Abstract
Micropropagation of oil palm by somatic embryogenesis produces a proportion of off-type individuals (approximately 5% overall) displaying a homeotically modified flower structure known as mantled. Transformation of the fertile or sterile androecium into carpel-like structures is observed in staminate and pistillate mantled flowers, respectively, resulting in lower oil yields in affected plantations. Given the epigenetic nature of the mantled condition, a gene expression-based approach was used rather than a genetic one to investigate its molecular basis. Suppression subtractive hybridisation (SSH) and macroarray hybridisation were used to compare transcriptome patterns between normal and mantled inflorescences. Two SSH libraries, enriched for complementary deoxyribonucleic acids (cDNAs) of either true-to-type or somaclonal variant material, were generated. Bioinformatic analysis of these two libraries allowed the identification of 1,350 unique sequences and their annotation by a gene ontology-based approach. Macroarray hybridisation was used to compare gene expression between normal and mantled inflorescences, and 32 genes were found to be differentially expressed. The temporal expression patterns of six genes were further investigated in more detail in relation to male and female inflorescence development. Full-length cDNAs were isolated and characterised for two of these genes, EgFB1 and EgRING1, both of which are down-regulated in the mantled inflorescences and both of which encode proteins associated with proteolytic signalling complexes. Our data shed light on gene expression changes associated with the mantled phenotype and have provided novel transcriptome markers which can help to distinguish the abnormal and wild-type inflorescences.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The oil palm (Elaeis guineensis Jacq.) is a major cultivated crop and the world’s largest source of edible vegetable oil. This monocotyledonous perennial plant belonging to the family Arecaceae originates from West Africa and is cultivated in the tropical areas of Africa, Asia and Latin America. It is monoecious, with alternating cycles of male and female inflorescence production. This makes the species essentially allogamous, and seed progenies of oil palm show a significant level of genetic variability. For this reason, and due to its long life cycle, there is a worldwide demand for clonally propagated palms in order to disseminate elite planting material in a shorter time frame. Tissue culture-based multiplication was developed in the 1980s; however, the occurrence of a somaclonal variant known as mantled (Corley et al. 1986) proved detrimental to the yield of clonal plantations. Indeed, about 5% of palms regenerated by tissue culture show an abnormal flower phenotype which results in partial or total sterility (with negative consequences on yield) and parthenocarpic fruits, depending on the severity of the phenotype (Rival 2007). The mantled abnormality is characterised by a homeotic transformation of stamens and staminodes (rudimentary stamens), in male and female flowers, respectively, into pseudocarpels (Adam et al. 2005). Field observations have shown that the mantled phenotype is heterogeneously distributed between and within clonal progenies. Furthermore, a significant degree of reversion to the normal phenotype has been observed in the field as well as a non-Mendelian transmission of the character through seeds (Durand-Gasselin et al. 1990; Rao and Donough 1990). These characters suggest that epigenetic deregulation of gene expression is the cause of the variant phenotype. This is partly supported by the absence of observable changes at the ploidy and deoxyribonucleic acid (DNA) sequence levels (Rival et al. 1997, 1998; Matthes et al. 2001). Moreover, both genome-wide DNA hypomethylation and sequence-specific methylation changes have been demonstrated in mantled palms when compared to their normal counterparts (Jaligot et al. 2000, 2002, 2004; Kubis et al. 2003). This decrease in DNA methylation cannot be explained by a change in the transcription level of any of the three major DNA methyltransferase genes (Rival et al. 2008), making it likely that those genes are not directly involved in the occurrence of the abnormal phenotype. In addition, Kubis et al. (2003) failed to detect in the mantled context any major rearrangement or change in methylation status of the transposable elements which are frequently associated with tissue culture-induced somaclonal variation (Peschke et al. 1987). Given the phenotypic resemblance between mantled flowers and those of floral B-class gene mutants in model plants (Coen and Meyerowitz 1991), parallel studies were performed to identify MADS-box organ identity genes in oil palm (Adam et al. 2006; Alwee et al. 2006). Using this approach, Adam et al. (2007) demonstrated that expression of both putative B-type genes (EgDEF1 and EgGLO2) was reduced in mantled flowers of both sexes.
In other studies, a messenger ribonucleic acid (mRNA)-based approach using differential display enabled the characterisation of genes displaying enhanced transcript accumulation in callus producing abnormal plants (Tregear et al. 2002; Morcillo et al. 2006). These genes have a potential utility as early markers for clonal conformity; however, the mechanisms which govern their differential expression and, more generally, the mantled phenotype remain unclear. Given that oil palm is only distantly related to existing model species, it is to be expected that divergences in gene function will be commonplace with respect to plants for which functional genomics tools are available. For this reason, it is desirable to search for genes of interest not only on the basis of their sequence affinities but also by an ‘anonymous’ approach on the basis of their expression patterns. In the last decade, advances in high-throughput technologies have provided the means to monitor simultaneously the expression of large subsets of genes involved in numerous aspects of plant development. Among these techniques, complementary DNA (cDNA) array-based approaches have led to gene discovery in a number of species (Aharoni et al. 2000; Nogueira et al. 2003; Tamaoki et al. 2004; Laitinen et al. 2007; Kim et al. 2009). In some cases, in order to maximise the chance of identifying genes involved in the process of interest, this technique was used in conjunction with suppression subtractive hybridisation (SSH) cDNA libraries. The SSH approach is a powerful tool to generate a population of cDNAs enriched for genes that are differentially expressed between two samples. This is achieved by combined steps of normalisation and subtraction which facilitate the identification of rare transcripts. This approach has been used to successfully isolate genes involved in floral development in Arabidopsis thaliana (Hu et al. 2003), in Acacia mangium (Wang et al. 2005) and in citrus (Zhang et al. 2009). In the present study, with the aim of understanding the molecular processes underlying flower development in the oil palm, a strategy combining SSH and cDNA macroarrays was used to identify on a large scale, genes displaying altered expression in the mantled inflorescence. This approach allowed the identification of 32 putative marker genes of the mantled phenotype, six of which were chosen for wider expression analysis by reverse transcription polymerase chain reaction (RT-PCR) and two of which were studied in further detail by the isolation of full-length cDNAs in order to provide information on their possible functions.
Materials and methods
Plant material
Immature inflorescences from oil palms obtained through in vitro somatic embryogenesis (Pannetier et al. 1981) were collected at the same time at the Tun Razak Research Station, Malaysia, by excision from axils and immediate freezing in liquid nitrogen. Normal and mantled inflorescences of both sexes were sampled from palms of the FC166 clonal line. For marker validation work, a second pair of palms was studied, the normal inflorescence being collected from a palm of clonal line FC1238 and the mantled inflorescence from a palm of clonal line FC1292 (Table S1). In each case, the abnormal palm was obtained by a supplementary cloning step from a mantled regenerant, a process which is known to strongly increase the prevalence and intensity of the abnormality. Spikelets from each inflorescence were fixed and analysed by light microscopy as described by Adam et al. (2005) in order to check their developmental stage. The first pair of material belongs to the same phenotype and was used for the initial analysis, the second pair of material comes from different genotypes in order to analyze the same sex and stage of floral development. Leaf and root material was harvested from seed-derived greenhouse plants (origin C1001F) grown in Montpellier, France. Oil palm embryogenic nodular calli (clonal line LMC 458) were obtained through the in vitro culture of leaf explants (Pannetier et al. 1981) sampled on a normal adult palm.
SSH and cDNA library construction
Total RNA was extracted from oil palm tissues as described previously (Morcillo et al. 2006). RT was carried out using the SMART-PCR cDNA synthesis kit (Clontech, USA). Two micrograms of RNA were used for suppressive subtractive hybridisation. Forward (normal male inflorescence as tester and mantled male inflorescence as driver) and reverse (mantled male inflorescence as tester and normal male inflorescence as driver) SSH cDNA libraries (referred to as INFSSHM1 and INFSSHM2, respectively) were constructed using the Clontech PCR-Select cDNA subtraction kit (Clontech) following the manufacturer’s instructions. Two microliters of purified secondary SSH PCR products were used to perform ligations into the pGEMT-Easy Vector System, followed by subsequent transformation into JM109 competent cells (Promega, USA), according to the manufacturer’s instructions. Each clone library was plated on lysogeny broth (LB) agar medium, and 960 colonies were then randomly selected for liquid culture. Bacterial clones were stored in 96-well microtitre plates as 10% (v/v) glycerol stocks.
EST assembly and annotation
A number of 960 clones from each library were single pass sequenced with the M13 primer by Cogenics (France). The sequence data were first passed through a custom-processing chain (Poncet et al. 2006) to remove vector sequences, primers, adaptors and sequences shorter than 50 bp or of low quality. PolyA ends and repeat sequences were masked by RepeatMasker (A.F.A. Smit, R. Hubley and P. Green RepeatMasker at http://repeatmasker.org). Clustering and assembling of valid sequences was performed using the TIGR gene indices clustering tools (TGICL) program (Pertea et al. 2003) with a threshold value of 96% sequence similarity per frame of 80 bases minimum. The BLASTX algorithm (Altschul et al. 1990) with an e-value cutoff at 10−5 was used to search for similarity in the resulting unigenes (singletons plus contigs) against the National Center for Biotechnology Information non-redundant ((NCBI nr) protein database. Gene ontology (GO) annotation of unigenes was carried out using the Blast2GO software (http://www.blast2go.de; Conesa and Gotz 2008) using default parameters and with an e-value threshold of 10−5 for the BLAST step. The annotation configuration was set on BLASTX results showing an e-value less than 10−6 with a GO weight of 5, a minimal alignment length of 6 and an annotation cutoff of 55. An annotation enrichment was also performed via an InterProScan comparison. Ultimately, GOslim annotations were generated using the plant GOslim (http://www.geneontology.org/GO.slims.shtml) mapping in order to simplify data visualisation. Relative abundance of GO term categories were compared in two libraries using Spearman correlation test (Statistica©, Statsoft, France). All sequences were deposited into Genbank dbEST under the accession numbers GT119120 to GT120724.
cDNA macroarray preparation and hybridisation
The cDNA inserts from the SSH libraries were PCR-amplified (from 2.5 μL of bacterial glycerol stock) in four independent reactions with the P1 and 2R primers (Clontech) used for the nested PCR step of the suppressive subtractive hybridisation procedure. The four PCR products were pooled and precipitated with a solution containing 95% (v/v) ethanol and 5% (v/v) sodium acetate at pH 5.2 before resuspension in 25 μL sterile water. The quality and quantity of the amplicons were checked by running an aliquot on a 2.5% agarose gel. PCR products were then spotted onto nylon membrane matrix (Hybond™-N+, Amersham Biosciences) using a Flexys plus apparatus (Genomic Solutions). All the cDNAs amplified from reverse and forward libraries were arrayed on the same membrane in a single print. The membranes were then soaked successively in a denaturing solution (0.5 M NaOH) then in a neutralising solution (1.5 M NaCl, 0.5 M Tris, pH 7.5, 1 mM EDTA) and finally rinsed with 2× SSC buffer. After air-drying overnight, the membranes were submitted to DNA cross-linking under UV light (70 mJ/cm2). Denatured total RNA (20 μg) was reverse-transcribed and radiolabelled for 2 h at 37°C using the LabelStar kit (Qiagen) in a 50 μL reaction mixture containing 5 μL of 10× RT buffer, 5 μL of dNTP Mix [dATP, dGTP, dTTP (20 mM) and dCTP (0.2 mM)], 0.5 μL of RNase inhibitor (40 units/μL), 5 μL of oligo-dT primer (20 μM), 5 μL of [α32P]dCTP (3000 Ci/mmol) and 2.5 μL of LabelStar reverse transcriptase. The membranes were pre-hybridised for 2 h at 65°C in 15 mL of hybridisation solution (5× SSC, 5× Denhart, 0.6% SDS, 60 μL of denatured salmon sperm DNA) before adding 25 μL of labelling solution. Overnight hybridisation was carried out at 65°C. The arrays were then washed at 65°C three times in 1× SSC, 0.1% SDS for 10 min and once in 0.1× SSC, 0.1% SDS, for 15 min. The membranes were wrapped still damp in Saran film and exposed to a Kodak storage phosphor screen (Amersham Biosciences) for 2 days. Image acquisition was performed using a Typhoon 9400 Phosphoimager (Amersham Biosciences) with 50-μM resolution in GEL format.
Macroarray data analysis
Quantification of spot signals was performed using the volume quantification method on the ArrayVision™ software (Imaging Research, Canada). For each membrane, an average background value was calculated from non-DNA-spotted areas on the membrane and was subtracted from the raw value of each spot. This corrected value was then normalised against the average corrected signal intensity value on the membrane. Each hybridisation experiment was carried out independently on four replicate membranes hybridised to four independent labelling reactions. For each spot position, the three normalised intensity values displaying the least variation were retained for further examination. A one-way analysis of variance (ANOVA) was then performed between the two sets of values of each spot. Differential expression was recognised when mean signals were significantly different (p value < 0.001) and when there was at least a twofold difference between normal and mantled values.
Semi-quantitative RT-PCR
RT was performed using ImProm-II™ RT system kit (Promega) from 1 μg of total RNA in the presence of oligo-dT primer. In order to reduce variations in RT reaction yields, each cDNA sample was derived from three independent RT reactions. For PCR reactions, 0.5 μL of cDNA was amplified in a final volume of 50 μL containing 5 μL of 10× PCR buffer, 0.5 μL of 2.5 mM dNTPs and 0.2 μL (5 U/μL) of Taq DNA polymerase (Promega), using 1 μL of a 10 mM solution for each of the gene-specific primers designed using the Amplify program (Wisconsin University, USA; Table S2). The oil palm elongation factor 1-alpha EgEF1-α1 (GenBank accession no. AY550990) was used as an internal control for each analysis. After optimisation of the number of amplification cycles, PCRs were performed at least twice and amplifications were checked by electrophoresis on 2% agarose gels. Finally, the amplified cDNA fragments were cloned and sequenced for identification.
Isolation of full-length cDNAs
In order to extend and complete the partial cDNA clones obtained from the SSH library, the SMART™ RACE cDNA amplification kit and Advantage® 2 PCR enzyme system (Clontech) were used according to the manufacturer recommendations. Of the total RNA, 1 μg was used as template for the synthesis of the first-strand cDNA. RACE amplifications were performed with gene-specific primers designed to generate the 5′ or 3′ cDNA fragments (Table S3). RACE products were cloned and sequenced for identification. The full-length cDNA sequences were assembled in silico and confirmed by end-to-end RT-PCR amplifications. The polypeptide sequence encoded by each full-length cDNA was deduced using the ‘Translate’ tool available on the ExPASy web site (http://www.expasy.org/tools/dna.html). Putative protein domains were identified by comparison with the Pfam database version 23.0 (http://pfam.sanger.ac.uk).
Results and discussion
Preparation, sequence analysis and functional annotation of ESTs from oil palm SSH libraries
In order to isolate genes which might be involved in determining the mantled phenotype, a SSH strategy was used for enriching the cDNA population for reverse transcripts of mRNAs differentially accumulated in normal and abnormal inflorescences. Male inflorescences were chosen as the source material since the phenotypic alterations observed are more drastic than those of female inflorescences (transformation of stamens to carpels as opposed to supernumary carpels). Forward and reverse subtractions were carried out between cDNAs from normal and mantled male inflorescences (clonal line FC166) and the corresponding SSH libraries INFSSHM1 and INFSSHM2 were generated. The studied inflorescences were from stage 3 (Adam et al. 2007), corresponding to the development of perianth organs and the initiation of reproductive organs. A total of 1,920 clones were sequenced, from which 1,594 valid ESTs were produced (Table 1). Of these, 717 and 877 valid EST sequences were obtained from the INFSSHM1 and INFSSHM2 libraries, respectively. Sequence comparisons were performed within each library, resulting in 597 unique sequences (77 contigs and 520 singletons) in library INFSSHM1 and 766 unigenes (60 contigs and 706 singletons) in library INFSSHM2 (Table 1). Sequence redundancy (ESTs in clusters/total ESTs) was 27% for INFSSHM1 and 19% for INFSSHM2, suggesting a higher degree of normalisation in the former library. The effectiveness of the cDNA subtraction was evaluated by comparing sequence data from the two libraries. A total of 1,350 unigenes, in the form of 1,204 singletons and 146 contigs, resulted from inter-library clustering analysis, demonstrating that the SSH strategy had generated a low redundancy overall (24.4%). Given that the whole genome sequence of Elaeis guineensis is not yet available, the data presented here will enrich the EST resources currently available (Jouannic et al. 2005; Ho et al. 2007; Low et al; 2008; Lin et al. 2009). Of the 145 clusters identified in and between the INFSSHM1 and INFSSHM2 libraries, 13 were derived from genes represented in both libraries (32 ESTs total) and constituted only 2% of the total number of sequences compared (Table 1). A large proportion of the clusters was specific to one library (50.3% and 40.7% of the total clusters for the libraries INFSSHM1 and INFSSHM2, respectively). This result reveals a dissimilar sequence composition of the two libraries and suggests that an enrichment for differentially expressed genes has occurred. Nevertheless, it should be noted that some of the cDNAs selected in this way could correspond to genes regulated independently of the mantled phenotype, for instance, by localized factors such as soil microenvironment.
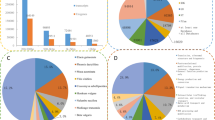
Subsequently, GO annotation was performed on each library set to assign functional GO terms to each unigene. During the first step, the unigene sets were compared to the NCBI nr database using the BLASTX program. For both libraries, approximately 70% of the sequences shared a significant similarity to existing sequences. After mapping, it proved possible to assign more than 50% of each gene set (323 sequences for INFSSHM1 and 397 for INFSSHM2) to slim plant GO terms. The number of unigenes classified within the three functional categories (cellular component, biological process and molecular function) was 197, 274 and 200, respectively, for the INFSSHM1 library and 245, 361 and 253, respectively, for the INFSSHM2 library, bearing in mind that a single sequence can be classified within more than one category (Table 2).
A comparison of GO term distributions within the two libraries did not reveal a statistically significant difference in relative abundances at the global level (Spearman R = 0.90, p < 0.005). However, a pairwise comparison of the number of ESTs in each GO class revealed a significant difference between the two distributions for two categories, namely, carbohydrate metabolic process (χ 2 = 5.64, p = 0.017) and lipid metabolic process (χ 2 = 4.33, p = 0.037), both of which contained about twice as many accessions in library INFSSHM2. This suggests that cellular activities of these two types are more intense in the mantled inflorescence. Overall, the GO annotation indicates differences in sequence composition between the forward and reverse libraries, but no major differences in functional categories.
Identification of genes differentially expressed between normal and mantled male inflorescence using macroarray hybridisation
A total of 32 EST clones hybridising to differentially accumulated transcripts were identified based on our selection criteria (probability value < 0.001, signal fold change ≥ 2) (Table 3); 16 of them gave a lower signal and the other 16 a higher signal in the mantled inflorescence. Of the 16 ESTs representing mantled down-regulated genes, only one (EGM21001C01) belonged to the reverse library (INFSSHM2). The 16 ESTs representing mantled up-regulated genes consisted of 4 ESTs from the INFSSHM1 library and 12 ESTs from the INFSSHM2 library. The library origins of the EST clones producing differential signals were thus in general agreement with the signal intensity ratios that they produced (corresponding to up- or down-regulation). The mantled vs. normal signal ratios ranged from 0.03 to 114. BLASTX analysis against the non-redundant protein database indicated that seven transcripts coded for proteins with either no significant similarity to database accessions or to proteins of unknown function (Table 3). For simplicity, we will refer to genes displaying a differential expression pattern between normal and mantled inflorescences as ‘marker genes’ hereafter in the text, bearing in mind that this designation refers to variations in mRNA accumulation and not to gene sequence.
Semi-quantitative PCR analysis of the differential gene expression
To confirm the differential expression patterns detected by macroarray analysis, 12 of the 32 candidate marker genes selected on the basis of their expression ratios (i.e. the most strongly differentially expressed) were subjected to semi-quantitative RT-PCR analysis. Primer pairs were designed for each candidate and tested on the total RNAs originally used for generating SSH libraries. Of the 12 transcripts, ten displayed differential accumulation of target transcripts (Table 3). However, for the two remaining candidate genes, represented by ESTs EGM21006D09 and EGM11002F9, no difference in transcript accumulation was observed. This is likely to be due to differences in gene specificity between the macroarray hybridisation and RT-PCR techniques. Nevertheless, the RT-PCR results overall showed a good validation rate (83%).
In order to evaluate the reliability of validated candidate genes, their expression was investigated on a second pair of normal and mantled male inflorescences of different genotypes. The genes which were validated on both pairs are shown in Fig. 1. For six of the ten initially validated candidate genes, the same differential expression pattern was observed with the second normal/mantled pair. For simplicity, these six genes were renamed according to their sequence similarities with the protein databases (see nomenclature in Table 3). The EgUK1 gene (short for UnKnown protein) was the only one for which transcript abundance was higher in the mantled inflorescence. The other genes, namely, EgUK2, EgPHOS1, EgFB1, EgRING1 and EgXG1, were more strongly expressed in normal inflorescence, as expected. For the four remaining candidate genes, the differential expression pattern observed in the clonal line FC166 was not confirmed with the other genotypes. In this case, it is presumed that other factors, such as environmental conditions, caused the original differential effect. Subsequent experiments therefore focused on the six candidate genes for which the differential expression was consistent for both genetic backgrounds.
Validation by semi-quantitative RT-PCR of the differential expression pattern of six candidate marker genes between normal (N) and mantled (M) male inflorescence of oil palm of two different genetic backgrounds A FC166, B FC1238/1292. The names of genes are indicated on the left (see Table 3). The EgEF1-α1 housekeeping gene was used as control
In order to determine whether the differential expression pattern of these six candidate genes is specific to developmental stage 3, transcript abundance was studied in normal and mantled male inflorescences at two other developmental stages (2 and 4) according to Adam et al. (2007). Figure 2a shows that the transcripts of EgPHOS1, EgFB1 and EgXG1 genes were only detected at stages 2 and 3 in normal inflorescence, whereas the EgUK2 and EgRING1 transcripts were detected at all stages. Overall, the differential expression patterns previously observed between normal and abnormal inflorescences were confirmed. Evidence for temporal shift in expression was noticed for some markers (EgUK2, EgPHOS1 and EgXG1). RT-PCR analysis of EgUK1, the only marker gene with higher expression in mantled material, revealed transcript accumulation at stages 3 and 4 in both phenotypes, with a slightly weaker signal in normal male inflorescence.
Analysis by semi-quantitative RT-PCR of the expression of candidate genes during early stages of normal and mantled male (a) and female (b) inflorescence development. Numbers correspond to different stages of inflorescence development according to Adam et al. (2007). Stage 2 corresponds to the initiation of perianth organs. Stage 3 corresponds to the development of perianth organs and the initiation of reproductive organs. Stage 4 corresponds to the development of reproductive organs. The names of genes are indicated on the left (see Table 3)
The expression patterns of the six candidate genes identified in the male inflorescence were further investigated in the female inflorescence (Fig. 2b). For EgPHOS1, EgFB1, EgRING1 and EgXG1, evidence of a temporal expression shift was observed between normal and mantled material. Intriguingly, the expression pattern of EgUK2 showed a weaker transcript accumulation at stage 3 compared to stages 2 and 4, both in normal and abnormal inflorescences. No difference in accumulation of EgUK1 transcripts was observed between normal and mantled material in the female inflorescence, in contrast to the situation observed in the male material. Overall, the expression patterns of the candidate genes were significantly different between male and female inflorescences.
To evaluate the tissue specificity of expression of the six candidate genes throughout the plant, further RT-PCR analyses were carried out on RNAs extracted from juvenile leaf, root and callus. The results (Fig. 3) revealed transcript accumulation in leaves and, to a lesser extent, in roots for all marker genes, with the exception of EgUK1. Transcripts were also detected in callus for EgFB1, EgXG1 and, more weakly, for EgUK2.
Our results indicate that the six studied genes are expressed at defined stages during reproductive development, with distinct expression patterns in male and female inflorescences and that their functions are not restricted to reproductive organs. Apart from EgUK1, stage-specific expression of candidate genes appears to be altered in the mantled phenotype of both sexes. The inflorescences are complex organs consisting of rachillae, bracts and flowers. Furthermore, the oil palm female flowers belong to a floral triad flanked by two abortive staminate flowers (Adam et al. 2005). Studies of the expression of the marker genes in excised flower tissues may help to shed light on this matter. The significance of the differential expression patterns of these genes in normal and mantled inflorescence development remains, for the moment, unclear.
Sequence and function analysis of candidate genes
EgRING1 and EgFB1 were selected for further study on the basis of strong differences of expression profiles between normal and mantled inflorescences.
During the SSH procedure, partial digestion of cDNAs with the restriction endonuclease RsaI results in truncation of reverse transcripts. Thus, in order to identify the complete protein sequences encoded by two of the differentially expressed genes, 5′ and 3′ RACE-PCR were carried out.
For EgRING1, the full-length cDNA obtained in this way was 961 bp in length (Genbank accession no. GQ914934) and encoded a predicted protein of 261 amino acid residues with a calculated molecular weight of 31.4 kDa and an isoelectric point of 6.6 (Fig. 4). EgRING1 belongs to the really interesting new gene (RING) protein family and encodes a protein sharing 41% and 37% identities with its putatives Arabidopsis and rice orthologues, respectively. A C3H2C3-type zinc–finger (RING finger) domain with a characteristic CX2CX9-39-CX1-3HX2-3HX2CX4-48CX2C signature (Joazeiro and Weissman 2000) was identified at the C-terminus of the protein. The RING domain contains four pairs of zinc ligands formed by cysteine and histidine residues, which bind two zinc ions in a cross-brace structure. The RING motif is often associated with coiled-coil domains or WD40 repeats which are involved in protein–protein interaction. None of the other domains previously identified in the members of the RING protein family could be detected in the EgRING1 protein sequence. It should be noted that in Arabidopsis, Stone et al. (2005) identified 469 RING domain-proteins, of which the majority contained only a RING domain. The RING domain has been proposed to be involved in the ubiquitination pathway by mediating the transfer of an ubiquitin moiety to the target protein which is then recognised by the 26S proteasome and degraded (Mazzucotelli et al. 2006). In this way, RING proteins play a crucial role in numerous biological processes including developmental, hormonal, abiotic and biotic stress responses. Specific functions have been attributed to some RING proteins, such as CIP8 and COP1 in photomorphogenesis (Hardtke et al. 2002; Osterlund et al. 2000), EL5 in rice root development (Koiwai et al. 2007), AIP2 in ABA signalling during seed development (Zhang et al. 2005), SHA1 in maintenance of shoot apical meristem (Sonoda et al. 2007) or SDIR1 in drought and salt stress responses through the ABA signalling pathway (Zhang et al. 2007).
Sequence alignment of the deduced EgRING1 amino sequence protein with close relatives from other plants including the following model species: Ricinus communis (EEF34882), Oryza sativa (ABF94555), Populus trichocarpa (XP002313898) and Arabidopsis thaliana (AAM64325). The position of the putative RING finger C3H2C3-type domain identified using Pfam database is indicated by a line above the sequence. Conservative substitutions at an amino acid position are shaded in gray and conserved residues in black. Asterisks indicate the conserved metal ligand Cys and His residues of the RING domain
For EgFB1, a full-length cDNA of 979 bp (Genbank accession no. GQ914933) was isolated by RACE-PCR, with an open-reading frame of 537 bp encoding a predicted protein of 178 amino acid residues with an estimated molecular mass of 19.4 kDa and a pI of 9.71 (Fig. 5). A database search revealed sequence similarities between this protein and members of the plant F-box protein family (47% and 42% identities shared with the putative rice and Arabidopsis orthologues, respectively). Figure 5 shows an alignment of the EgFB1 F-box protein along with those of closely related proteins in sorghum, rice, poplar and Arabidopsis. Several studies have indicated that the F-box domain which is about 50 amino acids length is not highly conserved (Kipreos and Pagano 2000). The EgFB1 protein contains an F-box domain that shares 68% identities with that of its rice relative (CAO94352) and 29% with the consensus F-box motif pfam00646 (http://pfam.sanger.ac.uk). Additional analysis of the deduced EgFB1 protein sequence did not reveal any other domains commonly found at the carboxy-terminus in this group (Jain et al. 2007). F-box proteins comprise a large family in a wide range of organisms. In plants, 964 potential F-box genes were identified in Arabidopsis and 687 in rice (Gagne et al. 2002; Jain et al. 2007). Most of these proteins form part of a SCF protein complex which is the major type of E3 protein ligase involved in the ubiquitin–proteasome pathway. The generic SCF complex consists of four major subunits, namely, homologues of CULLIN 1 (CUL1), suppressor of kinetochore protein 1 (SKP1), RING-box 1 (RBX1) and F-box protein (Ni et al. 2004; Ho et al. 2006). Within the complex, the F-box protein confers target specificity by recruiting the protein substrate. F-box proteins therefore play a crucial role in different aspect of plant regulation and development. For instance, the F-box proteins TIR1, GID2, COI1 are involved in plant hormone responses (Yu et al. 2007), FKF1 controls flowering (Sawa et al. 2007), ZTL is implicated in the circadian rhythm (Kim et al. 2007), OSDRF1 plays a role in disease resistance (Cao et al. 2008), SKP2A regulates cell division (Jurado et al. 2008) and MAX2 regulates shoot branching in Arabidopsis (Stirnberg et al. 2007). Interestingly, the F-box protein unusual floral organs (UFO) has previously been shown to be involved in floral development in Arabidopsis (Samach et al. 1999). Defects in UFO which acts upstream of the B-class homeotic genes (Coen and Meyerowitz 1991) result notably in homeotic changes in the second and third whorls that are reminiscent of the oil palm mantled phenotype (Levin and Meyerowitz 1995; Wilkinson and Haughn 1995). UFO positively regulates, in conjunction with the transcription factor LEAFY (LFY), the expression of the B-class organ identity gene, APETALA3 (AP3) by direct binding to the AP3 promoter (Samach et al. 1999; Ni et al. 2004; Chae et al. 2008). Likewise, in Anthirrhinum, a mutation of the gene FIMBRIATA (an orthologue of UFO gene) affects floral organ identity, consistent with a reduction in B and C genes activity (Ingram et al. 1997). To date, no clear orthologue of UFO has been identified in monocots.
Sequence alignment of the deduced EgFB1 amino sequence protein with close relatives from other plants including the following model species: Sorghum bicolor (EER99544), Oryza sativa (CAO9435), Populus trichocarpa (EEE6166), and Arabidopsis thaliana (AAO39934). The F-box domain, identified according to consensus sequence pFam00646 (http://pfam.sanger.ac.uk), is indicated by a line above the sequence. Conservative substitutions at an amino acid position are shaded in grey and conserved residues in black. The alignment was generated using the CLUSTALW program
Conclusion
Overall, our results have shown that the EgFB1 and EgRING1 genes, belonging to the F-box and RING families, respectively, are deregulated during the development of mantled inflorescences of both sexes. EgFB1 and EgRING1 encode proteins which could potentially act in the same signalling pathway although this can only be speculated at present given the lack of functional evidence. Nevertheless, it is interesting to note the similarity of the expression profiles observed for EgFB1 and EgRING1, both of which display reduced expression in the mantled inflorescence. Both genes belong to large multigene families, implying the existence of a large number of distinct signalling modules used for a wide variety of purposes in the plant (Vierstra 2009).
In earlier studies, oil palm MADS box genes of putative class B function were isolated and their expression was shown to decrease in the mantled flower (Adam et al. 2006, 2007). It remains to be determined how these genes are regulated in oil palm and why the tissue culture process induces a modification of their expression. The genes identified in the current study should contribute to the understanding of the molecular pathways affected in the mantled phenotype. One important aspect to be addressed is the link between the gene expression changes noted in this and previous studies (Adam et al. 2007) and the epigenetic nature of the mantled abnormality, the somaclonal variant being associated with a genome-wide DNA hypomethylation (Jaligot et al. 2000). The involvement of some components of the ubiquitin–proteasome system in the control of chromatin structure in plants was recently demonstrated (Vierstra 2009). Indeed, it has been established that the Arabidopsis protein ORTH2/VIM1, which has two RING domains, possesses an E3 ligase activity and is involved in determining DNA methylation status (Kraft et al. 2008). The knowledge gained from this work can help to determine the nature of the epigenetic deregulation occurring in mantled palms and provide a basis for the development of an early conformity test to identify and eliminate mantled individuals.
References
Adam H, Jouannic S, Escoute J, Duval Y, Verdeil JL, Tregear JW (2005) Reproductive developmental complexity in the African oil palm (Elaeis guineensis). Am J Bot 92:1836–1852
Adam H, Jouannic S, Morcillo F, Richaud F, Duval Y, Tregear JW (2006) MADS box genes in oil palm (Elaeis guineensis): patterns in the evolution of the squamosa, deficiens, globosa, agamous, and sepallata subfamilies. J Mol Evol 62:15–31
Adam H, Jouannic S, Orieux Y, Morcillo F, Richaud F, Duval Y, Tregear JW (2007) Functional characterization of MADS box genes involved in the determination of oil palm flower structure. J Exp Bot 58:1245–1259
Aharoni A, Keizer LC, Bouwmeester HJ, Sun Z, Alvarez-Huerta M et al (2000) Identification of the SAAT gene involved in strawberry flavor biogenesis by use of DNA microarrays. Plant Cell 12:647–662
Altschul SF et al (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Alwee SS, Van der Linden CG, Van der Schoot J, de Folter S, Angenent GC, Cheah SC, Smulders MJM (2006) Characterization of oil palm MADS box genes in relation to the mantled flower abnormality. Plant Cell Tissue Organ Cult 85:331–344
Cao Y, Yang Y, Zhang H, Li D, Zheng Z, Song F (2008) Overexpression of a rice defense-related F-box protein gene OsDRF1 in tobacco improves disease resistance through potentiation of defense gene expression. Physiol Plant 134:440–452
Chae E, Tan QK, Hill TA, Irish VF (2008) An Arabidopsis F-box protein acts as a transcriptional co-factor to regulate floral development. Development 135:1235–1245
Coen ES, Meyerowitz EM (1991) The war of the whorls: genetic interactions controlling flower development. Nature 353:31–37
Conesa A, Gotz S (2008) Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int J Plant Genomics 2008:619832
Corley RHV, Lee CH, Law IM, Wong CY (1986) Abnormal flower development in oil palm clones. Planter 62:233–240
Durand-Gasselin T, Le Guen VL, Konan E, Duval Y (1990) Oil palm (Elaeis guineensis Jacq.) plantations in Côte d’Ivoire obtained through in vitro culture-first results. Oléagineux 45:1–11
Gagne JM, Downes BP, Shiu SH, Durski AM, Vierstra RD (2002) The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc Natl Acad Sci USA 99:11519–11524
Hardtke CS, Okamoto H, Stoop-Myer C, Deng XW (2002) Biochemical evidence for ubiquitin ligase activity of the Arabidopsis COP1 interacting protein 8 (CIP8). Plant J 30:385–394
Ho MS, Tsai PI, Chien CT (2006) F-box proteins: the key to protein degradation. J Biomed Sci 13:181–191
Ho CL, Kwan YY, Choi MC, Tee SS, Ng WH et al (2007) Analysis and functional annotation of expressed sequence tags (ESTs) from multiple tissues of oil palm (Elaeis guineensis Jacq.). BMC Genomics 8:381
Hu W, Wang Y, Bowers C, Ma H (2003) Isolation, sequence analysis, and expression studies of florally expressed cDNAs in Arabidopsis. Plant Mol Biol 53:545–563
Ingram GC, Doyle S, Carpenter R, Schultz EA, Simon R, Coen ES (1997) Dual role for fimbriata in regulating floral homeotic genes and cell division in Antirrhinum. EMBO J 16:6521–6534
Jain M, Nijhawan A, Arora R, Agarwal P, Ray S et al (2007) F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol 143:1467–1483
Jaligot E, Rival A, Beulé T, Dussert S, Verdeil JL (2000) Somaclonal variation in oil palm (Elaeis guineensis Jacq.): the DNA methylation hypothesis. Plant Cell Rep 19:684–690
Jaligot E, Beulé T, Rival A (2002) Methylation-sensitive RFLPs: characterisation of two oil palm markers showing somaclonal variation-associated polymorphism. Theor Appl Genet 104:1263–1269
Jaligot E, Beulé T, Baurens FC, Billotte N, Rival A (2004) Search for methylation-sensitive amplification polymorphisms associated with the mantled variant phenotype in oil palm (Elaeis guineensis Jacq). Genome 47:224–228
Joazeiro CA, Weissman AM (2000) Ring finger proteins: mediators of ubiquitin ligase activity. Cell 102:549–552
Jouannic S, Argout X, Lechauve F, Fizames C, Borgel A et al (2005) Analysis of expressed sequence tags from oil palm (Elaeis guineensis). FEBS Lett 579:2709–2714
Jurado S, Diaz-Trivino S, Abraham Z, Manzano C, Gutierrez C, del Pozo C (2008) SKP2A, an F-box protein that regulates cell division, is degraded via the ubiquitin pathway. Plant J 53:828–841
Kim WY, Fujiwara S, Suh SS, Kim J, Kim Y et al (2007) Zeitlupe is a circadian photoreceptor stabilized by gigantea in blue light. Nature 449:356–360
Kim C, Lemke C, Paterson AH (2009) Functional dissection of drought-responsive gene expression patterns in Cynodon dactylon L. Plant Mol Biol 70:1–16
Kipreos ET, Pagano M (2000) The F-box protein family. Genome Biol 1: REVIEWS3002
Koiwai H, Tagiri A, Katoh S, Katoh E, Ichikawa H et al (2007) RING-H2 type ubiquitin ligase EL5 is involved in root development through the maintenance of cell viability in rice. Plant J 51:92–104
Kraft E, Bostick M, Jacobsen SE, Callis J (2008) ORTH/VIM proteins that regulate DNA methylation are functional ubiquitin E3 ligases. Plant J 56:704–715
Kubis SE, Castilho AM, Vershinin AV, Heslop-Harrison JS (2003) Retroelements, transposons and methylation status in the genome of oil palm (Elaeis guineensis) and the relationship to somaclonal variation. Plant Mol Biol 52:69–79
Laitinen RA, Pollanen E, Teeri TH, Elomaa P, Kotilainen M (2007) Transcriptional analysis of petal organogenesis in Gerbera hybrida. Planta 226:347–360
Levin JZ, Meyerowitz EM (1995) UFO: an Arabidopsis gene involved in both floral meristem and floral organ development. Plant Cell 7:529–548
Lin HC, Morcillo F, Dussert S, Tranchant-Dubreuil C, Tregear JW, Tranbarger TJ (2009) Transcriptome analysis during somatic embryogenesis of the tropical monocot Elaeis guineensis: evidence for conserved gene functions in early development. Plant Mol Biol 70:173–192
Low ET, Alias H, Boon SH, Shariff EM, Tan CY et al (2008) Oil palm (Elaeis guineensis Jacq.) tissue culture ESTs: identifying genes associated with callogenesis and embryogenesis. BMC Plant Biol 8:62
Matthes M, Singh R, Cheah SC, Karp A (2001) Variation in oil palm (Elaeis guineensis Jacq.) tissue culture-derived regenerants revealed by AFLPs with methylation-sensitive enzymes. Theor Appl Genet 102:971–979
Mazzucotelli E, Belloni S, Marone D, De Leonardis A, Guerra D et al (2006) The e3 ubiquitin ligase gene family in plants: regulation by degradation. Curr Genomics 7:509–522
Morcillo F, Gagneur C, Adam H, Richaud F, Singh R et al (2006) Somaclonal variation in micropropagated oil palm. Characterization of two novel genes with enhanced expression in epigenetically abnormal cell lines and in response to auxin. Tree Physiol 26:585–594
Ni W, Xie D, Hobbie L, Feng B, Zhao D et al (2004) Regulation of flower development in Arabidopsis by SCF complexes. Plant Physiol 134:1574–1585
Nogueira FT, De Rosa VE, Jr MM, Ulian EC, Arruda P (2003) RNA expression profiles and data mining of sugarcane response to low temperature. Plant Physiol 132:1811–1824
Osterlund MT, Hardtke CS, Wei N, Deng XW (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405:462–466
Pannetier C, Arthuis P, Lievoux D (1981) Neoformation of young Elaeis guineensis plants from primary calluses obtained on leaf fragments in vitro. Oleagineux 36:119–122
Pertea G, Huang X, Liang F, Antonescu V, Sultana R, Karamycheva S, Lee Y, White J, Cheung F, Parvizi B et al (2003) TIGR gene indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics 19:651–652
Peschke VM, Phillips RL, Bengenbach BG (1987) Discovery of transposable element activity among progeny of tissue culture-derived maize plants. Science 228:804–807
Poncet V, Rondeau M, Tranchant C, Cayrel A, Hamon S et al (2006) SSR mining in coffee tree EST databases: potential use of EST-SSRs as markers for the Coffea genus. Mol Genet Genomics 276:436–449
Rao V, Donough CR (1990) Preliminary evidence of genetic cause for the floral abnormalities in some oil palm ramets. Elaeis 2:199–207
Rival A (2007) The oil palm. In: Pua EC, Davey MR (eds) Biotechnology in agriculture and forestry, vol 61. Transgenic crops VI. Springer-Verlag, Berlin, pp 59–80
Rival A, Beulé T, Barre P, Hamon S, Duval Y, Noirot M (1997) Comparative flow cytometric estimation of nuclear DNA content in oil palm (Elaeis guineensis Jacq) tissue cultures and seed-derived plants. Plant Cell Rep 16:884–887
Rival A, Bertrand L, Beulé T, Combes MC, Trouslot P, Lashermes P (1998) Suitability of RAPD analysis for the detection of somaclonal variants in oil palm (Elaeis guineensis Jacq). Plant Breed 117:73–76
Rival A, Jaligot E, Beulé T, Finnegan EJ (2008) Isolation and expression analysis of genes encoding MET, CMT, and DRM methyltransferases in oil palm (Elaeis guineensis Jacq.) in relation to the ‘mantled’ somaclonal variation. J Exp Bot 59:3271–3281
Samach A, Klenz JE, Kohalmi SE, Risseeuw E, Haughn GW, Crosby WL (1999) The unusual floral organs gene of Arabidopsis thaliana is an F-box protein required for normal patterning and growth in the floral meristem. Plant J 20:433–445
Sawa M, Nusinow DA, Kay SA, Imaizumi T (2007) FKF1 and gigantea complex formation is required for day-length measurement in Arabidopsis. Science 318:261–265
Sonoda Y, Yao SG, Sako K, Sato T, Kato W et al (2007) SHA1, a novel RING finger protein, functions in shoot apical meristem maintenance in Arabidopsis. Plant J 50:586–596
Stirnberg P, Furner IJ, Ottoline Leyser HM (2007) MAX2 participates in an SCF complex which acts locally at the node to suppress shoot branching. Plant J 50:80–94
Stone SL, Hauksdottir H, Troy A, Herschleb J, Kraft E, Callis J (2005) Functional analysis of the ring-type ubiquitin ligase family of Arabidopsis. Plant Physiol 137:13–30
Tamaoki M, Matsuyama T, Nakajima N, Aono M, Kubo A, Saji H (2004) A method for diagnosis of plant environmental stresses by gene expression profiling using a cDNA macroarray. Environ Pollut 131:137–145
Tregear JW, Morcillo F, Richaud F, Berger A, Singh R et al (2002) Characterization of a defensin gene expressed in oil palm inflorescences: induction during tissue culture and possible association with epigenetic somaclonal variation events. J Exp Bot 53:1387–1396
Vierstra RD (2009) The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 10:385–397
Wang XJ, Cao XL, Hong Y (2005) Isolation and characterization of flower-specific transcripts in Acacia mangium. Tree Physiol 25:167–178
Wilkinson MD, Haughn GW (1995) Unusual floral organs controls meristem identity and organ primordia fate in Arabidopsis. Plant Cell 7:1485–1499
Yu H, Wu J, Xu N, Peng M (2007) Roles of F-box proteins in plant hormone responses. Acta Biochim Biophys Sin (Shanghai) 39:915–922
Zhang X, Garreton V, Chua NH (2005) The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes Dev 19:1532–1543
Zhang Y, Yang C, Li Y, Zheng N, Chen H et al (2007) SDIR1 is a RING finger E3 ligase that positively regulates stress-responsive abscisic acid signaling in Arabidopsis. Plant Cell 19:1912–1929
Zhang JZ, Li ZM, Yao JL, Hu CG (2009) Identification of flowering-related genes between early flowering trifoliate orange mutant and wild-type trifoliate orange (Poncirus trifoliata L. Raf.) by suppression subtraction hybridization (SSH) and macroarray. Gene 430:95–104
Acknowledgements
The authors are grateful to colleagues at Felda Agricultural Services (FASSB) for their valuable help, to Dr Helene Adam for the supply of inflorescence RNA and expert advice on inflorescence development, to Ivanna Fuentes for skilled technical support and to Myriam Collin for excellent assistance in histology and microscopy studies. We thank Drs. Alain Rival and Fabienne Morcillo for the critical reading of the manuscript. This research was supported by institutional funding from IRD and CIRAD.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J. Dean
Rights and permissions
About this article
Cite this article
Beulé, T., Camps, C., Debiesse, S. et al. Transcriptome analysis reveals differentially expressed genes associated with the mantled homeotic flowering abnormality in oil palm (Elaeis guineensis). Tree Genetics & Genomes 7, 169–182 (2011). https://doi.org/10.1007/s11295-010-0323-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-010-0323-9