Abstract
Rosy apple aphid (Dysaphis plantaginea), is one of the major insect pests of apple, causing serious physical and economic damage to fruit production. A dominant resistance gene Dp-fl was previously mapped at the bottom of linkage group LG8 from the cultivar ‘Florina’, linked to the SSR CH01h10. The development of additional genetic markers mapping closer to Dp-fl was needed to position the gene accurately and to improve the effectiveness of marker-assisted breeding (MAB). The aims of this study were to identify single nucleotide polymorphisms (SNPs) in the region of Dp-fl and to position these SNPs relative to Dp-fl. To generate a fine map of the Dp-fl interval, a total of 191 plants segregating for resistance and derived from four different populations were tested with temperature-switch PCR (TSP) markers developed for SNPs located in the region of CH01h10. All the plants were phenotypically evaluated for aphid resistance and those data compared with the genetic data. These efforts resulted in positioning the Dp-fl resistance locus in a genetic interval corresponding to a physical distance of about 330 kb on the ‘Golden Delicious’ genome. The new markers were tested on several apple founder cultivars in order to test the specificity of the SNPs and, thus, the best markers for the MAB were identified. Finally, the 330-kb interval was analyzed for the identification of coding sequences and putative candidate genes for D. plantaginea resistance were identified.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rosy apple aphid (RAA), Dysaphis plantaginea (Passerini) (Hemiptera: Aphididae), one of the aphid species regularly infesting apple trees (Malus × domestica), is considered one of the major insect pests of apple orchards in Europe and North America (Brown and Mathews 2007; Miñarro and Dapena 2008; Brown and Myers 2010; Parisi et al. 2013). RAA causes shoot distortion and leaf rolling, and in the case of severe infestation, the fruit remains smaller and become deformed, hence yields and its commercial value are reduced (De Berardinis et al. 1994). Control of RAA relies mainly on pesticide sprays since naturally occurring antagonists are not effective enough to control aphid populations (Miñarro et al. 2005; Brown and Mathews 2007). However, new alternative strategies to control RAA should be adopted for a more sustainable apple production. Cultivar resistance may be a good alternative strategy to manage RAA, considering that significant differences among cultivars in aphid abundance and damage level have already been well documented (Angeli and Simoni 2006; Arnaoudov and Kutinkova 2006; Miñarro and Dapena 2007, 2008; Razmjou et al. 2014). The presence of single dominant resistance (R) genes controlling aphid resistances in fruits and other crops, such as cereals and vegetables, has been reported. This process may involve the gene-for-gene recognition of aphid-derived elicitors by plant resistance genes (Smith and Boyko 2007). The cloning and identification of some aphid R genes in different plant species support this hypothesis. For instance, Rossi et al. (1998) found that the nematode resistance gene Mi of tomato provides resistance to the potato aphid, Macrosiphum euphorbiae (Thomas). Mi is a member of the nucleotide-binding site and leucine-rich region (NBS-LRR) class II family, and a model for Mi interaction with elicitors from the aphid was proposed by Kaloshian (2004). Several NBS-LRR sequences putatively involved in aphid resistance have been cloned and mapped in barley, wheat, sorghum, melon, and Medicago truncatula, too (Smith and Boyko 2007). Sequences from M. × domestica with a significant degree of similarity to NBS-LRR genes were also retrieved during the development of the BAC contig spanning the region of Dysaphis devecta (Walker) resistance locus Sd-1 (Cevik and King 2002).
Different parameters describe the way a plant resists to insects, such as antixenosis, antibiosis, and tolerance. Antixenosis deters or reduces colonization by insects, whereas antibiosis causes adverse effects on insect developmental traits. Tolerance is the ability of the plant to grow with little or no damage despite infestation (Hesler and Tharp 2005). Apple accessions may differ in their characteristics associated with RAA resistance, as has been described for woolly aphid resistance (Sandanayaka et al. 2003, 2005). ‘Florina’ is one of the best known cultivars resistant to RAA. It is considered tolerant to the aphid because it does not show the typical foliar or shoot deformations caused by RAA, despite possible small aphid development. ‘Florina’ RAA resistance has also been classified as antibiosis since RAA is less fecund and has a high mortality after feeding on ‘Florina’ leaves (Rat-Morris 1993). In addition, Marchetti et al. (2009) demonstrated that aphids on ‘Florina’ need a longer period before the first probe and do not show signs of complete phloem sap ingestion, proving that surface features and phloem factors are involved in the cultivar resistance, which may in turn indicate antixenosis mechanisms (Hesler and Tharp 2005).
In a preliminary study, ‘Florina’ RAA resistance was shown to be under the control of a single resistance gene, named Dp-fl, located at the distal end of linkage group (LG) 8, at about 6 cM below the SSR marker CH01h10 (Charles-Eric Durel, unpublished data). The high heritability of RAA resistance (Rat-Morris and Lespinasse 1995; Miñarro and Dapena 2004, 2008) based on its simple dominant genetic determinism was confirmed by phenotypic and genotypic evaluations on several ‘Florina’ progenies, in which the genetic linkage between Dp-fl and the SSR marker CH01h10 was re-confirmed, too (Dapena et al. 2009). Thus, ‘Florina’ is considered a good parent for the transmission of this resistance to D. plantaginea, which was inherited from its ancestor, the wild apple genotype Malus floribunda 821. Some scab-resistant cultivars derived from this accession and its derivative F2-26829-2-2, such as ‘Galarina’, ‘Golden Orange’, ‘GoldRush’, and ‘Liberty’, showed resistance to RAA, whereas others, for example, ‘Ariane’, ‘Redfree’, and ‘Topaz’, do not. Many other cultivars have been classified as susceptible, such as ‘Golden Delicious’ and ‘Melrose’ as well as various local cultivars, such as ‘De la Riega’, ‘Perico’, and ‘Raxao’ (Angeli and Simoni 2006; Arnaoudov and Kutinkova 2006; Miñarro and Dapena 2007, 2008). Further sources of RAA resistance have also been described in other genetic backgrounds and a Malus robusta derivative was shown to carry a single dominant gene for hypersensitivity to RAA, the gene Smh (Alston and Briggs 1970), but to date, this gene has not been mapped. Furthermore, a QTL for RAA resistance was reported on LG17 of ‘Fiesta’ (Stoeckli et al. 2008). Two major genes for woolly aphid resistance on LG8 have also been reported, Er1 and Er3, but were mapped closer to the opposite end of the chromosome (Bus et al. 2008).
The phenotypic evaluation for RAA resistance is time-consuming and expensive in terms of money and labor costs. Other ways of selecting resistant genetic material are therefore needed, and the use of molecular markers is a good option, as already proposed by Bus et al. (2008) with markers for three major resistance genes to woolly apple aphid. To date, CH01h10 has been a useful tool for the screening of Dp-fl resistance (Dapena et al. 2009), but the identification of new markers more closely linked to the gene will be crucial for more efficient marker-assisted breeding (MAB) for resistance. SNPs are good candidates for marker development, as they constitute the most common source of DNA sequence variations found in genomes of most organisms, including apple. Moreover, they are found in both non-coding and coding regions, thus rendering them effective markers for mapping genes (Newcomb et al. 2006; Chagné et al. 2008; Jänsch et al. 2015). Finally, the availability of three high-density SNP arrays greatly facilitates genome analyses in M. × domestica (Chagné et al. 2012; Bianco et al. 2014, 2016).
Thus, the overall goals of our work were to construct a fine map of the Dp-fl region on ‘Florina’ LG8 through the development of specific and reliable SNP markers tightly linked to the resistance locus and to select the most effective markers for the implementation of MAB screening for D. plantaginea resistance. Finally, a search for candidate genes in the Dp-fl region was performed on the apple genome sequence (Velasco et al. 2010).
Materials and methods
Plant material and genomic DNA extraction
A panel of 191 plants derived from four progenies of ‘Florina’ was included in this work for the fine mapping of the region of interest. The resistant cultivar ‘Florina’ was either used as the female parent with the American cultivar ‘Melrose’, or as the male parent with the three local cider cultivars ‘Perico’, ‘Raxao’, and ‘De la Riega’ (Rat-Morris and Lespinasse 1995; Miñarro and Dapena 2004), resulting in 28, 79, 41, and 43 seedlings, respectively.
A panel of seven apple “founder” cultivars was analyzed to evaluate the specificity of the newly identified SNP markers: ‘Braeburn’, ‘Cox’s Orange Pippin’, ‘Delicious’, ‘Golden Delicious’, ‘Granny Smith’, ‘Jonathan’, and ‘McIntosh’ (Durel et al. 1998; Salvi et al. 2014). Furthermore, they were tested on the progenitor M. floribunda 821, on its derivative F2-26829-2-2 from which ‘Florina’ has been bred, and on ‘GoldRush’, another known resistant cultivar. Another 25 apple accessions were also analyzed: four resistant to RAA, four susceptible, and 17 of unknown reaction. Among these, 21 directly derive from M. floribunda 821 and were selected for the Rvi6 (Vf) gene for resistance to apple scab (Venturia inaequalis) (Online Resource 1).
For all the samples, total genomic DNA was isolated from young leaf tissues, using the Qiagen DNeasy Plant mini kit (Qiagen) following the manufacturer’s protocol. DNA quantity and quality were measured spectrophotometrically with a Nanodrop ND-8000® (Thermo Scientific, USA).
Phenotypic evaluation
The response to RAA was evaluated on apple plants after infestation with aphids in two different locations: the three crossing populations ‘Raxao’ × ‘Florina’, ‘Perico’ × ‘Florina’, and ‘De la Riega’ × ‘Florina’ were evaluated in a greenhouse at SERIDA, Villaviciosa, Asturias, Spain, and the population ‘Florina’ × ‘Melrose’ was evaluated both in a greenhouse (until their shoot was 50–80 cm high) and in an unsprayed orchard at INRA, Angers, France.
At SERIDA, the number of replicates per individual seedling varied from three to eight depending on the cross. The same numbers of plants of ‘Florina’ and ‘Golden Delicious’ were used as resistant and susceptible controls, respectively. Plants were grafted on M.7 rootstocks and kept outdoors in 4-L pots. Plants were fertilized with 8 g of Osmocote plus. In mid-June, when new shoots were about 20 cm, plants were introduced into the greenhouse and randomly distributed. They were irrigated three times a week and maintained at a temperature below 22 °C using an automatically regulated cooling system. Plants were infested when new shoots were about 30–60 cm, and aphid movement from one plant to another was prevented by putting the pots in dishes filled with water. Secondary shoots were periodically pruned to keep the aphids on the principal stem. There were no overlapping branches.
Methodology at INRA was similar with same greenhouse conditions, but the number of replicates was two or three. Ten replicates were used in the case of the resistant (‘Florina’) and susceptible (‘Melrose’) controls. The plants were infested for two consecutive years, twice each year. Plants were then transferred into an experimental orchard, and field assessments were carried out for two consecutive years following natural infestation.
Aphids
Infestation was performed by placing young adult apterous virginiparous females on the upper side of the first leaf beneath the terminal cluster with a paint brush. At SERIDA, aphids were collected in the field from different apple cultivars to capture some of the natural variability. Individuals from each cultivar were reared separately on susceptible apple plants. Thus, several distinct populations of RAA were maintained in the laboratory. Four adults, each from a different population, were placed on each plant. Re-infestation was performed when necessary during the first 4 days to ensure aphid settlement. At INRA, aphids derived from a clonal aphid line of RAA obtained from one founder collected in the field and raised on grafted ‘Golden Delicious’ plants were used. Five adults were placed on each plant. Plants were considered as infested and the experiment as successful when a second generation was produced, i.e., more than five aphids were observed on the plants.
Damage assessment
The plants were inspected regularly for damage and scored 21 days after the placement of the aphids for tolerance (level of leaf distortion) and antibiosis (number of aphids on leaves). A clear relationship between antibiosis and tolerance was observed in the progenies although some tolerant plants developed aphid populations (75 % of tolerant plants were infested by fewer than five aphids while 79 % of the susceptible individuals were infested by more than five aphids or a colony, data not shown). Therefore, the leaf roll reaction was chosen as the criterion for damage assessment. The following scoring scale was used for tolerance: 0 = no leaf distortion; 1 = leaf very slightly curled; 2 = leaf slightly curled; and 3 = typical leaf roll (i.e., susceptible). The most distorted leaf was used for classifying the plant, and any plant showing just one distorted leaf was classified as susceptible. Each genotype was classified with the highest damage level recorded for any of the replicates (e.g., for a given genotype, if only one of the replicates showed typical leaf roll symptoms, the genotype was scored as 3 and considered susceptible). Because the scoring was performed independently by different people and at different times at SERIDA and INRA, class 2 was not scored exactly in the same way at both places. Consequently, at SERIDA, plants exhibiting shoot damage classes of 0, 1, or 2 were considered tolerant and class 3 susceptible, whereas at INRA, plants scored as classes 0 and 1 were considered tolerant and plants scored as classes 2 and 3 were considered susceptible both in the greenhouse and in the orchard. Regarding antibiosis, the scale used at SERIDA was described by six indices: 0 = no aphids; 1 = 1 to 5 aphids; 2 = 6 to 25 aphids; 3 = 26 to 125 aphids; 4 = 126 to 625 aphids; and 5 = more than 625 aphids. At INRA, the scale was described by four indices: 0 = no aphids; 1 = 1 to 5 aphids; 2 = more than 5 aphids, no reproduction; and 3 = aphid colony.
SSR genotyping and Dp-fl mapping
Four apple SSRs spanning LG8 already available from the literature and heterozygous in the resistant parent ‘Florina’ were chosen to create the backbone of the genetic map of the chromosome: CH01h10, CH01c06, Ch01f09, and C13470 (http://www.hidras.unimi.it; Liebhard et al. 2002; Wang et al. 2012) in the largest progeny, ‘Perico’ × ‘Florina’. The SSR primers were synthesized with a generic non-complementary nucleotide sequence at their 5′-end, with the forward and reverse primer for each marker synthesized with the nucleotide sequence 5′-ACGACGTTGTAAAA-3′ and 5′-CATTAAGTTCCCATTA-3′, respectively (Hayden et al. 2008). Two generic tag primers, tagF and tagR with the sequences 5′-ACGACGTTGTAAAA-3′ and 5′-CATTAAGTTCCCATTA-3′, respectively, were also synthesized. The tagF primer was labelled at its 5′-end with either the VIC or the FAM fluorescent dye (Applied Biosystems, Vienna, Austria). The amplification of SSR markers was performed using the PCR protocol as described by Hayden et al. (2008), and the thermal cycling conditions were the same as those reported by Liang et al. (2015).
Single amplification products (1 μL) of each SSR were multiplexed post-PCR with 0.25 μL of internal lane standard, Gene ScanTM–500 LIZ® Size Standard and 9.75 μL of Hi-Di formamide (Applied Biosystems). Separation was performed using a 3730 DNA Analyzer (Applied Biosystems). Data analysis was carried out using the Peak Scanner™ Software v1 (Applied Biosystems).
For the resistance gene mapping, the RAA susceptible plants were classified as recessive (aa), while the resistant plants were coded as heterozygous for the presence of the resistance gene (Aa). The genetic map of LG8 was calculated using the JoinMap 4.1 software (Van Ooijen 2006).
SNPs identification in the Dp-fl region and TSP marker development
First, the physical position of SSR CH01h10 was determined through the blast tool on ‘Golden Delicious’ genome (http://www.rosaceae.org/tools/ncbi_blast; Velasco et al. 2010). Then, in order to identify SNPs linked to the Dp-fl gene, the 8K and 20K apple Infinium® SNP chips (Chagné et al. 2012; Bianco et al. 2014) were exploited. The parents ‘Florina’ and ‘Melrose’, as well as M. floribunda 821 and its derivative F2-26829-2-2, were genotyped with the two Illumina arrays following the standard Illumina protocol (www.illumina.com). The SNP data were analyzed using GenomeStudio Data Analysis software (Illumina Inc.) with a GenCall threshold of 0.15. All the SNPs with GenTrain score >0.6 in the region of the CH01h10 physical position were checked and the SNPs heterozygous in ‘Florina’, M. floribunda 821 and F2-26829-2-2, and homozygous in ‘Melrose’ were selected. The homozygous condition in ‘Melrose’ was preferred for the SNP to be more informatory.
For each SNP marker selected as described above, the temperature-switch PCR (TSP) assay was designed to perform the genotyping in the parents and segregating progenies, as described by Hayden et al. (2009) and Tabone et al. (2009). All the primers were designed using Primer3 Software (www.bioinformatics.nl/primer3plus). Primer sequences and PCR product sizes are provided in Online Resource 2. Some technical details were modified from the protocol of Hayden et al. (2009) and Tabone et al. (2009), including standard PCR reagents and the addition of 50 ng diluted DNA instead of 20 ng dessicated DNA. PCR assays were performed using 1 U Taq DNA Polymerase (Thermo Fisher Scientific), 5 ng of bovine serum albumin Fraction V (Thermo Fisher Scientific), 1× reaction buffer, 1.5 mM MgCl2, 0.2 mM dNTPs (Sigma), 0.1 μM each of forward and reverse LS primer, 0.5 μM of forward/reverse AS primer, and 50 ng genomic DNA in a total reaction volume of 10 μl. PCRs were performed in an Applied Biosystems 2720 thermal cycler. After the test with standard TSP thermal cycling program, specific optimization of PCR conditions was needed for each SNP assay in order to ensure accurate and clear SNP genotyping. In particular, the number of cycles and the Tm were specifically adjusted (Online Resource 3). The PCR products were visualized on an Image Station 440 CF (Kodak, Rochester, NY, USA) after electrophoresis on 1.5 % (w/v) agarose gels and GelRed™ (Biotium) in precast.
Once the best TSP amplification system was set up for each assay, the parents and the four crossing populations were tested in order to detect: (i) which SNPs were informative (‘Aaxaa’ or ‘AaxAa’) and (ii) whether the seedlings did or did not have the allele of ‘Florina’ at the analyzed locus in each population. The graphical genotyping approach was used in order to visualize the genotype of individuals to detect the recombinant plants easily (Young and Tanksley 1989). This approach allowed the identification of the markers delimiting the Dp-fl gene region.
Identification of candidate genes in the Dp-fl interval region
As long as the Dp-fl interval region was well defined, the putative CDSs included in it were traced back from the ‘Golden Delicious’ genome sequence, version 1.0 (Velasco et al. 2010). The nucleotide sequences of these CDS, their physical position, and their biological function descriptions were retrieved from M. × domestica Gbrowse (http://www.rosaceae.org/) and NCBI database (http://www.ncbi.nlm.nih.gov/).
Results and discussion
Segregation for resistance and Dp-fl mapping
The results observed on ‘Florina’ (0 for tolerance to RAA, no damage, and 2 for antibiosis), ‘Melrose’ (classified 3 for tolerance and 3 for antibiosis, i.e., typical leaf roll and thriving colonies), and their progeny showed a clear relationship between antibiosis and tolerance although a little proportion of tolerant plants developed aphid populations like susceptible plants (i.e., they do not show antibiosis). These plants did not show damages, and therefore, it is possible to conclude that the damage in tolerant plants is independent from the number of aphids. Tolerance, our main criterion for resistance, is considered the most important from an agronomic and economic point of view. Moreover, the field tests conducted in the population ‘Florina’ × ‘Melrose’ at INRA showed a good correlation between results of greenhouse and field tests (data not shown).
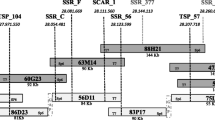
The observed RAA resistance segregations fit the 1:1 model in all the progenies except the ‘Perico’ × ‘Florina’ one (Table 1). The skewing of the segregation towards susceptibility of this progeny had already been partially reported by Miñarro and Dapena (2004). In order to create the backbone of the genetic map of LG8 of ‘Florina’, four SSRs spanning the whole LG8 were tested on the ‘Perico’ × ‘Florina’ progeny (Table 2 and Fig. 1). The resulting map confirmed the previously reported position of the Dp-fl gene at the bottom of LG8 (Charles-Eric Durel, unpublished data). It mapped between the SSR markers CH01h010 and C13470, at 4.3 and 1.2 cM, respectively. The two SSRs located at the top of the linkage group (CH01c06 and CH01f09) showed a 1:1 segregation, while the Dp-fl flanking SSRs (Ch01h10 and C13470) were distorted. Therefore, we can conclude that the distortion in the ‘Perico’ × ‘Florina’ progeny is more likely related to a general skewness of the whole Dp-fl region instead of a different inheritance model. Segregation distortions are common in apple and, i.e., they were reported also for the two major genes for woolly aphid resistance, Er1 and Er3, on LG8 (Bus et al. 2008).
Fine mapping of the Dp-fl locus
The genotyping of the parents ‘Florina’ and ‘Melrose’ with the 8K and 20K Illumina SNP chip resulted in the identification of 16 SNPs in the region of SSR CH01h10, according to their physical position on the ‘Golden Delicious’ genome. These 16 SNPs were distributed in a region of about 6 million bp at the bottom of LG8. They were chosen because of their heterozygous state in ‘Florina’ to fit with its R/S heterozygous state for Dp-fl. Thus, depending on the genotype of the susceptible parent at each locus, two types of segregation were expected: ‘Aaxaa’ and ‘AaxAa’. Initially, TSP assays were developed for the 16 SNPs in order to obtain an efficient and specific method of SNP genotyping. TSP provides several advances, including simplified assay design, being cheap and easy-to-use because they are based on endpoint PCR and the detection of polymorphisms on agarose gel (Hayden et al. 2009). Then, the TSP assays were tested on the parents and the protocol was specifically optimized for each SNP (Online Resources 2 and 3). Among the 16 SNPs, 10 were further selected because of their homozygous state in at least one of the susceptible parents, giving fully informative segregation (‘Aaxaa’, Table 3). The remaining six TSP markers were discarded because they did not perform well and were not accurately scored on agarose gel. Finally, the 10 selected TSPs were tested on the four populations. The mapping of these TSP markers confirmed their positions in the region of SSR CH01h10 with all of them linked with each other and showed that the genetic position of the SNPs was in full agreement with the physical order in the ‘Golden Delicious’ genome sequence (Table 3 and Fig. 2).
Graphical genotyping of the Dp-fl region in the apple cultivar ‘Florina’. Molecular and phenotypic data are reported for the recombinant plants of the four populations: PF ‘Perico’ × ‘Florina’, FM ‘Florina’ × ‘Melrose’, RF ‘Raxao’ × ‘Florina’, and DR ‘De la Riega’ × ‘Florina’. The white letters are used for the SNPs segregating in an ‘AaxAa’ pattern in some of the progenies (not fully informative meiosis). This indicates that the genotype was inferred from the surrounding samples; the “a-” is used when it is not possible to deduce the full genotype from the scoring. R resistant, S susceptible
The Dp-fl resistance locus could be easily integrated into the SSR/SNPs linkage map of the distal part of ‘Florina’ LG8 thanks to the comparison of molecular and phenotypic data for RAA resistance in the four segregating populations. A total of 14 plants showing recombination in the Dp-fl region were identified, and therefore, the Dp-fl gene was fine mapped in a small genetic window (Fig. 2). The graphical genotypes of two recombinants (FM_F145 from ‘Florina’ × ‘Melrose’ and RF_X-9104-8 from ‘Raxao’ × ‘Florina’) made it possible to identify the borders of the Dp-fl window between SNP_104 and SNP_585, considerably narrowing down the physical interval to about 330 Kb.
As reported in Fig. 2, three SNPs (SNP_205, SNP_783, and SNP_585) exhibited a fully informative segregation pattern (‘Aaxaa’) in all the populations. For the other SNPs, an ‘AaxAa’ segregation prevented assignment of the exact genotype to all seedlings because only the ‘aa’ genotypes could be unambiguously assigned. For almost all of these plants, the genotype could be imputed according to the scoring of the surrounding markers. Only a few genotypes remained uncertain because they were directly flanking a recombination event, as was the case for PF_X-9504-07 with markers SNP_494 and SNP_104, and RF_89-13 and DF_X-9401-4 with marker SNP_494.
Molecular markers for MAB
To evaluate the applicability of the newly identified SNP markers in marker-assisted breeding (MAB) for the RAA resistance in apple, they were tested on seven apple breeding founder varieties and on the ‘Florina’ progenitor M. floribunda 821 and its direct derivative F2-26829-2-2 (Table 4).
Besides ‘Florina’, the whole Dp-fl haplotype was also present in M. floribunda 821 and F2-26829-2-2, which is consistent with the expected strong linkage disequilibrium in a genomic region introgressed from a wild progenitor into a cultivated species. The alleles linked to resistance of three SNPs (SNP_585, SNP_205, and SNP_398) appeared highly specific to the M. floribunda 821 genome (Table 4). Of course, the specificity has to be evaluated case by case in each cross because a lack of informativeness of these SNPs in other genetic backgrounds cannot be excluded. Besides SNP_585, the other flanking marker, SNP_104, has a good level of specificity as well, but its resistance-linked allele is also present in ‘Braeburn’ and ‘Cox’s O.P.’, which are susceptible to RAA; therefore, this SNP is not fully informative in crosses with these cultivars and their derivatives. Interestingly, ‘Braeburn’ is reported to carry the Sd1 gene for resistance to D. devecta (Alston and Briggs 1970).
Unfortunately, for the other SNPs (SNP_875, SNP_494, SNP_783, SNP_171, SNP_496, and SNP_013), the allele coupled with resistance is also present in some known susceptible cultivars. Therefore, these SNPs are not fully suitable for MAB. Conversely, using a set of these SNPs and also considering the haplotype should be an efficient way to perform MAB for RAA in apple.
The efficiency of the two closest Dp-fl markers (SNP_104 and SNP_585) was also demonstrated by scoring them in another set of 25 apple cultivars (Table 5), mostly of unknown reaction. The molecular and phenotypic data were in agreement for the eight genotypes of known resistance/susceptibility to RAA (‘Ariane’, ‘Gala’, ‘Galarina’, ‘GoldRush’, ‘Golden Orange’, ‘Liberty’, ‘Redfree’, and ‘Topaz’). Of the 17 cultivars of unknown reaction, seven cultivars (‘Primiera’, ‘Modì’, ‘Delorina’, ‘Brina’, PRI-Coop 17, PRI-Coop 39, and PRI-Coop 15), all deriving from M. floribunda 821, are postulated to carry the Dp-fl gene on the basis of the molecular analysis, making them putatively interesting genotypes for breeding. Conversely, several other cultivars deriving from M. floribunda 821 are not carrying the Dp-fl gene (‘Doriane’, ‘Enterprise’, ‘Initial’, and ‘MacFree’). The results on the efficiency of these two Dp-fl flanking markers in MAB are promising, and a survey for the presence of the Dp-fl gene in scab-resistant accessions can thus be performed since the donor of the Dp-fl gene (M. floribunda 821) is well known also as the source of the Rvi6 (Vf) scab resistance gene and has been widely used in breeding worldwide (Gessler and Pertot 2012). RAA resistance-associated markers are particularly desirable because resistant cultivars are an efficient and sustainable method to control RAA and reduce chemical inputs in apple orchards. In fact, apple genotype strongly affects RAA population and damage levels (Miñarro and Dapena 2007, 2008; Razmjou et al. 2014).
Identification of the genomic region spanning the Dp-fl locus in the apple genome sequence
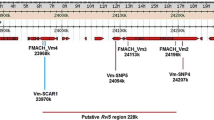
Our efforts to fine map the Dp-fl locus greatly facilitated the molecular identification of putative functional candidate R genes. The sequences of the SNP_104 and SNP_585 markers were aligned to the ‘Golden Delicious’ genome sequence version 1.0 (Velasco et al. 2010). The survey of the 330 Kb region between the two flanking markers resulted in 35 principal contigs, organized mainly in two clusters because of the presence of three gaps (Fig. 3).
Image of the Dp-fl window on LG8 of ‘Golden Delicious’ from GDR Gbrowse, Malus × domestica v1.0 (http://www.rosaceae.org). The flanking TSP markers and the gaps in the sequence are reported with black and gray boxes, respectively
A total of 54 CDSs were retrieved from the 35 contigs (Online Resource 4). The annotation of these predicted genes showed variability in their biological functions, spanning from transporter activity, cell division activity to DNA binding function. Among these CDSs, some were identified as putative candidate genes thanks to the gene ontology (GO) annotation. Indeed, for 19 CDSs putative involvement in the RAA was postulated (indicated in bold in Online Resource 4). Out of these 19, twelve genes might be directly related to M. × domestica-D. plantaginea specific interaction by acting as R genes. More precisely, MDP0000399704 is homologous to a Defensin Ec-AMP-D2-like gene and, according to the GO annotation, it encodes for a protein involved in reactions triggered in response to the presence of a foreign body (biological process: defense response). Two CDSs, MDP0000129202 and MDP0000255502, showed a leucine-rich repeats (LRRs) domain. NBS-LRR genes are one of the largest plant R gene classes and have already been identified in members of the Rosaceae family (Zhong et al. 2015). Most interestingly, sequences from M. × domestica with a significant degree of similarity to NBS-LRR genes have already been found in the region of locus Sd-1, the resistance gene for another aphid, D. devecta (Cevik and King 2002). Eight CDSs showed homology with a TMV resistance protein (MDP0000283149, MDP0000285698, MDP0000273990, MDP0000234409, MDP0000210537, MDP0000292484, MDP0000642458, and MDP0000122099), and their GO annotation imputes a role for them in defense response, transmembrane signaling receptor activity, innate immune response, and apoptotic process. Moreover, these predicted genes contain an LRR conserved domain, putatively important in plant response as explained above. It is known that R gene families possess a higher proportion of duplicate genes than other gene families (Meyers et al. 2005), and the rapid gene expansion or contraction of these families might be a survival strategy to combat rapidly-evolving, species-specific pathogens (Chen et al. 2010). Thus, the cluster organization of these last eight CDSs supports the hypothesis of their involvement in RAA resistance. Finally, MDP0000756536 is a protein belonging to the pectin acetylesterase family which is responsible for catalyzing the deacetylation of pectin. It might be important for the defense mechanism since it has been found to be up-regulated in ‘Florina’ after RAA infestation and not in the susceptible cultivar ‘Topaz’ (Qubbaj et al. 2005). By employing the cDNA-AFLP method, Qubbaj et al. (2005) showed that several defense-related genes are specifically up- or down-regulated after RAA infestation, such as the pectin acetylesterase and also a vacuolar H(+)-ATPase subunit-like protein and an ADP-ribosylating enzyme. Indeed, this comparison of differentially expressed genes in resistant and susceptible apple cultivars after RAA infection suggested that the resistance might be regulated by signal transduction mechanisms similar to other biotic and abiotic stresses.
Specific aphid recognition by R genes is not the only possible resistance mechanism of apple, and other processes could be involved in the elicitation of a general stress response. Aphid saliva secreted into host leaves while feeding contains peroxidases, β-glucosidases, and other potential signal-generating enzymes able to alter the expression of inducible plant physiological factors and to trigger a general stress response similar to those involved in defense against pathogens, for instance, PR proteins (Smith and Boyko 2007). In fact, besides the 12 candidate R genes, the other seven genes are homologous to genes coding for serine/threonine kinase proteins and they may be involved in non-specific plant biotic/abiotic stress responses by acting in the signaling pathways (see Online Resource 4). Moreover, the Pto-like serine/theronine kinase gene was classified as a good candidate for the resistance gene in wheat for another aphid, Diuraphis noxia (Boyko et al. 2006).
With all the information acquired in this work about candidate genes for RAA resistance, the finding and validation of the R gene will be greatly facilitated.
Conclusions
A fine map of the region containing the locus responsible for the resistance of apple to the aphid D. plantaginea was developed and several easy-to-use SNP markers tightly linked to the Dp-fl locus were identified. The mapping efforts were greatly accelerated by the availability of two high-density SNP arrays that were a good starting point for the identification of SNPs in the Dp-fl gene region segregating in the four populations involved in the study. Each SNP was then tested on different genotypes with a cheap but efficient approach, the TSP method. This panel of SNPs, tightly linked among them and with the resistant allele of the Dp-fl gene, will be useful to introgress the resistance to the aphid D. plantaginea.
The characterization of founders and cultivars with the new markers developed in the region of the Dp-fl gene was useful to identify the best markers for MAB, indeed the most specific markers, in coupling with the resistance gene and present only in resistant genotypes. This will help breeders to identify the best parents for breeding programs, and also to pyramid Dp-fl with other RAA resistance genes such as Smh from Malus robusta (Alston and Briggs 1970) when markers are available.
Moreover, our high-resolution genetic map of the Dp-fl locus allowed the identification of interesting candidate genes in the small genomic region enclosed by the two most closely linked SNP markers, thus paving the way for the cloning of Dp-fl. The full sequencing of the Dp-fl region is essential for the identification of the R gene. It should be noted that the candidate genes identified in this work are from ‘Golden Delicious’, which is susceptible. Thus, we cannot exclude a different set of genes in this region in a resistant cultivar such as ‘Florina’. The identification of the R gene will contribute to a better understanding of the functional mechanism involved in apple aphid resistance.
References
Alston FH, Briggs JB (1970) Inheritance of hypersensitivity to rosy apple aphid Dysaphis plantaginea in apple. Can J Genet Cytol 12:257–258
Angeli G, Simoni S (2006) Apple cultivars acceptance by Dysaphis plantaginea Passerini (Homoptera: Aphididae). J Pest Sci 79:175–179
Arnaoudov V, Kutinkova H (2006) Susceptibility of some apple cultivars to infestation by the rosy apple aphid (Dysaphis plantaginea Pass., Homoptera: Aphididae). J Fruit Ornam Plant Res 14:137–142
Bianco L, Cestaro A, Sargent DJ, Banchi E, Derdak S, Di Guardo M, Salvi S, Jansen J, Viola R, Gut I, Laurens F, Chagné D, Velasco R, van de Weg E, Troggio M (2014) Development and validation of a 20K single nucleotide polymorphism (SNP) whole genome genotyping array for apple (Malus × domestica Borkh). PLoS One 9(10), e110377. doi:10.1371/journal.pone.0110377
Bianco L, Cestaro A, Linsmith G, Muranty H, Denancé C, Théron A, Poncet C, Micheletti D, Kerschbamer E, Di Pierro EA, Larger S, Pindo M, van de Weg WE, Davassi A, Laurens F, Velasco R, Durel CE, Troggio M (2016) Development and validation of the Axiom®Apple480K SNP genotyping array. Plant J. doi:10.1111/tpj.13145
Boyko EV, Smith CM, Thara VK, Bruno JM, Deng Y, Starkey SR, Klaahsen DL (2006) Molecular basis of plant gene expression during aphid invasion: wheat Pto- and Pti-like sequences are involved in interactions between wheat and Russian wheat aphid (Homoptera: Aphididae). J Econ Entomol 99:1430–1445
Briggs JB, Alston FH (1969) Sources of pest resistance in apple cultivars. Rep East Malling Res Stat 1968:159–162
Brown MW, Mathews CR (2007) Conservation biological control of rosy apple aphid, Dysaphis plantaginea (Passerini), in eastern North America. Environ Entomol 36:1131–1139
Brown A, Myers JH (2010) Temporal and spatial variability of rosy apple aphid Dysaphis plantaginea populations: is there a role of the alternate host plant Plantago major? Agric Forest Entomol 12:333–341
Bus VGM, Changné D, Bassett HCM, Bowatte D, Calenge F, Celton JM, Durel CE, Malone MT, Patocchi A, Ranatunga AC, Rikkerink EHA, Tustin DS, Zhou J, Gardiner SE (2008) Genome mapping of three major resistance genes to woolly apple aphid (Eriosoma lanigerum Hausm.). Tree Genet Genomes 4:223–236
Cevik V, King GJ (2002) High-resolution genetic analysis of the Sd-1 aphid resistance locus in Malus spp. Theor Appl Genet 105:346–354
Chagné D, Gasic K, Crowhurst RN, Han Y, Bassett HC, Bowatte DR, Lawrence TJ, Rikkerink EHA, Gardiner SE, Korban SS (2008) Development of a set of SNP markers present in expressed genes of the apple. Genomics 92:353–358
Chagné D, Crowhurst RN, Troggio M, Davey MW, Gilmore B, Lawley C, Vanderzande S, Hellens RP, Kumar S, Cestaro A, Velasco R, Main D, Rees JD, Iezzoni A, Mockler T, Wilhelm L, Van de Weg E, Gardiner SE, Bassil N, Peace C (2012) Genome-wide SNP detection, validation, and development of an 8K SNP array for apple. PLoS One 7(2), e31745. doi:10.1371/journal.pone.0031745
Chen QH, Han ZX, Jiang HY, Tian DC, Yang SH (2010) Strong positive selection rives rapid diversification of R-genes in Arabidopsis relatives. J Mol Evol 70:137–148
Dapena E, Miñarro M, Blazquez MD (2009) Evaluation of the resistance to the rosy apple aphid using a genetic marker. ISHS Acta Hortic 814:787–790. doi:10.17660/ActaHortic.2009.814.133
De Berardinis E, Baronio P, Baumgärtner J (1994) The effect of aphid (Dysaphis plantaginea Pass., Hom., dAphididae) feeding on apple fruit growth. Ecol Model 72:115–127
Durel CE, Laurens F, Fouillet A, Lespinasse Y (1998) Utilization of pedigree information to estimate genetic parameters from large unbalanced data sets in apple. Theor Appl Genet 96:1077–1085
Gessler C, Pertot I (2012) Vf scab resistance of Malus. Trees 26:95–108
Hayden MJ, Nguyen TM, Waterman A, McMichael GL, Chalmers KJ (2008) Application of multiplex-ready PCR for fluorescence-based SSR genotyping in barley and wheat. Mol Breeding 21:271–281
Hayden M, Tabone T, Mather D (2009) Development and assessment of simple PCR markers for SNP genotyping in barley. Theor Appl Genet 119:939–951
Hesler LS, Tharp CI (2005) Antibiosis and antixenosis to Rhopalosiphum padi among triticale accessions. Euphytica 143:153–160
Jänsch M, Broggini GAM, Weger J, Bus VGM, Gardiner SE, Bassett H, Patocchi A (2015) Identification of SNPs linked to eight apple disease resistance loci. Mol Breeding 35:45
Kaloshian I (2004) Gene-for-gene disease resistance: bridging insect pest and pathogen defense. J Chem Ecol 30:2419–2438
Liang W, Dondini L, De Franceschi P, Paris R, Sansavini S, Tartarini S (2015) Genetic diversity, population structure and construction of a core collection of apple cultivars from Italian germplasm. Plant Mol Biol Rep 33:458–473
Liebhard R, Gianfranceschi L, Koller B, Ryder CD, Tarchini R, van de Weg E, Gessler C (2002) Development and characterization of 140 new microsatellites in apple (Malus × domestica Borkh.). Mol Breed 10:217–241
Marchetti E, Civolani S, Leis M, Chicca M, Tjallingii WF, Pasqualini E, Baronio P (2009) Tissue location of resistance in apple to the rosy apple aphid established by electrical penetration graphs. Bull Insectology 62:203–208. ISSN 1721–8861
Meyers BC, Kaushik S, Nandety RS (2005) Evolving disease resistance genes. Curr Opin Plant Biol 8:129–134
Miñarro M, Dapena E (2004) Inheritance of the tolerance to the rosy apple aphid of the cv. ‘Florina’. Acta Hortic 663:261–264. doi:10.17660/ActaHortic.2004.663.42
Miñarro M, Dapena E (2007) Resistance of apple cultivars to Dysaphis plantaginea (Hemiptera: Aphididae): role of tree phenology in infestation avoidance. Environ Entomol 36:1206–1211
Miñarro M, Dapena E (2008) Tolerance of some scab-resistant apple cultivars to the rosy apple aphid, Dysaphis plantaginea. Crop Prot 27:391–395
Miñarro M, Hemptinne JL, Dapena E (2005) Colonization of apple orchards by predators of Dysaphis plantaginea: sequential arrival, response to prey abundance and consequences for biological control. BioControl 50:403–414
Newcomb RD, Crowhurst RN, Gleave AP, Rikkerink EHA, Allan AC, Beuning LL, Bowen JH, Gera E, Jamieson KR, Janssen BJ et al (2006) Analyses of expressed sequence tags from apple. Plant Physiol 141:147–166
Parisi L, Gros C, Combe F, Parveaud CE, Gomez C, Brun L (2013) Impact of a cultivar mixture on scab, powdery mildew and rosy aphid in an organic apple orchard. Crop Prot 43:207–212
Qubbaj T, Reineke A, Zebitz CPW (2005) Molecular interactions between rosy apple aphids, Dysaphis plantaginea, and resistant and susceptible cultivars of its primary host Malus domestica. Entomol Exp Appl 115:145–152
Rat-Morris E (1993) Development of the rosy apple aphid Dysaphis plantaginea Pass. on a tolerant apple cultivar ‘Florina’. Bulletin OILB/SROP 16:91–100
Rat-Morris E, Lespinasse Y (1995) Pommier: la résistance au puceron cendré associée à la résistance à la tavelure. Phytoma - La Défense des végétaux 471:15–17
Razmjou J, Changizi M, Golizadeh A, Khiavi HK, Asghar Fathi SA, Mottaghinia L (2014) Evaluation of resistance in seven apple cultivars to rosy apple aphid, Dysaphis plantaginea (Hemiptera: Aphididae) under greenhouse and field conditions. J Crop Prot 3:173–180
Rossi M, Goggin FL, Milligan SB, Klaoshian I, Ullman DE, Williamson VM (1998) The nematode resistance gene Mi of tomato confers resistance against the potato aphid. Proc Natl Acad Sci U S A 95:9750–9754
Salvi S, Micheletti D, Magnago P, Fontanari M, Viola R, Pindo M, Velasco R (2014) One-step reconstruction of multigeneration pedigree networks in apple (Malus × domestica Borkh.) and the parentage of Golden Delicious. Mol Breed 34:511–524. doi:10.1007/s11032-014-0054-y
Sandanayaka WRM, Bus VGM, Connolly P, Newcomb R (2003) Characteristics associated with woolly apple aphid Eriosoma lanigerum, resistance of three apple rootstocks. Entomol Exp Appl 109:63–67
Sandanayaka WRM, Bus VGM, Connolly P (2005) Mechanisms of woolly aphid [Eriosoma lanigerum (Hausm.)] resistance in apple. J Appl Entomol 129:534–541
Smith CM, Boyko EV (2007) The molecular bases of plant resistance and defense responses to aphid feeding: current status. Entomol Exp Appl 122:1–16
Stoeckli S, Mody K, Gessler C, Patocchi A, Jermini M, Dorn S (2008) QTL analysis for aphid resistance and growth traits in apple. Tree Genet Genomes 4:833–847
Tabone T, Mather DE, Hayden MJ (2009) Temperature Switch PCR (TSP): robust assay design for reliable amplification and genotyping of SNPs. BMC Genomics 10:580
Van Ooijen J (2006) JoinMap 4, Software for the calculation of genetic linkage maps in experimental populations. Kyazma B.V, Wageningen, Netherlands
Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A et al (2010) The genome of the domesticated apple (Malus × domestica Borkh.). Nat Genet 42:833–839
Wang A, Aldwinckle H, Forsline P, Main D, Fazio G, Brown S, Xu K (2012) EST contig-based SSR linkage maps for Malus × domestica cv Royal Gala and an apple scab resistant accession of M. sieversii, the progenitor species of domestic apple. Mol Breed 29:379–397. doi:10.1007/s11032-011-9554-1
Young ND, Tanksley SD (1989) Restriction fragment length polymorphism maps and the concept of graphical genotypes. Theor Appl Genet 77:95–101
Zhong Y, Yin H, Sargent DJ, Malnoy M, Cheng ZM (2015) Species-specific duplications driving the recent expansion of NBS-LRR genes in five Rosaceae species. BMC Genomics 16:77. doi:10.1186/s12864-015-1291-0
Acknowledgments
This work has been funded under the EU Seventh Framework Programme by the FruitBreedomics Project No. 265582: Integrated Approach for Increasing Breeding Efficiency in Fruit Tree Crop, INIA and FEDER (Projects RTA2012-00118-C03-01, RTA2008-00120-0000, and RTA04-147-C02-01). The views expressed in this work are the sole responsibility of the authors and do not necessarily reflect the views of the European Commission.
Conflict of interest
The authors declare that they have no competing interests.
Data archiving statement
The SSR and TSP data for this research are available (tfGDR1027) at the Genome Database for Rosaceae (www.rosaceae.org).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by E. Dirlewanger
Electronic supplementary material
Below is the link to the electronic supplementary material.
Online Resource 1
List of 25 apple accessions included in the analysis of markers for the Dp-fl locus for the resistance to RAA. (PDF 316 kb)
Online Resource 2
Primers for the development of the TSP assays for the 10 SNPs in the region of SSR CH01h10. These SNPs were selected as heterozygous in ‘Florina’ and homozygous in at least one of the susceptible parents, thus giving fully informative segregation (‘Aaxaa’). The specific segregating nucleotide for each assay is indicated in bold and the non-complementary 2–3 bp nucleotide tails added to the 5′ end region are underlined. (PDF 144 kb)
Online Resource 3
Amplification protocols optimized for each TSP assay developed in this study. In order to increase the specificity of each assay, annealing temperature and extension time were specifically calculated. (PDF 146 kb)
Online Resource 4
List of the predicted CDS sequences in the Dp-fl region on LG8 from ‘Golden Delicious’ genome. For the CDS, the annotation with the BASTN approach and Gene Ontology approach is reported. The candidate genes with a possible involvement in Malus-D. plantaginea interaction are indicated in bold. (PDF 245 kb)
Rights and permissions
About this article
Cite this article
Pagliarani, G., Dapena, E., Miñarro, M. et al. Fine mapping of the rosy apple aphid resistance locus Dp-fl on linkage group 8 of the apple cultivar ‘Florina’. Tree Genetics & Genomes 12, 56 (2016). https://doi.org/10.1007/s11295-016-1015-x
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-016-1015-x