with often low population densities in undisturbed forests, pioneer species have unique life histories and population structures (Alvarez-Buylla and Garay 1994). Different theoretical models have been set up to explain the genetic structure of pioneers, either postulating strong population substructuring due to continuous founder effects (Wright 1940) or reduced interpopulational differentiation due to elevated gene flow rates (Slatkin 1985). Previous studies on tropical pioneer trees generally revealed low levels of genetic structure, thus favoring Slatkin’s (1985) hypothesis (Alvarez-Buylla and Garay 1994; Muloko-Ntoutoume et al. 2000; Cavers et al. 2005; Litrico et al. 2005).
The total haplotype diversity of M. gigantea and M. pearsonii was high (h T 0.915 and 0.894, h S 0.283 and 0.371, respectively). Studies in other tropical tree species generally found lower levels of total haplotype diversity (e.g., Caron et al. 2000; Dutech et al. 2000; Cavers et al. 2003; Salgueiro et al. 2004). However, as most of these studies were based on cpDNA polymerase chain reaction–restriction fragment length polymorphisms (PCR–RFLP) that possibly show lower mutation rates than cpDNA microsatellites analyzed in the present study, a direct comparison of the values may be misleading. Population subdivision was moderate to high, especially when taking into account genetic distances among haplotypes (G ST 0.69 and 0.59, N ST 0.86 and 0.88, in M. gigantea and M. pearsonii, respectively). However, a certain bias may have been introduced by the broad sampling over two or more geographical regions separated by mountain barriers or the sea. When considering only populations from the lowlands of Sabah, i.e., on a much smaller geographic scale, G ST values of 0.42 and 0.52 were obtained for the two species. A G ST in a similar range (0.54) was also found for another tropical pioneer tree species, Aucoumea klaineana, in Gabon, and was considered low by the authors (Muloko-Ntoutoume et al. 2000).
In a parallel study, we found that nuclear microsatellite data show hardly any differentiation (F ST < 0.1) among populations of species of Macaranga section Pruinosae from within geographic regions (to be published elsewhere). Low overall F ST values were also detected in the neotropical pioneer tree species Cecropia obtusifolia, using allozyme markers (Alvarez-Buylla and Garay 1994). A lack of genetic differentiation observed with nuclear markers often indicates efficient pollen flow. The main pollinators of Macaranga are thrips (Moog et al. 2002; Moog 2002), and long-distance pollen dispersal might be facilitated by wind drift of these tiny insects (Lewis 1973, 1997; Setsuko et al. 2007). We may conclude that extensive pollen and/or seed dispersal apparently has maintained genetic connectivity among populations in M. gigantea and M. pearsonii and that our study is in line with Slatkin’s (1985) hypothesis of reduced interpopulational differentiation in pioneer trees.
Ant-associated pioneer trees
It is noteworthy that two of the most successful tropical pioneer tree genera worldwide, i.e., the neotropical genus Cecropia and the palaeotropical genus Macaranga, are ant-associated (Berg et al. 2005; Whitmore 2008). The association with ants may therefore be interpreted as a key innovation that facilitates the colonization of pioneer habitats, probably because the ants protect their host plants efficiently against competing herbivores and vines. On the other hand, one could hypothesize that the obligate association with ants might restrain dispersal abilities in obligate Macaranga ant-plants. Under natural conditions, Macaranga myrmecophytes are dependent on the presence of their ant partner to survive to the reproductive stage, and, vice versa, the ant queens need the trees to found their colonies (Fiala et al. 1989; Heil et al. 2001). There is good evidence from small-scale population genetic studies that dispersal of Macaranga-associated ant queens is restricted (Türke et al. 2010). Therefore, myrmecophytes need a suitable partner ant to establish a new population at a remote site, whereas in non-myrmecophytes, the successful recruitment is mainly restricted by seed dispersal efficiency. One may therefore hypothesize that myrmecophytic Macaranga species are less efficient long-distance colonizers than non-myrmecophytes, with negative consequences for maintaining gene flow between geographically separated populations. If this assumption is correct, myrmecophytes may be expected to show stronger patterns of population differentiation than non-myrmecophytes.
In the present study, we found no significant differences in population parameters of the myrmecophytic M. pearsonii and the non-myrmecophyte M. gigantea. Both species exhibited similar numbers of cpDNA haplotypes and similar levels of genetic diversity. Overall population differentiation was even higher in M. gigantea than in M. pearsonii (Table 3). Likewise, we found no significant differences between myrmecophytic and non-myrmecophytic species of Macaranga section Pachystemon (Weising et al. 2010). These results argue against a strong negative effect of the symbiotic ant partners on colonization abilities in obligate ant-plants.
On the other hand, we found some indications for isolation by distance in M. pearsonii within the lowlands of Sabah, whereas no such effect became apparent in M. gigantea (Fig. 3). This observation is compatible with our hypothesis that myrmecophytes only disperse continuously whereas non-myrmecophytes are also capable of long-distance dispersals. Further support for more restricted dispersal abilities of myrmecophytes as compared with non-myrmecophytes comes from the observation that M. gigantea also grows outside the forest and even close to or within cities, whereas M. pearsonii trees are always found close to forested areas. However, to finally clarify whether the obligate association with ants may influence dispersal abilities in their host plants and if so, at what geographical scale this would become apparent, other factors would need to be considered, including seed dormancy capability, natural population abundances, habitat preferences, and seed dispersal distances. Furthermore, we currently do not know how colonization of secondary habitats, as a consequence of anthropogenic destruction of primary forests, may influence population patterns of pioneer trees in the future. Conversion of primary or secondary forest into agricultural land and the establishment of large oil palm plantations unsuitable for Macaranga are likely to restrict gene flow among fragmented populations much more efficiently than in the past.
References
Alvarez-Buylla ER, Garay AA (1994) Population genetic structure of Cecropia obtusifolia, a tropical pioneer tree species. Evolution 48:437–453
Bänfer G, Fiala B, Weising K (2004) AFLP analysis of phylogenetic relationships among myrmecophytic species of Macaranga (Euphorbiaceae) and their allies. Plant Syst Evol 248:213–231
Bänfer G, Moog U, Fiala B, Mohamed M, Weising K, Blattner FR (2006) A chloroplast genealogy of myrmecophytic Macaranga species (Euphorbiaceae) in southeast Asia reveals hybridization, vicariance and long distance dispersals. Mol Ecol 15:4409–4424
Berg CC, Rosselli PF, Davidson DW (2005) Cecropia. The New York Botanical Garden, Bronx
Blattner FR, Weising K, Bänfer G, Maschwitz U, Fiala B (2001) Molecular analysis of phylogenetic relationships among myrmecophytic Macaranga species (Euphorbiaceae). Mol Phyl Evol 19:331–344
Born C, Kjellberg F, Chevallier M-H, Vignes H, Dikangadissi J-T, Sanguié J, Wickings EJ, Hossaert-McKey M (2008) Colonization processes and the maintenance of genetic diversity: insights from a pioneer rainforest tree, Aucoumea klaineana. Proc R Soc Lond B 275:2171–2179
Brooks TM, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Rylands AB, Komstant WR, Flick P, Pilgrim J, Oldfield S, Magin G, Hilton-Taylor C (2002) Habitat loss and extinction in the hotspots of biodiversity. Conserv Biol 16:909–923
Burban C, Petit RJ, Carcreff E, Jactel H (1999) Rangewide variation of the maritime pine bast scale Matsucoccus feytaudi Duc. (Homoptera: Matsucoccidae) in relation to the genetic structure of its host. Mol Ecol 8:1593–1602
Cannon CH, Manos PS (2003) Phylogeography of the Southeast Asian stone oaks (Lithocarpus). J Biogeogr 30:211–226
Cannon CH, Morley RJ, Bush ABG (2009) The current refugial rainforests of Sundaland are unrepresentative of their biogeographic past and highly vulnerable to disturbance. Proc Natl Acad Sci USA 106:11188–11193
Caron H, Dumas S, Marque G, Messier C, Bandou E, Petit RJ, Kremer A (2000) Spatial and temporal distribution of chloroplast DNA polymorphism in a tropical tree species. Mol Ecol 9:1089–1098
Cavers S, Navarro C, Lowe AJ (2003) Chloroplast DNA phylogeography reveals colonization history of a Neotropical tree, Cedrela odorata L., in Mesoamerica. Mol Ecol 12:1451–1460
Cavers S, Navarro C, Hopkins P, Ennos RA, Lowe AJ (2005) Regional and population-scale influences on genetic diversity partitioning within Costa Rican populations of the pioneer tree Vochysia ferruginea Mart. Silvae Genet 54:258–264
Clement M, Posada D, Crandall KA (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9:1657–1660
Crandall KA, Templeton AR (1993) Empirical tests of some predictions from coalescent theory with applications to intraspecific phylogeny reconstruction. Genetics 134:959–969
Davies SJ (2001) Systematics of Macaranga sects Pachystemon and Pruinosae (Euphorbiaceae). Harv Pap Bot 6:371–448
Davies SJ, Lum SKY, Chan RKG, Wang LK (2001) Evolution of myrmecophytism in western Malesian Macaranga (Euphorbiaceae). Evolution 55:1542–1559
Dick CW, Heuertz M (2008) The complex biogeographic history of a widespread tropical tree species. Evolution 62:2760–2774
Doyle JJ, Morgante M, Tingey SV, Powell W (1998) Size homoplasy in chloroplast microsatellites of wild perennial relatives of soybean (Glycine subgenus Glycine). Mol Biol Evol 15:215–218
Dutech C, Maggia L, Joly HI (2000) Chloroplast diversity in Vouacapoua americana (Caesalpiniaceae), a neotropical forest tree. Mol Ecol 9:1427–14
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinforma Online 1:47–50
FAO (2006) Global Forest Resource Assessment 2005. FAO, Rome
Fiala B, Maschwitz U, Tho YP, Helbig AJ (1989) Studies of a South East Asian ant–plant association: protection of Macaranga trees by Crematogaster borneensis. Oecologia 79:463–470
Fiala B, Jakob A, Maschwitz U, Linsenmair KE (1999) Diversity, evolutionary specialization and geographic distribution of a mutualistic ant–plant complex: Macaranga and Crematogaster in South East Asia. Biol J Linn Soc 66:305–331
Gathorne-Hardy FJ, Syaukani DRG, Eggleton P, Jones DT (2002) Quaternary rainforest refugia in Southeast Asia: using termites (Isoptera) as indicators. Biol J Linn Soc 75:453–466
Grivet D, Petit RJ (2002) Phylogeography of the common ivy (Hedera sp.) in Europe: genetic differentiation through space and time. Mol Ecol 11:1351–1362
Guicking D, Kröger-Kilian T, Weising K, Blattner FR (2008) Single nucleotide sequence analysis: a cost- and time-effective protocol for the analysis of microsatellite- and indel-rich chloroplast DNA regions. Mol Ecol Res 8:62–65
Hall R (2009) Southeast Asia’s changing palaeogeography. Blumea 54:158–161
Heil M, Fiala B, Maschwitz U, Linsenmair KE (2001) On the benefits of indirect defence: short- and long-term studies in antiherbivore protection via mutualistic ants. Oecologia 126:394–403
Jakob SS, Blattner FR (2006) A chloroplast genealogy of Hordeum (Poaceae): long-term persisting haplotypes, incomplete lineage sorting, regional extinction, and the consequences for phylogenetic inference. Mol Biol Evol 23:1602–1612
Kulju KKM, Sierra SEC, Draisma SGA, Samuel R, van Welzen PC (2007) Molecular phylogeny of Macaranga, Mallotus, and related genera. Am J Bot 94:1726–1743
Lewis T (1973) Thrips: their biology, ecology and economic importance. Academic, New York
Lewis T (1997) Flight and dispersal. In: Lewis T (ed) Thrips as crop pests. CAB International, Oxon, pp 175–196
Litrico I, Ronfort R, Verlaque R, Thompson JD (2005) Spatial structure of genetic variation and primary succession in the pioneer tree species Antirhea borbonica on La Réunion. Mol Ecol 14:1576–1584
Lowe AJ, Harris SA, Ashton P (2004) Ecological genetics: design, analysis and application. Blackwell, Oxford
Moog U (2002) Die Reproduktion von Macaranga (Euphorbiaceae) in Südostasien: Bestäubung durch Thripse und Kastration durch Pflanzenameisen. University of Frankfurt, Dissertation
Moog U, Fiala B, Federle W, Maschwitz U (2002) Thrips pollination of the dioecious ant plant Macaranga hullettii (Euphorbiaceae) in Southeast Asia. Am J Bot 89:50–59
Morley RJ (2000) Origin and evolution of tropical rain forests. Wiley, Chichester
Moyle RG, Schilthuizen M, Rahman MA, Sheldon FH (2005) Molecular phylogenetic analysis of the white-crowned forktail Enicurus leschenaulti in Borneo. J Avian Biol 36:96–101
Muloko-Ntoutoume N, Petit RJ, White L, Abernethy K (2000) Chloroplast DNA variation in a rainforest tree (Aucoumea klaineana, Burseraceae) in Gabon. Mol Ecol 9:359–363
Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J (2000) Biodiversity hotspots for conservation priorities. Nature 403:853–858
Ng KKS, Lee SL, Koh CL (2004) Spatial structure and genetic diversity of two tropical tree species with contrasting breeding systems and different ploidy levels. Mol Ecol 13:657–669
Petit RJ, Csaikl UM, Bordács S (2002) Chloroplast DNA variation in European white oaks: phylogeography and patterns of diversity based on data from over 2600 populations. For Ecol Manag 156:5–26
Pons O, Petit RJ (1995) Estimation, variance and optimal sampling of gene diversity. I. Haploid locus. Theor Appl Genet 90:462–470
Pons O, Petit RJ (1996) Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics 144:1237–1245
Quek S-P, Davies SJ, Ashton PS, Itino T, Pierce NE (2007) The geography of diversification in mutualistic ants: a gene’s eye view into the Neogene history of Sundaland rain forests. Mol Ecol 16:2045–2062
Raes N, Roos MC, Slik F, van Loon EE, ter Stenge H (2009) Botanical richness and endemicity patterns in Borneo derived from species distribution models. Ecography 32:180–192
Salgueiro F, Felix D, Caldas JF, Margis-Pinheiro M, Margis R (2004) Even population differentiation for maternal and biparental gene markers in Eugenia uniflora, a widely distributed species from the Brazilian coastal Atlantic rain forest. Divers Distrib 10:201–210
Setsuko S, Ishida K, Ueno S, Tsumura Y, Tomaru N (2007) Population differentiation and gene flow within a metapopulation of a threatened tree, Magnolia stellata (Magnoliaceae). Am J Bot 94:128–136
Slatkin M (1985) Gene flow in natural populations. Annu Rev Ecol Syst 18:474–511
Slik JWF, Poulsen AD, Ashton PS et al (2003) A floristic analysis of the lowland dipterocarp forests of Borneo. J Biogeogr 30:1517–1531
Sodhi NS, Koh LP, Brook BW, Ng PKL (2004) Southeast Asian biodiversity: an impending disaster. Trends Ecol Evol 19:654–660
Türke M, Fiala B, Linsenmair KE, Feldhaar H (2010) Estimation of dispersal distances of the obligately plant-associated ant Crematogaster decamera. Ecol Entomol 35:662–671
Vogel M, Bänfer G, Moog U, Weising K (2003) Development and characterization of chloroplast microsatellite markers in Macaranga (Euphorbiaceae). Genome 46:845–857
Weising K, Gardner RG (1999) A set of conserved PCR primers for the analysis of simple sequence repeat polymorphisms in chloroplast genomes of dicotyledonous angiosperms. Genome 42:9–19
Weising K, Guicking D, Fey-Wagner C et al (2010) Mechanisms of speciation in Southeast Asian ant-plants of the genus Macaranga (Euphorbiaceae). In: Glaubrecht M (ed) Evolution in action—adaptive radiations and the origins of biodiversity. Springer, Berlin, pp 169–191
Whitmore TC (2008) The genus Macaranga. A Prodromus. Kew, Kew
Woodruff DS (2010) Biogeography and conservation in Southeast Asia: how 2.7 million years of repeated environmental fluctuations affect today’s patterns and the future of the remaining refugial-phase biodiversity. Biodivers Conserv 19:919–941
Wright S (1940) Breeding structure of populations in relation to speciation. Am Nat 74:232–248
Acknowledgements
The authors thank the German Research Foundation (DFG) for financial support within priority program 1127 (Biological Radiations, grants We 1830/4, Fi 606/5, and Bl 462/2). Permission to conduct research in Malaysia was kindly granted by the Economic Planning Unit (EPU) of the Prime Minister’s Office, Kuala Lumpur, and EPU in Kota Kinabalu, Sabah, as well as by the Danum Valley Management Committee and the Forest Department, Sarawak. Permit to work in Brunei was obtained by the Universiti Brunei Darussalam and the Brunei Museum. We thank our counterparts and colleagues in Malaysia and Brunei for their cooperation and support, especially Dr. Rosli bin Hashim, Dr. Kamariah Abu Salim, and members of Sabah Parks, namely Dr. Jamili Nais and Dr. Maklarin bin Lakim. Logistic support and help in many other ways by Prof. Dr. K. Eduard Linsenmair is gratefully acknowledged. We thank Prof. Dr. Ulrich Maschwitz for the kind supply of plant material. Reviewers’ comments on earlier versions of this manuscript are acknowledged.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Y. Tsumura
An erratum to this article can be found at http://dx.doi.org/10.1007/s11295-011-0415-1
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table S1
Geographic distribution of cpDNA haplotypes in five species of Macaranga section Pruinosae. *M. pearsonii haplotype. (DOC 377 kb)
Table S2
Definition of cpDNA haplotypes for Macaranga section Pruinosae. Polymorphic sites and their first position in base pairs (with reference to GenBank accessions EF565903, EF565927, and EF565917) are indicated. (DOC 106 kb)
Rights and permissions
About this article
Cite this article
Guicking, D., Fiala, B., Blattner, F.R. et al. Comparative chloroplast DNA phylogeography of two tropical pioneer trees, Macaranga gigantea and Macaranga pearsonii (Euphorbiaceae). Tree Genetics & Genomes 7, 573–585 (2011). https://doi.org/10.1007/s11295-010-0357-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-010-0357-z