Abstract
Members of the Euphorbiaceae are ecologically important elements of Southeast Asian forests. Species of the pioneer tree genus Macaranga, which is also known for its association with ants, are often abundant in disturbed areas. Phylogenetic studies suggested a recent radiation of section Pachystemon which comprises the majority of obligate myrmecophytes within Macaranga. In the present study, we analyzed the genetic structure of three closely related species of this section (M. constricta, M. griffithiana, and M. motleyana) with the aim of resolving their controversial taxonomy and historical biogeography. Chloroplast DNA haplotypes proved to be species-specific and showed a strong phylogeographic pattern. Nuclear microsatellite data supported the taxonomic distinctness of M. griffithiana and M. motleyana, but gave ambiguous results for M. constricta. Genetic differentiation was stronger each within M. griffithiana and M. motleyana than between M. constricta and M. griffithiana, highlighting problems of defining species boundaries. We found no indication for introgression or hybridization events. The high intraspecific morphological variation of the Bornean endemic M. motleyana was partly reflected by similar patterns of genetic variation. The pronounced genetic structure indicates a comparatively long diversification for this species. In contrast, the weak genetic differentiation within M. griffithiana, as well as the widespread distribution of its most common chloroplast haplotype from peninsular Malaysia up to Indochina, indicates a recent expansion in this area. Despite their morphological similarity and close relatedness, all species possess their own specific ant-partners with a corresponding distribution pattern.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Members of the Euphorbiaceae are important elements of Southeast Asian forests. Ranking second behind the dominant Dipterocarpaceae in abundance in Borneo (Slik et al. 2003a), euphorbs make a large contribution to the rainforest ecosystem, especially in secondary forests. Many pioneer species of the genus Macaranga are indicators for forest disturbance (Slik et al. 2003b) and belong to the most abundant trees in disturbed areas. A conspicuous feature of Macaranga in Sundaland is their close mutualistic association with ants, mainly from genus Crematogaster (Fiala et al. 1999). About 30 Macaranga species are involved in the highly specific obligate myrmecophytic associations that are confined to the everwet dipterocarp rainforests and do usually not extend into the more seasonal monsoon forest regions (e.g., Fiala et al. 1999; Davies 2001). The ants protect their host trees against herbivore damage and vine cover and provide an important ecological advantage, probably facilitating colonization of newly available pioneer habitats (Fiala et al. 1989, 1994). The tight association is assumed to have contributed to the diversification of both ant and plant partners (Feldhaar et al. 2010; Weising et al. 2010).
The genus Macaranga is organized in four sections and 14 informal species groups (Whitmore 2008). Myrmecophytism is mainly confined to sections Pruinosae, Winklerianae, and Pachystemon, the latter of which was further subdivided into four monophyletic groups (Blattner et al. 2001; Davies et al. 2001). The so-called bancana-clade (Bänfer et al. 2004, 2006; corresponding to Pachystemon group 1 in Davies 2001) is the largest group with at present 19 species, of which 15 are obligatorily associated with ants. Phylogenetic studies based on nuclear ribosomal ITS sequences revealed low genetic variation among species of the bancana-clade, suggesting that this group has undergone a recent and rapid radiation (Blattner et al. 2001; Davies et al. 2001). High levels of hybridization and introgression further argue for a young age of the species of this group. Chloroplast DNA (cpDNA) haplotypes were frequently found to be shared among species and followed a geographic pattern rather than reflecting taxonomic boundaries (Bänfer et al. 2006). The existing phylogenies do not unambiguously resolve the position of the species, and even the delimitation of some species is not well supported.
In this study, we focus on three closely related species of the bancana-clade: M. motleyana (Müll. Arg.) Müll. Arg. 1866, M. constricta Whitmore and Airy Shaw 1971, and M. griffithiana Müll. Arg. 1866. These species are morphologically similar and form a weakly supported sister group to the remainder of the bancana-clade in ITS phylogenies (Blattner et al. 2001). The relationships among the three study species remained largely unresolved, although some support was found for M. motleyana being sister group to M. griffithiana plus M. constricta. The taxonomy of the study species has a controversial history, especially regarding the species status of M. griffithiana (see Materials and Methods Section). Macaranga griffithiana also has a remarkably disjunct distribution; it occurs throughout the southern part of the Thai-Malay Peninsula to Sumatra but also in very isolated populations in the Chanthaburi pocket in east Thailand and Cambodia (Whitmore 1984; Maschwitz et al. 2004). It is thus the only myrmecophytic Macaranga species known from the seasonal monsoon zone. Whereas M. constricta is restricted to a small area of peninsular Malaysia, M. motleyana is endemic to Borneo. The latter species shows a considerable morphological variation and is one of the few species of section Pachystemon which is found in almost all parts of Borneo. Its haplotypes take a central position in a cpDNA network of the bancana-clade (Bänfer et al. 2006), suggesting an ancestral state and a probable evolutionary origin of myrmecophytism in this group. Despite their phenotypic similarity and close phylogenetic relationship, M. motleyana, M. constricta, and M. griffithiana appear to be associated with different partner ants which also vary in distribution, age, and genetic structure (Fiala et al. 1999; Feldhaar et al. 2003b, 2010; Quek et al. 2004, 2007).
Taken together, the three species are of particular interest in terms of questions of taxonomic delimitations, gene flow, biogeographic history, and evolution of ant–plant relationships. In the present study, we use cpDNA sequences and nuclear microsatellite markers to (i) test whether molecular data support the morphological distinctness of the three closely related taxa, (ii) determine their genetic diversity and structure, and (iii) reconstruct phylogeographic patterns within each of the species and compare these with the respective patterns found in the associated ants.
Materials and methods
Study species
All three species have 2–3 stamens per flower and a conspicuous wax coating of the stem (Online Resource 1) that is mostly absent in the remaining species of the bancana-clade. Macaranga constricta is a large, early successional tree with a restricted occurrence in forest on dry hillsides in the east of peninsular Malaysia (Fig. 1), where it was discovered as late as 1967 during a logging operation. The species was first described by Whitmore and Airy Shaw (1971) but Whitmore doubted of whether it could actually be distinguished from M. griffithiana. Davies (2001), however kept its species status in his revision due to e.g., “much longer and erect inflorescences, and entire acuminate staminate bracteoles.”
Macaranga griffithiana is light demanding and most common as a bushy treelet in swampy secondary forest. It occurs in Sumatra, peninsular Malaysia, and southeastern peninsular Thailand but was also reported from Cambodia and southern Vietnam (T. Kenzo, M. Nuraliev, H. Sovanly, I. Theilade, pers. com.). Whitmore (1974) reduced M. griffithiana to a geographical form of M. motleyana and established two subspecies: M. motleyana subsp. griffithiana (Müll. Arg.) Whitmore and M. motleyana (Müll. Arg) Muell. Arg. subsp. motleyana. Davies (2001), however re-established the species status of M. griffithiana. The leaves of M. griffithiana are shallowly dissected, whereas they are long and ascending with deep narrow sinuses in M. motleyana. The bracteoles of the staminate inflorescences are only shortly and irregularly toothed in M. griffithiana, whereas they are deeply pectinate in M. motleyana (for further characteristics see Davies 2001).
Macaranga motleyana is widespread over Borneo but was not collected from west Kalimantan and only recently found by us in the coastal forest of Similajau, northern Sarawak. In Sabah, we recognized two morphotypes (A and B) which differed from the general appearance of M. motleyana. We did not yet come across fertile specimens of these morphotypes. Morphotype A was only found in heavily disturbed habitat near the Tawai Forest Reserve near Telupid. It had long, acuminate stipules and oval, tough leaves which were—atypical for the species—completely undissected (Online Resource 1 E–G). Older leaves were dull green, and young leaves brownish red on the adaxial side and bright glaucous on the abaxial side. Some plants had conspicuous patterns of herbivore damage that were never found on the “normal” type of M. motleyana along the same logging road a few kilometers away. Morphotype B was characterized by a thin, soft stem and dark green leaves which were papery thin, especially when dry. This morphotype occurred in lowland forest in Tawau Hills and in Kinabalu Park (Poring) as well as in Deramakot F.R. where it was found sympatrically with the “normal type” which grew along logging roads.
Ant-associates
Myrmecophytic Macaranga species are obligatorily associated with ten only partly described (morpho)species of the ant genus Crematogaster (Myrmicinae) (Fiala et al. 1999; Feldhaar et al. 2003a, b, 2010). Additional genetic lineages of associated ants were established by Quek et al. (2004, 2007) based on COI and COII sequences of the mitochondrial DNA (see Online Resource 2 for an overview of synonyms). The patterns of ant-association are complex but important for the understanding of the evolution of this system. There is extensive data available on the general biology, diversity, and distribution of the associations from earlier publications (e.g., Fiala et al. 1999; Feldhaar et al. 2003a, 2010), so we refer to these for details and here summarize only the most relevant information concerning the associations of our study species. Macaranga motleyana and M. constricta have mainly been found with Crematogaster msp. 7/lineage A and C. msp. 9/lineage B, respectively, which came out as early-branching and thus perhaps most ancient partners in mitochondrial DNA-based phylogenies of the Macaranga-associated ants (Feldhaar et al. 2003a; Quek et al. 2004, 2007). Macaranga grifftihiana had been reported to be colonized by C. borneensis (formerly C. msp. 1) which is the most generalistic ant species with respect to host plant use and occurs in the whole distribution area of myrmecophytic Macaranga from south Thailand to Borneo (Fiala et al. 1999; Feldhaar et al. 2003b, Quek et al. 2007; see Online Resource 2).
Sampling
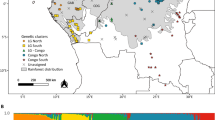
Collection sites are shown in Fig. 1 and summarized in Table 1. We collected four specimens of the rare M. constricta from its only known locality, 51 specimens of M. griffithiana from ten sites on Sumatra and the Malay Peninsula, and 137 specimens of M. motleyana from 14 sites on Borneo. As the forest has been largely cleared in these regions, we mainly sampled in forest remnants that were separated by a matrix of open land or plantations. No balanced sampling was possible due to the patchy occurrence of the species at different sites. We were able to cover the known distribution area (Davies 2001) of all species relatively well with the exception of south and central Kalimantan and the east coast of Sumatra. Two leaves of each specimen were collected from small trees, silica-dried, and stored at −20 °C until further analysis. Voucher specimens were deposited at the National Herbarium Leiden (L), the Universiti Brunei Darussalam Herbarium (UBDH), and the University of Würzburg. Leaf material of analyzed specimens has been stored at the University of Kassel. Ant-associates collected from sampled trees are stored with the first author at the University of Würzburg and with H. Feldhaar at the University of Bayreuth, Germany.
Molecular analyses
DNA was isolated following a modified cetyl trimethyl ammonium bromide (CTAB) protocol (Bänfer et al. 2006). Plastid haplotypes were defined from the combined information at three plastid microsatellite (simple sequence repeat; SSR) loci, using the universal primer pairs ccmp2, ccmp5, and ccmp6 (Weising and Gardner 1999; Vogel et al. 2003). The ccmp2 locus was characterized by fragment length analysis. Sequencing of 20 polymerase chain reaction (PCR) products from M. motleyana and M. griffithiana specimens (GenBank accession numbers KT449838-KT449865) revealed that length differences at the ccmp2 locus (294–313 bp) were attributable to three indels. Two of these are due to simple insertion–deletion mutations, the third one is more complex with four different variants (see Online Resource 3). The ccmp5 (71–75 bp) and the ccmp6 (62–76 bp) loci were genotyped by single nucleotide sequencing analysis (Guicking et al. 2008). Plastid haplotypes were rewritten as pseudo-sequences with a total length of 13 nucleotide positions (Online Resource 3). Indels were coded as single changes in the final alignment. Poly (A/T) and poly (CT) microsatellites were coded according to the stepwise mutation model (Kimura and Ohta 1978). A cpDNA haplotype network was created using statistical parsimony as implemented in TCS 1.21 (Clement et al. 2000).
Seven nuclear SSR loci were used in this study (Online Resource 4). All loci were originally developed from other Macaranga spp. (Guicking et al. 2006; Baier et al. 2009), but cross-amplified easily in our study species. PCR amplifications were performed in final volumes of 25 μL, containing approximately 50 ng of genomic DNA, 1.5 mM magnesium chloride (MgCl2), 20 mM Tris-hydrochloride (HCl) (pH 8.0), 50 mM potassium chloride (KCl), 0.2 mM of each deoxyribonucleotide (dNTP), 5 μg bovine serum albumin (BSA), 0.5 units Taq DNA polymerase (Invitrogen), and 10 pmol of each primer. One primer of each pair was 5′-labeled with IRDye700 or IRDye800 fluorescent dyes. After initial denaturation (94 °C, 6 min), samples were submitted to 35 cycles of a touch-down PCR protocol in a Biometra thermocycler (Biometra, Germany). Each cycle consisted of 45 s at 94 °C (denaturation), 30 s at 65°–54 °C (annealing), and 45 s at 72 °C (elongation). The initial annealing temperature of 65 °C was decreased by 1 °C per cycle until 54 °C were reached. After final elongation (2 min, 72 °C), fragments were denatured and separated on a Li-Cor 4200 IR2 sequencer (Li-Cor Biosciences, Germany). Allele sizes were determined by visual examination, using an external size standard and reference samples loaded on each gel.
For each nuclear SSR locus, the numbers and sizes of alleles, Nei’s gene diversity H S (Nei 1987), and G IS (an estimator of Wrighťs inbreeding coefficient F IS) were calculated with the program FSTAT 2.9.3 (Goudet 2002). All loci were tested for departures from Hardy–Weinberg equilibrium (HWE) and for linkage disequilibrium between loci using Genepop 3.4 (Raymond and Rousset 1995) (Online Resource 4). We used the infinite alleles model for all nuclear SSR-based calculations, because some loci contained strongly interrupted or composed microsatellite motifs (as revealed by sequence analysis), and frequent deviations of the allele patterns from the expected regular length differences prohibited the assumption of a stepwise mutation model.
Analysis of population structure
Analyses of molecular variance (AMOVA; Excoffier et al. 1992) were performed with the program Arlequin 3.11 (Excoffier et al. 2005) to quantify the relative contribution of variation at different hierarchical levels. For intraspecific analyses, regions were defined for M. griffithiana as (1) Thai-Malay Peninsula (MP) and (2) Sumatra (SU), and for M. motleyana as (1) East Kalimantan (EK), (2) Sabah (SAB), and (3) Sarawak (SAR). Pairwise differences were used as the distance method.
Estimates of genetic diversity and population structure were calculated for each species, for the separate regions. For cpDNA, levels of within-population diversity were calculated as H (haplotype diversity) and π (nucleotide diversity) using Arlequin 3.11. For nuclear SSR data, FSTAT 2.9.3 (Goudet 2002) was used to calculate the numbers of alleles and allelic richness (number of alleles standardized for sample size; El Mousadik and Petit 1996) within each species and region. Observed (H o) and expected heterozygosity (H e; Nei 1987) at nuclear SSR loci were calculated with Arlequin 3.11.
Bayesian cluster analysis of nuclear SSR genotypes
To estimate the number of genetic units among and within the three species, Bayesian cluster analyses based on nuclear SSR data were performed with the program Structure 2.3 (Pritchard et al. 2000). The number of genetic clusters (K) was varied from one to ten. The burn-in was set to 100,000, and the run length was set to 1,000,000 iterations. For each number of genetic clusters, the program was run ten times. To test for hybridization among species, the “use population information” option was applied. In all other analyses, the admixture model was used, and allele frequencies were allowed to be correlated among clusters (Falush et al. 2003). The best number of genetic clusters was chosen by calculating the ΔK statistic of Evanno et al. (2005), using Structure Harvester v. 0.6.94 (Earl and vonHoldt 2012).
Dated phylogeny of myrmecophytic Macaranga
To estimate the age and divergence times of our study species, we extended the only available dated phylogeny of Macaranga and its sister genus Mallotus provided by van Welzen et al. (2014) by including additional ITS sequences. Because none of our study species and only two species of the bancana-group were included by van Welzen et al. (2014), we used the ITS sequence alignment of Kulju et al. (2007) that was also used by van Welzen et al. (2014) and complemented this with available sequences of myrmecophytic Macaranga species (Blattner et al. 2001; Davies et al. 2001). We used BEAST 1.82 (Drummond et al. 2012) and the same parameter settings as van Welzen et al. (2014) to generate a dated phylogeny from the complemented data set.
Results
Species delimitation
A total of 19 different cpDNA haplotypes were found among the 192 samples from the combined pseudo-sequences of three cpDNA loci (ccmp2, ccmp5, and ccmp6) (HT 0 to HT 18; Online Resource 3). Two haplotypes were found in M. constricta, six in M. griffithiana, and 11 in M. motleyana. None of the haplotypes were shared between species (Table 1, Fig. 1; Online Resource 3). The relationships among cpDNA haplotypes expressed in mutational steps are illustrated in the statistical parsimony network depicted in Fig. 2. All species proved to be separated from each other by a minimum of three mutational steps (see Fig. 2 for a list of alternative three-step connections). Most cpDNA haplotypes formed region-specific groups within species. For example, haplotypes 11 to 16 were exclusively found in M. motleyana plants from Sabah.
Statistical parsimony network based on cpDNA haplotype data, generated with the TCS program (Clement et al. 2000). Each circle represents a unique haplotype (indicated by numbers). Circle sizes are proportional to the number of individuals carrying the respective haplotype. Lines connecting the haplotypes represent mutational steps (thin lines for plastid microsatellite mutations, bold lines for indels and base substitutions). Small circles represent missing (uncollected or extinct) haplotypes
In the nuclear SSR analysis, a maximum of two alleles per locus per individual was observed, supporting that all species are diploid. No indication for linkage disequilibrium was found. Deviation from HWE was found in some populations and some loci (Online Resource 4). For example, locus B10 showed a significant deviation from HWE in nine out of 13 M. motleyana populations. Eight of these had a positive F IS value (equivalent to a heterozygote deficit), suggesting that a null allele might have been present at this locus, in which case genetic diversity may be underestimated.
While all cpDNA haplotypes in our analysis were species-specific, the nuclear SSR (ncSSR) data revealed a more complex pattern of species delimitation. In the Structure analysis, the optimum number of genetic clusters according to Evanno et al. (2005) was found for K = 2 (Online Resource 5a, b) and K = 8 (Fig. 3), both with the “use population information” and the “admixture model option,” although the latter detected an additional peak for K = 5 (Online Resource 5d). For K = 2, M. constricta and M. griffithiana plus one particular M. motleyana sample from Bako (study site 25) were generally differentiated from all remaining M. motleyana specimens (Online Resource 5a, b). However, under the admixture model, a similarly good likelihood value was found when M. motleyana samples from Danum Valley, Tawai, and Poring (sites 18, 19, and 22) grouped together with the M. constricta-M. griffithiana-Bako-cluster (Online Resource 5c). At this K, the output of the structure program is therefore ambiguous.
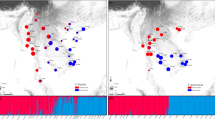
Genetic differentiation among 192 specimens from three Macaranga species collected at 25 localities, based on a Bayesian analysis with the program Structure 2.3 (Pritchard et al. 2000). Four specimens of the rare M. constricta were collected from the only known locality (1) in peninsular Malaysia, 51 specimens of M. griffithiana from eight localities (2–9) in peninsular Malaysia and two from Sumatra (10–11), and 137 specimens of M. motleyana from 14 sites on Borneo. Bar plots for K = 8 clusters obtained by analyzing all samples of all three study species are shown using the “use populations information” option (three “populations” here referring to the three species) (a) and the “admixture model” option (b). Each individual is represented as a vertical line, which is partitioned into colored segments that indicate the individual’s membership fractions in the assumed species. The numbers in between the plots refer to sample localities as indicated in Fig. 4
At K = 8, the three species were clearly differentiated in all runs, and further clusters showed intraspecific differentiation in M. griffithiana and in M. motleyana. The aberrant M. motleyana sample from Bako (see above) either formed a separate cluster (population information option, Fig. 3a) or grouped with M. griffithiana from Sumatra (admixture model, Fig. 3b). For K = 5, Macaranga constricta and M. griffithiana samples from the Malay Peninsula were grouped into the same cluster, whereas the M. griffithiana samples from Sumatra formed a separate cluster that also included the M. motleyana sample from Bako, further underlining the special status of this plant. Taken together, these results revealed a stronger intraspecific differentiation within M. griffithiana as well as within M. motleyana than between M. griffithiana and M. constricta (Online Resource 5d). Accordingly, an AMOVA based on ncSSR data attributed a much lower proportion (22.2 %) of the total genetic variation to the species level as compared with an AMOVA based on cpDNA data (66.5 %) (Table 2a).
Genetic diversity and intraspecific variation
All estimates of genetic diversity derived from cpDNA and ncSSR data were highest for M. motleyana which also had the largest sample size (Table 3). Almost all cpDNA haplotypes were region-specific for either Sabah, East Kalimantan, or Sarawak. Only HT 9 occurred both in northern East Kalimantan and eastern Sabah (Figs. 1 and 2). Notably, the M. motleyana morphotype A from Tawai (site 19) had two specific haplotypes (HT 14 and HT 15; Fig. 2). In the three populations that included morphotype B individuals of M. motleyana (Poring, Deramakot, and Tawau; Table 1), HT 12 was most abundant, but this haplotype is widely distributed and was also found in the “normal” M. motleyana morphotype (Telupid, Danum). Thus, there was no strict association of a particular haplotype with the morphotype B. The observed large differences between H o and H e calculated from ncSSR data in M. griffithiana and M. motleyana (Table 3a) and for M. motleyana in Sabah (Table 3b) are attributable to the pronounced population substructure in these species and regions.
A species-specific Bayesian cluster analysis based on ncSSR data (performed without the sample from Bako) underlined the strong intraspecific variation in M. motleyana (Fig. 4). When the optimum number of clusters (K = 4) was applied, a clear differentiation was found between East Kalimantan, Sabah, and Sarawak (Fig. 4). Specimens from population Berau (site 15, northern East Kalimantan) showed an affiliation to one cluster from Sabah but also to East Kalimantan (Fig. 4). In Kubah (site 24, west Sarawak), some influence from East Kalimantan was also observed (Fig. 4). Within Sabah, two genetic clusters were found, one including all individuals from Tawai (morphotype A) plus most from Poring and some from Danum, whereas the second cluster comprised all remaining individuals from Sabah (Fig. 4). The first cluster was even more differentiated in the Structure analysis run with the “use population information” option (Fig. 3a). Under these settings, the Tawai population that comprised all morphotype A individuals became almost perfectly separated from all other M. motleyana specimens.
Results of species-specific Bayesian structure analyses of M. griffithiana (optimal K = 2) and M. motleyana (optimal K = 4). Bar plots were obtained with the admixture model and are illustrated on the map. Numbers in parentheses are locality numbers as used in Fig. 3
The results of the intraspecific AMOVA in M. motleyana differed strongly between cpDNA and ncSSR data (Table 2). While the highest percentage of variation in cpDNA data was attributed to the “among regions” level and the lowest to the “within populations” level, ncSSR data showed by far the highest percentage of variation within populations and the lowest percentage among regions.
In M. griffithiana, four cpDNA haplotypes exclusively occurred on the Malay Peninsula and two on Sumatra (Figs. 1 and 2). The most widespread haplotype HT 6 was distributed from the southern Malay Peninsula up to eastern Thailand, whereas another, closely related haplotype (HT 7) was confined to the isolated northern populations in Chanthaburi and Kampong Thom in Cambodia (Fig. 1). A strong differentiation between M. griffithiana specimens from the Thai-Malay Peninsula and those from Sumatra was indicated by a species-specific Bayesian Structure analysis of ncSSR data with K = 2 (Fig. 4). Finally, a strong regional structure in M. griffithiana was likewise revealed by an AMOVA, in which cpDNA and ncSSR data both attributed more than 40 % of the total variation to the “among regions” level (Table 2b).
Estimation of age and divergence times
The dated phylogeny of our BEAST analysis is provided in Online Resource 6. Despite some inconsistencies (see Discussion Section), the analysis provided a rough estimate for the age of our study species. All M. motleyana, M. griffithiana, and M. constricta samples sequenced by Blattner et al. (2001) cluster together in one group that diverged from its sister group (notably containing the M. motleyana from Davies et al. 2001) between 5 and 10 million years BP (mya) (Online Resource 6). Divergence of M. motleyana, M. griffithiana, and M. constricta accessions from each other is dated to less than 5 mya (95 % highest posterior density interval 1.53–5.57 mya).
Distribution of associated ants
The main ant-partner of M. motleyana proved to be Crematogaster msp. 7, which like its host plant is restricted to Borneo (Fig. 5). This morphospecies was occasionally found on other host plants of section Pachystemon, mostly on the likewise waxy and widespread M. hypoleuca which is normally inhabited by C. decamera (see Online Resource 2). The latter ant species also occurred on M. motleyana, especially in East Kalimantan and Sarawak but was found only once on M. griffithiana in peninsular Malaysia (Fig. 5). Crematogaster borneensis was the main colonizer in all M. griffithiana saplings on peninsular Malaysia and Sumatra (very rarely C. msp. 2), and was only occasionally found in M. motleyana in Borneo. Macaranga constricta was colonized by a further species, C. msp. 9. This ant species was also found in a few M. hypoleuca saplings together with C. decamera at a disturbed road site some kilometers from the next M. constricta.
Distribution of the different ant species on their respective host plants M. constricta, M. griffithiana, and M. motleyana (including data from Quek et al. 2007 = lineages A, B, D, K, and G/H)
Discussion
Species delimitation and intraspecific variation
In the present study, all cpDNA haplotypes proved to be species-specific, suggesting a clear distinction of the plastid genome among species. This is a remarkable finding, given that an earlier molecular study of section Pachystemon had revealed a mainly geographical rather than taxonomical pattern of cpDNA haplotype distribution for most species of the bancana-clade, but not for M. motleyana and M. griffithiana (Bänfer et al. 2006). In the case of these two taxa, species boundaries and geographic patterns appear to be congruent with each other, so geographical isolation and taxonomic differentiation may have proceeded hand in hand. Macaranga motleyana and M. griffithiana may therefore represent a disjunct species pair having originated in allopatry, as it was previously suggested by Bänfer et al. (2006). The clear distinction between M. griffithiana and M. constricta haplotypes also suggests genetic independence as both species grow in close proximity. Overall, we found no indication for hybridization or introgression among the three species. The only exception may be the single specimen from Bako (Sarawak) which morphologically resembles M. motleyana but was grouped genetically with M. griffithiana from Sumatra. The Bako area has also been identified as being floristically rather distinct within Borneo and shows strong affinities to both the Malay Peninsula and Sumatra (Slik et al. 2003a).
For delimiting closely related species, ncSSRs have been reported to overcome problems associated with the use of cpDNA or ITS sequences (such as, e.g., incomplete lineage sorting, chloroplast capture, or insufficient among species variation) and have been applied for a number of tropical tree taxa (e.g., Duminil et al. 2012). The ncSSR data in our study supported the genetic distinctness of M. griffithiana and M. motleyana, whereas the results for M. constricta were ambiguous, depending on the model used. All three species were found to be colonized by their own specific ant-partner, also indicating distinctive features of the species. The main pollinators differ as well between M. griffithiana and M. motleyana (Fiala et al. 2011). As a further ecological trait, M. griffithiana was found to be associated with a specific endophytic scale insect probably endemic to peninsular Malaya (Heckroth et al. 1998). Finally, the typical habitat characteristics of the species appear different. Whereas M. motleyana occurs on dry as well as on wetter soils, M. griffithiana is found in swampy areas only, and M. constricta seems to be restricted to dryland forests (Davies 2001, and own observations). We always found M. constricta on hill slopes with good drainage and M. griffithiana on the swampy basis of the hills. These two types of habitat were sometimes only separated by 1–2 km at our sampling sites, so gene flow between the habitats by pollen or seeds is certainly possible.
Interestingly, the ncSSRs in our study showed stronger differentiation within M. griffithiana (peninsular Malaysia vs. Sumatra) and within M. motleyana, than between peninsular Malaysian M. constricta and M. griffithiana. Defining criteria for species delimitation is particularly challenging in M. motleyana which showed very high intraspecific morphological as well as genetic variation. The ncSSR data suggest that notwithstanding their morphological distinction, M. motleyana and morphotype B are the same, despite some differences in AFLP (Bänfer et al. 2004) and ITS data (Blattner et al. 2001). Morphotype A, on the other hand, was separated from the remainder of M. motleyana by cpDNA haplotype data as well as in part by the nuclear data. Therefore, what is currently named as M. motleyana might actually comprise different (sub)taxa or be a single widespread and polymorphic taxon, depending on species definition. The high internal diversity in M. motleyana is apparently not due to hybridization, although it grows sympatrically with other species from the same section (probably sharing pollinators, Fiala et al. 2011).
A plausible cause of the high genetic differentiation within this species might be a relatively long history of diversification. This would be in accordance with the internal (and therefore presumably ancient) position of M. motleyana and their allies in the haplotype network of Bänfer et al. (2006), as well as with the relatively early-branching position of M. motleyana from the remainder of the bancana-clade in the ITS phylogeny of Blattner et al. (2001) and in a strict consensus tree from the combined analysis of molecular and morphological characters in Davies et al. (2001). In the latter study, the ITS data of M. motleyana alone resulted in an unresolved polytomy together with other species of the bancana-clade (Pachystemon group 1). In our extension of the BEAST analysis of van Welzen et al. (2014), it is striking that all sequences from Blattner et al. (2001) cluster together, whereas two identical sequences from the samples used in Davies et al. (2001) came out in a separate position (Online Resource 6). Whether this difference reflects true sequence differences (possibly due to different sampling localities?) or whether the sequences from different laboratories are not compatible is unknown. The resolution of section Pachystemon in general and of the bancana-group in particular appears insufficient when based on ITS sequences only. This is illustrated by several instances in our tree in which conspecific accessions do not cluster together (whereas they do in species from other sections, e.g., M. winkleri, Online Resource 6), and internal nodes are found in less than 50 % of the resulting trees (at such nodes, no error bars are provided for the age estimate). The section Pachystemon was also poorly resolved in Kulju et al. (2007), and van Welzen et al. (2014) reported reconstructing the phylogeny of the Macaranga group 1 from Kulju et al. 2007 with BEAST to be difficult.
The M. griffithiana specimens from the Malay Peninsula showed little intraspecific differentiation, and even the specimens from Thailand and Cambodia were not separated by the ncSSR data, despite the large geographical distance. The weak genetic differentiation of M. griffithiana, as well as the wide distribution of its most common haplotype from peninsular Malaysia up to Indochina most probably indicates a recent expansion in this area. Studies on the genus Shorea also reported signatures of bottlenecks and subsequent population expansion on peninsular Malaysia (Kamiya et al. 2012; Ohtani et al. 2013). As myrmecophytic Macaranga species are confined to dipterocarp lowland forests, their distribution range may have been similar, and their life history strategy as pioneer trees may later have promoted an even more rapid dispersal. The M. griffithiana populations in eastern Thailand and Cambodia might be a recent remnant of surviving pockets of stable rainforest in the area (Whitmore 1984). Another possible explanation would be extant long-distance gene flow, e.g., by wind-driven thrips pollinators (see Guicking et al. 2013 for M. hypoleuca). Given that the respective populations are separated by more than 1000 km distance, we consider the latter hypothesis as rather unlikely.
In contrast to the lack of differentiation on the mainland, M. griffithiana specimens from Sumatra formed a clearly separate cluster in the nuclear data and also had its own “private” cpDNA haplotypes (i.e., unique haplotypes occurring in only one population). Macaranga griffithiana may therefore actually comprise two different varieties, one in peninsular Malaysia and the Asian mainland and one in Sumatra. Whitmore (1982) already mentioned some differences in the bracteoles of Malayan material of this species compared to those from Sumatra. Slik et al. (2011) found little floristic affinities between forests in peninsular Malaysia and west Sumatra, indicating that they differ in vegetation as well. Our sample size of M. constricta was too small for an intraspecific Bayesian Structure analysis; but nevertheless, this species proved to harbor two unique cpDNA haplotypes, pointing to a considerable differentiation despite its small distribution area.
Many tropical tree species have poorly delimited taxonomic boundaries and contain cryptic species as well as a high proportion of unrecognized intraspecific diversity (Rosell et al. 2010). Our results highlight the problems of defining species boundaries solely based on genetic data. If we used the extent of intraspecific differentiation as major criterion for taxon delimitation, it might be justified to split M. griffithiana and M. motleyana into two or more taxa each. It is, however, questionable whether such splitting of rather well-defined ecological units would be reasonable. Further investigations comprising more species of the bancana-clade and additional molecular data are needed to provide more insight into the relevance of morphological and genetic data for defining exact taxonomic boundaries in Macaranga.
Differentiation across Sundaland
Macaranga motleyana/M. griffithiana is at present the only myrmecophytic case in the genus where the genetic separation of populations between Borneo and Sumatra/Peninsula Malaysia seems to have resulted in allopatric speciation (Bänfer et al. 2006; Guicking et al. 2011, 2013; Weising et al. 2010). All other members of sections Pachystemon as well as Pruinosae (Davies 2001) in peninsular Malaysia also occur on Borneo, and several of them also show large morphological variation that has, however, not been given species status in the different regions (e.g., M. hullettii, M. pruinosa, see also Davies 2001). Genetic differences between these regions were also detected in Shorea curtisii (Kamiya et al. 2012) but have not been found in other widespread dipterocarp species like S. parvifolia and S. leprosula (Iwanaga et al. 2012; Ohtani et al. 2013).
Although the dated phylogeny reconstruction of available Macaranga ITS sequences (Online Resource 6) revealed several inconsistencies of the data in the bancana-clade, it gave some rough insights into the temporal evolution of the group. Divergence times of the distinct motleyana-group of Blattner et al. (2001) from its sister group could be dated to less than 10 mya with the BEAST approach, and divergence of M. motleyana from M. griffithiana plus M. constricta from this study to between 5.57 and 1.5 mya, and might therefore have predated Pleistocenic events (see also Chomicki and Renner 2015, Fig. S9). The last common ancestor of the only two members of the bancana-clade (M. hullettii and M. triloba) included in van Welzen et al. (2014) was there dated to 4.6–0.2 mya. Sumatra and Borneo were connected to mainland Asia until about 5 mya, and since then became separated by the sea for more than half of the time (e.g., Voris 2000; Cannon et al. 2009; Raes et al. 2014). A deeper history of vicariant evolution between the Asian mainland and the Sundaland islands dating back to the Pliocene was observed for many other plant and animal groups (e.g., Bendiksby et al. 2010; Leonard et al. 2015, and references therein). Without a more comprehensive sampling that includes samples from different geographical localities per species (and possibly more genes), it cannot yet be clarified whether M. motleyana is paraphyletic and M. griffithiana evolved after a long-distance dispersal of some ancient M. motleyana to the Malay Peninsula and Sumatra, or whether the two species are sister species that evolved allopatrically from a former widespread precursor.
Comparative phylogeography of ant-associates
Crematogaster msp. 7/lineage A on M. motleyana is presumed to have originated between 9 and 6 mya (Quek et al. 2004, 2007), which would match a probable origin of a common ancestor of the motleyana-group of less than 10 mya. This ant species exhibits large within-species genetic variation (up to 14 % in COI in Sabah alone, Feldhaar et al. 2010). It therefore mirrors the intraspecific variation found in its host, although gene flow appears to be even more restricted. Thus, ant specimens from M. motleyana trees found in Poring and Tawau were separated by six missing mtDNA haplotypes, although their host plants shared the same cpDNA haplotype (Feldhaar and Fiala, unpubl. results). Both ants and plants are restricted to Borneo, and it could indeed represent an old association which has been shaped by similar historical events, leading to high substructuring of both partners. A specific endophytic scale insect associated with C. msp. 7/lineage A also came out as the oldest coccid lineage (L6) of the species studied in Borneo, likewise pointing to old associations (Ueda et al. 2008).
Continuous rainforest seems to have covered large parts of Sundaland for a long time (Cannon et al. 2009; Raes et al. 2014) but for parts of Sumatra and peninsular Malaysia drier, and seasonal conditions predominated during the Pleistocene (Morley 2000). In this period, forest may have become restricted to refugial areas. Macaranga constricta and its specific ant-partner (C. msp. 9/lineage B) are both endemic on peninsular Malaysia where the ant was assumed to represent a relictual lineage, suggesting massive extinctions in its history (Quek et al. 2007). In contrast, Crematogaster borneensis/lineage K on M. griffithiana came out as rather derived and little differentiated in the phylogenies, and its origin was dated to about 2 mya (Blaimer 2012). Quek et al. (2007) assumed that an expansion of the ant lineage K took place in lowlands of Sumatra and peninsular Malaysia in the Pleistocene, possibly during one of the interglacials. It mirrors the fast expansion scenario for its host M. griffithiana in the area. A possible scenario for a relatively recent association between M. griffithiana and the generalist ant species C. borneensis might be that this ant species found an empty niche on M. griffithiana after possible original ant-associates had gone extinct during the Pleistocene. It may, however, not have reached the disjunct relict area of M. griffithiana in eastern Thailand and Cambodia, where the species is associated with ants from the genus Camponotus (Formicinae) instead (Maschwitz et al. 2004, and unpublished data).
Conclusions
Few genetic studies have focused on pioneer plant species in Sundaland; however, the ecological relevance of genera that grow successfully in secondary forest has tremendously increased following anthropogenic disturbance of the rainforests. The comparison of nuclear and plastid DNA in our study provided insights into the genetic delimitation of the study species and characterized their genetic structure across a large distribution area. However, our results point out problems of species delimitations/definitions which might be common in tropical trees in general. The congruence of biogeographic and genetic patterns of plants and their ants indicates a common history of the mutualistic associations, but also represents very different examples of this symbiosis: a widespread and presumably ancient plant species on Borneo, a vicariant, recently expanded species on Sumatra and the Asian mainland and a very local relict on peninsular Malaysia. The phylogeographical and ecological results would thus support the present status of these three closely related Macaranga taxa as separate species.
References
Baier, C., Guicking, D., Prinz, K., Fey-Wagner, C., Wöhrmann, T., Weising, K., Debener, T., et al. (2009). Isolation and characterization of 11 new microsatellite markers for Macaranga (Euphorbiaceae). Molecular Ecology Resources, 9, 1049–1052.
Bänfer, G., Fiala, B., & Weising, K. (2004). AFLP analysis of phylogenetic relationships among myrmecophytic species of Macaranga (Euphorbiaceae) and their allies. Plant Systematics and Evolution, 249, 213–231.
Bänfer, G., Moog, U., Fiala, B., Mohamed, M., Weising, K., & Blattner, F. R. (2006). A chloroplast genealogy of myrmecophytic Macaranga species (Euphorbiaceae) in Southeast Asia reveals hybridization, vicariance and long-distance dispersals. Molecular Ecology, 15, 4409–4424.
Bendiksby, M., Schumacher, T., Gussarova, G., Nais, J., Mat-Salleh, K., Sofiyanti, N., Madulid, D., et al. (2010). Elucidating the evolutionary history of the Southeast Asian holoparasitic, giant-flowered Rafflesiaceae: Pliocene vicariance, morphological convergence and character displacement. Molecular Phylogenetics and Evolution, 57, 620–633.
Blaimer, B. B. (2012). Acrobat ants go global: origin, evolution and systematics of the genus Crematogaster (Hymenoptera: Formicidae). Molecular Phylogenetics and Evolution, 65, 421–436.
Blattner, F. R., Weising, K., Bänfer, G., Maschwitz, U., & Fiala, B. (2001). Molecular analysis of phylogenetic relationships among myrmecophytic Macaranga species (Euphorbiaceae). Molecular Phylogenetics and Evolution, 19, 331–344.
Cannon, C. H., Morley, R. J., & Bush, A. B. G. (2009). The current refugial rainforests of Sundaland are unrepresentative of their biogeographic past and highly vulnerable to disturbance. Proceedings of the National Academy of Sciences of the United States of Ameica, 106, 11188–11193.
Chomicki, G., & Renner, S. S. (2015). Phylogenetics and molecular clocks reveal the repeated evolution of ant-plants after the late Miocene in Africa and the early Miocene in Australasia and the Neotropics. New Phytologist, 20, 411–424.
Clement, M., Posada, D., & Crandall, K. A. (2000). TCS: a computer program to estimate gene genealogies. Molecular Ecology, 9, 1657–1659.
Davies, S. J. (2001). Systematics of Macaranga sections Pachystemon and Pruinosae (Euphorbiaceae). Harvard Papers in Botany, 6, 371–448.
Davies, S. J., Lum, S. K. Y., Chan, R., & Wang, L. K. (2001). Evolution of myrmecophytism in western Malesian Macaranga (Euphorbiaceae). Evolution, 55, 1542–1559.
Drummond, A. J., Suchard, M. A., Xie, D., & Rambaut, A. (2012). Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology and Evolution, 29, 1969–1973.
Duminil, J., Kenfack, D., Viscosi, V., Grumiau, L., & Hardy, O. J. (2012). Testing species delimitation in sympatric species complexes: the case of an African tropical tree, Carapa spp. (Meliaceae). Molecular Phylogenetics and Evolution, 62, 275–285.
Earl, D. A., & von Holdt, B. M. (2012). STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources, 4, 359–361.
El Mousadik, A., & Petit, R. J. (1996). High level of genetic differentiation for allelic richness among populations of the argan tree (Argania spinosa (L) Skeels) endemic to Morocco. Theoretical and Applied Genetics, 9, 832–839.
Evanno, G., Regnaut, S., & Goudet, J. (2005). Detecting the number of clusters of individuals using the software structure: a simulation study. Molecular Ecology, 14, 2611–2620.
Excoffier, L., Smouse, P. E., & Quattro, J. M. (1992). Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics, 131, 479–491.
Excoffier, L., Laval, G., & Schneider, S. (2005). Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online, 1, 47–50.
Falush, D., Stephens, M., & Pritchard, J. K. (2003). Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics, 16, 1567–1587.
Feldhaar, H., Fiala, B., Gadau, J., Mohamed, M., & Maschwitz, U. (2003a). Molecular phylogeny of Crematogaster subgenus Decacrema ants (Hymenoptera: Formicidae) and the colonization of Macaranga (Euphorbiaceae) trees. Molecular Phylogenetics and Evolution, 27, 441–452.
Feldhaar, H., Fiala, B., Hashim, R., & Maschwitz, U. (2003b). Patterns of the Crematogaster-Macaranga association: the ant partner makes the difference. Insectes Sociaux, 50, 9–19.
Feldhaar, H., Gadau, J., & Fiala, B. (2010). Speciation in obligately plant-associated Crematogaster ants: host distribution rather than adaption towards specific hosts drives the process. In M. Glaubrecht (Ed.), Evolution in action—adaptive radiations and the origins of biodiversity (pp. 193–213). Heidelberg: Springer.
Fiala, B., Maschwitz, U., Tho, Y. P., & Helbig, A. J. (1989). Studies of a Southeast Asian ant-plant association: protection of Macaranga trees by Crematogaster borneensis. Oecologia, 79, 463–470.
Fiala, B., Grunsky, H., Maschwitz, U., & Linsenmair, K. E. (1994). Diversity of ant-plant interactions: protective efficacy in Macaranga species with different degrees of ant association. Oecologia, 97, 186–192.
Fiala, B., Jakob, A., Maschwitz, U., & Linsenmair, K. E. (1999). Diversity, evolutionary specialization and geographic distribution of a mutualistic ant-plant complex: Macaranga and Crematogaster in South East Asia. Biological Journal of the Linnean Society, 66, 305–331.
Fiala, B., Meyer, U., Hashim, R., & Maschwitz, U. (2011). Pollination systems in pioneer trees of the genus Macaranga (Euphorbiaceae) in Malaysian rainforests. Biological Journal of the Linnean Society, 103, 935–953.
Goudet, J. (2002). FSTAT, a program to estimate and test gene diversities and fixation indices version 2.9.3.2. http://www2.unil.ch/popgen/softwares/fstat.htm.
Guicking, D., Rana, T. S., Blattner, F. R., & Weising, K. (2006). Microsatellite markers for the palaeotropic pioneer tree genus Macaranga (Euphorbiaceae) and their cross-species transferability. Molecular Ecology Notes, 6, 245–248.
Guicking, D., Kröger-Kilian, T., Weising, K., & Blattner, F. R. (2008). Single nucleotide sequence analysis: a cost- and time-effective protocol for the analysis of microsatellite- and indel-rich chloroplast DNA regions. Molecular Ecology Resources, 8, 62–65.
Guicking, D., Fiala, B., Blattner, F. R., Slik, F., Mohamed, M., & Weising, K. (2011). Comparative chloroplast DNA phylogeography of two tropical pioneer trees, Macaranga gigantea and Macaranga pearsonii (Euphorbiaceae). Tree Genetics & Genomes, 7, 573–583.
Guicking, D., Fiala, B., Kröger-Kilian, T., Mohamed, M., & Weising, K. (2013). High gene flow in two thrips-pollinated Southeast Asian pioneer trees: genetic diversity and population structure of Macaranga hypoleuca and Macaranga beccariana (Euphorbiaceae). Botanical Journal of the Linnean Society, 173, 606–621.
Heckroth, H. P., Fiala, B., Gullan, P. J., Idris, A. H., & Maschwitz, U. (1998). The soft scale (Coccidae) associates of Malaysian ant-plants. Journal of Tropical Ecology, 14, 427–443.
Iwanaga, H., Teshima, K. M., Khatab, I. A., Inomata, N., Finkeldey, R., Siregar, I. Z., Siregar, U. J., & Szmidt, A. E. (2012). Population structure and demographic history of a tropical lowland rainforest tree species Shorea parvifolia (Dipterocarpaceae) from Southeastern Asia. Ecology and Evolution, 2, 1663–1675.
Kamiya, K., Nanami, S., Kenzo, T., Yoneda, R., Diway, B., Chong, L., Azani, M. A., et al. (2012). Demographic history of Shorea curtisii (Dipterocarpaceae) inferred from chloroplast DNA sequence variations. Biotropica, 44, 577–585.
Kimura, M., & Ohta, T. (1978). Stepwise mutation model and distribution of allelic frequencies in a finite population. Proceedings of the National Academy of Sciences of the United States of Ameica, 75, 2868–2872.
Kulju, K. K. M., Sierra, S. E. C., Draisma, S. G. A., Samuel, R., & van Welzen, P. C. (2007). Molecular phylogeny of Macaranga, Mallotus, and related genera (Euphorbiaceae s.s.): insights from plastid and nuclear DNA sequence data. American Journal of Botany, 94, 1726–1743.
Leonard, J. A., den Tex, R.-J., Hawkins, M. T. R., Muñoz-Fuentes, V., Thorington, R., & Maldonado, J. E. (2015). Phylogeography of vertebrates on the Sunda Shelf: a multi-species comparison. Journal of Biogeography, 42, 871–879.
Maschwitz, U., Fiala, B., & Dumpert, K. (2004). An unusual myrmecophytic Macaranga association, occurring in a disjunct area in the monsoon zone of South-East Asia: phenology and the description of a new ant species. Ecotropica, 10, 33–49.
Morley, R. J. (2000). Origin and evolution of tropical rain forests. Chichester: Wiley.
Nei, M. (1987). Molecular evolutionary genetics. New York: Columbia University Press.
Ohtani, M., Kondo, T., Tani, N., Ueno, S., Lee, L. S., Ng, K., et al. (2013). Nuclear and chloroplast DNA phylogeography reveals Pleistocene divergence and subsequent secondary contact of two genetic lineages of the tropical rainforest tree species Shorea leprosula (Dipterocarpaceae) in South-East Asia. Molecular Ecology, 22, 2264–2279.
Pritchard, J. K., Stephens, M., & Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics, 155, 945–959.
Quek, S. P., Davies, S. J., Itino, T., & Pierce, N. E. (2004). Codiversification in an ant-plant mutualism: stem texture and the evolution of host use in Crematogaster (Formicidae: Myrmicinae) inhabitants of Macaranga (Euphorbiaceae). Evolution, 58, 554–570.
Quek, S. P., Davies, S. J., Ashton, P. S., Itino, T., & Pierce, N. E. (2007). The geography of diversification in mutualistic ants: a gene’s-eye view into the Neogene history of Sundaland rain forests. Molecular Ecology, 16, 2045–2062.
Raes, N., Cannon, C. H., Hijmans, R. J., Piessens, T., Saw, L. G., van Welzen, P. C., & Slik, J. W. F. (2014). Historical distribution of Sundaland’s Dipterocarp rainforests at Quaternary glacial maxima. Proceedings of the National Academy of Sciences of the United States of Ameica, 111, 16790–16795.
Raymond, M., & Rousset, F. (1995). Genepop (version 1.2): population genetics software for exact tests and ecumenicism. Journal of Heredity, 86, 248–249.
Rosell, J. A., Olson, M. E., Weeks, A., De-Nova, J. A., Lemos, R. M., Camacho, J. P., Feria, T. P., et al. (2010). Diversification in species complexes: tests of species origin and delimitation in the Bursera simaruba clade of tropical trees (Burseraceae). Molecular Phylogenetics and Evolution, 57, 798–811.
Slik, J. W. F., Poulsen, A. D., Ashton, P. S., Cannon, C. H., Eichhorn, K. A. O., Kartawinata, K., Lanniari, L., et al. (2003a). A floristic analysis of the lowland dipterocarp forests of Borneo. Journal of Biogeography, 30, 1517–1531.
Slik, J. W. F., Keßler, P. J. A., & van Welzen, P. C. (2003b). Macaranga and Mallotus species (Euphorbiaceae) as indicators for disturbance in the mixed lowland dipterocarp forest of East Kalimantan (Indonesia). Ecological Indicators, 2, 311–324.
Slik, J. W. F., Aiba, S.-I., Bastian, M., Brearley, F. Q., Cannon, C. H., Eichhorn, K. A. O., Fredriksson, G., et al. (2011). Soils on exposed Sunda Shelf shaped biogeographic patterns in the equatorial forests of Southeast Asia. Proceedings of the National Academy of Sciences of the United States of Ameica, 108, 12343–12347.
Ueda, S., Quek, S. P., Itioka, T., Inamori, K., Sato, Y., Murase, K., & Itino, T. (2008). An ancient tripartite symbiosis of plants, ants and scale insects. Proceedings of the Royal Society of London B, 275, 2319–2326.
van Welzen, P. C., Strijk, J. S., van Konijnenburg-van Cittert, J. H. A., Nucete, M., & Merckx, V. S. F. T. (2014). Dated phylogenies of the sister genera Macaranga and Mallotus (Euphorbiaceae): congruence in historical biogeographic patterns? PloS One, 9(1), e85713. doi:10.1371/journal.pone.0085713.
Vogel, M., Bänfer, G., Moog, U., & Weising, K. (2003). Development and characterization of chloroplast microsatellite markers in Macaranga (Euphorbiaceae). Genome, 46, 845–857.
Voris, H. K. (2000). Maps of Pleistocene sea levels in Southeast Asia: shorelines, river systems and time durations. Journal of Biogeography, 27, 1153–1167.
Weising, K., & Gardner, R. C. (1999). A set of conserved PCR primers for the analysis of simple sequence repeat polymorphisms in chloroplast genomes of dicotyledonous angiosperms. Genome, 42, 9–19.
Weising, K., Guicking, D., Fey-Wagner, C., Kröger-Kilian, T., Wöhrmann, T., Dorstewitz, W., Bänfer, G., et al. (2010). Mechanisms of speciation in Southeast Asian ant-plants of the genus Macaranga (Euphorbiaceae). In M. Glaubrecht (Ed.), Evolution in action—adaptive radiations and the origins of biodiversity (pp. 169–191). Berlin: Springer.
Whitmore, T. C. (1974). Studies in Macaranga VI. Novelties form Borneo and a reduction in Malaya. Kew Bulletin, 29, 445–450.
Whitmore, T. C. (1982). Macaranga Thou. Kew Bulletin, 36, 312–323.
Whitmore, T. C. (1984). Tropical rain forests of the Far East (2nd ed.). Oxford: Oxford University Press.
Whitmore, T. C. (2008). The genus Macaranga: a prodromus. Kew: Royal Botanic Gardens.
Whitmore, T. C., & Airy Shaw, H. K. (1971). Studies in Macaranga VI. New and notable records for Malaya. Kew Bulletin, 25, 238.
Acknowledgments
Permission to conduct research in Malaysia was kindly granted by the Economic Planning Unit (EPU), Kuala Lumpur and EPU in Kota Kinabalu, Sabah, as well as by the Danum Valley Management Committee and the Forest Department, Sarawak. We thank our colleagues in Malaysia, especially Dr. Rosli bin Hashim, Dr. Jamili Nais, Dr. Maklarin bin Lakim, and Dr. G. Reynolds for their support. We also thank LIPI for providing research permits for East Kalimantan, and our counterpart Kade Sidiyasa, who sadly passed away recently, and the Tropenbos Foundation for logistic help on site. Financial support by the German Research Foundation is gratefully acknowledged (grant We 1830/4, Fi 606/5). We thank Prof. Dr. K. E. Linsenmair for manifold support, Dr. K. Khoshbakht for the help with the molecular analyses, and Dr. N. Ruppert, M. Zirpel, Dr. T. Kenzo, Forestry and Forest Products Research Institute, and the Forest-Wildlife Research and Development Institute, Cambodia, for the supply of plant material. Dr. S. P. Quek kindly provided a photograph. We thank two anonymous referees for their constructive and valuable comments on a previous version of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Brigitte Fiala and Daniela Guicking contributed equally to this work.
Rights and permissions
About this article
Cite this article
Fiala, B., Slik, F., Weising, K. et al. Phylogeography of three closely related myrmecophytic pioneer tree species in SE Asia: implications for species delimitation. Org Divers Evol 16, 39–52 (2016). https://doi.org/10.1007/s13127-015-0254-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13127-015-0254-2