Abstract
Biofilm formation and antibiotic efflux are two determinant factors in the development of drug resistance phenotype by Pseudomonas aeruginosa. Non-steroid anti-inflammatory drugs have shown the antimicrobial potential to be used in combination with antibiotics against bacterial pathogens. In this work, the effect of ibuprofen alone and in combination with ciprofloxacin on some virulence traits and the expression of the alginate synthesis and efflux pump genes of clinical isolates of P. aeruginosa was investigated. The checkerboard titration assay was used to evaluate the synergism of the drugs. P. aeruginosa strains were grown in the presence of sub-inhibitory concentrations of the drug and their biofilm formation level, swarming, swimming, and hemolytic activity were assessed. Also, the relative expression of the alg44, algT/U, mexB, and oprM genes was determined by qPCR assay. The MIC of ibuprofen and ciprofloxacin were measured 2048 and 32 µg/mL and the drugs showed synergic antibacterial activity (FIC = 0.4). Moreover, ibuprofen alone and in combination with ciprofloxacin, significantly reduced the expression of alg44 (0.22 and 0.25 folds) and algT/U (0.26 and 0.37 folds) genes, while increased the expression of the mexB (1.64 and 1.83 folds) and oprM (1.36 and 1.92 folds) genes. Simultaneous treatment of bacterial cells with ibuprofen and ciprofloxacin significantly decreased bacterial biofilm formation (65%), swimming, swarming, and hemolytic activity (85%), compared with the control. This work suggests that ibuprofen has considerable anti-virulence potential against P. aeruginosa and could be employed for combination therapy with antibiotics after further characterizations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pseudomonas aeruginosa, is an opportunistic Gram-negative bacterium associated with a variety of human infections, including urinary tract, respiratory, wound, and burn infections. The P. aeruginosa infections could develop into life-threatening diseases in immune-compromised and cystic fibrosis patients (Dai et al. 2019). Bacterial virulent traits and pathogenesis are controlled by the bacterial Quorum sensing (QS) systems and depend on bacterial cell density. In P. aeruginosa, the main QS system consists of two LuxIR circuits, termed LasIR and RhlIR. LasI and RhlI are autoinducer synthases that produce 3-oxo-C12-homoserine lactone and C4-homoserine lactone, respectively which bind to their cognate receptors, LasR and RhlI. Both QS systems play significant roles in the virulence and pathogenicity of P. aeruginosa. The autoinducer-receptor compound encodes many virulence factors such as exoenzymes, rhamnolipid, pyocyanin and pyoverdin, elastase, protease, swarming and twitching motility, and also biofilm formation (Karatuna and Yagci 2010).
Biofilm is a sessile association of bacterial cells that interact with each other in a self-synthesized extracellular polymeric matrix and are attached to a surface. Biofilm formation is regarded as a major pathogenicity determinant in many bacterial species, including P. aeruginosa. Also, biofilm formation is associated with antibiotic resistance development through the reduced penetration of the drug into the biofilm matrix. The biofilm of P. aeruginosa consists of three major types of polysaccharides, including Alginate, Pel, and Psl (Rasamiravaka et al. 2015). Alginate consists of β-D-mannuronic acid and α-L-guluronic acid and is overproduced in mucoid variants and during biofilm establishment.
Several regulatory systems at transcriptional, post-transcriptional, and post-translational levels control the synthesis of P. aeruginosa alginate. At the transcriptional level, biofilm formation is controlled by a sigma factor σ22 (AlgT/U). In response to the environmental factors or stress conditions, an inner membrane protease (called AlgW) leads to a cascade of regulated intramembrane proteolysis (RIP) that results in the rapid degradation of MucA and eventually the release of AlgT/U from the cytoplasmic membrane that regulates the transcription of a group of associated genes (Wood and Ohman 2015). At the post-translational level, the synthesis of exo-polysaccharides is controlled by the secondary messenger, bis-(3′–5′)-cyclic dimeric guanosine monophosphate (c-di-GMP). The interacting membrane-anchored protein, Alg8 and Alg44, which are considered alginate polymerases, are activated by binding of the dimeric c-di-GMP to the PilZ domain of Alg44 (Moradali and Rehm 2019).
Ciprofloxacin is a fluorinated quinolone, which has bactericidal activity on a variety of bacterial pathogens via the inhibition of bacterial DNA gyrase II (LeBel 1988). It is regarded as one of the most widely prescribed and most efficient antibiotics to treat P. aeruginosa infections. However, due to the extensive use, the prevalence of ciprofloxacin resistant P. aeruginosa strains has been increasing (Rehman et al. 2019). Bacterial strains employ several mechanisms of drug resistance, such as reducing drug permeability, activation of efflux systems, production of the enzymes that inactivate antibiotics, and modification of antibiotic targets (Bassetti et al. 2018). Efflux is a process in which bacterial cells extrude antimicrobial compounds from the cytoplasm to protect from the lethal concentration of the drug. The MexAB-OprM is a main P. aeruginosa efflux system that belongs to the resistance-nodulation-division (DNR) family and is involved with antibiotic resistance, especially resistance to the β-lactam and fluoroquinolone class of antibiotics (Bassetti et al. 2018). Structurally, the efflux pump includes three parts: the inner cytoplasmic membrane protein of MexB, the periplasmic lipoprotein of MexA that is attached to the bacterial inner membrane, and also the outer membrane lipoprotein, OprM (Askoura et al. 2011). Also, the MexAB-OprM transporter exports the QS system inducers, acyl-homoserine lactones, which initiate the expression of QS-dependent virulence factors, including proteases, rhamnolipids, exotoxin A, exoenzyme S, and pyocyanin (Pearson 1999).
The rapid and global spread of antibiotic-resistant bacteria highlights the urgent need for new treatments, such as the combination of conventional antibiotics with non-steroidal anti-inflammatory drugs (NSAIDs) (Chen and Wen 2011). NSAIDs exert strong antimicrobial properties against a variety of bacterial strains in both planktonic and biofilm growth. Ibuprofen, a widely used NSAID, has inhibitory effects on the growth of P. aeruginosa and also, inhibits the bacterial QS system, which could prevent biofilm formation and attenuate bacterial virulence (Dai et al. 2019). Bacterial infections may cause the accumulation of inflammatory cells and cytokines in the infection site and result in undesirable immune responses. The inflammatory responses could result in local tissue damage and complicate the treatment process (Čulić et al 2001). Therefore, the use of anti-inflammatory drugs, including ibuprofen, for adjuvant therapy with antibiotics could be an effective approach for treating bacterial infection (Chmiel et al. 2013). Due to the considerable antibiofilm and anti-QS potentials of ibuprofen against pathogenic P. aeruginosa strains and also, the anti-inflammatory property of this drug, the combination of ibuprofen with antibiotics could be considered as a novel therapeutic strategy to treat P. aeruginosa infections. Therefore, the current work was conducted to evaluate the effect of combination therapy using ibuprofen and ciprofloxacin on some virulence traits of clinical isolates of P. aeruginosa and the expression of bacterial efflux pump and alginate synthesis genes.
Materials and methods
Bacterial strains
Clinical strains of P. aeruginosa, isolated from clinical specimens including urine, wound secretion, and blood, were used in this study. Bacterial identification was performed using the biochemical assays, including the growth in cetrimide agar medium, growth at 42 °C, oxidase and catalase, Gram staining, etc. The P. aeruginosa ATCC 85,327 was also used as the standard strain. The resistance of the isolates to ciprofloxacin was screened by disc diffusion assay according to the Clinical Laboratory Standard Institute (CLSI) recommendation (CLSI 2021). The strains with a zone of inhibition ≤ 18 mm against the ciprofloxacin disk (5 µg) were considered ciprofloxacin resistant.
Determination of MIC and MBC
The minimum inhibitory concentration (MIC) of ibuprofen and ciprofloxacin was determined using the method described in CLSI guideline (CLSI 2021). A concentration range of ibuprofen (128–8192 µg/mL) and ciprofloxacin (16–1024 µg/mL) was prepared in 96-well plates and 100 µL of bacterial suspension (1.5 × 106 CFU/mL) was added to the wells. The plates were incubated at 37 °C for 24 h. The minimum concentration of each drug that inhibited bacterial growth was considered as the MIC value. To determine the minimum bactericidal concentration (MBC), samples from the wells that had no bacterial growth were inoculated in Muller Hinton agar plates and bacterial growth was monitored after incubation at 37 °C for 24 h. The minimum concentration of the drug with bactericidal activity was considered the MBC value.
Checkerboard titration assay
The checkerboard titration assay was used to determine the synergistic effect of ibuprofen and ciprofloxacin against P. aeruginosa. The test was performed in microtiter 96-well plates according to the method described previously (Bajaksouzian et al. 1997). Based on MIC concentrations, a concentration range (from 1/8 to 8 × MIC) of ibuprofen and ciprofloxacin were dispensed in the rows and columns of the plate, respectively. Then, 50 µL of bacterial suspension was added to each well and the plate was incubated at 37 °C overnight. Fractional inhibitory concentration (FIC) was calculated by the following formula:
where ibuprofen and ciprofloxacin were considered the drugs A and B. The FICtotal of less than 0.05 was considered synergistic interaction between the drugs.
Evaluation of the expression of alg44, algT/U, mexB and oprM genes
To survey the effect of ibuprofen and ciprofloxacin on the expression of alg44, algT/U, mexB, and oprM genes, bacterial cells were inoculated in TSB media containing 512 μg/mL ibuprofen (equivalent to the 1/4 MIC), 8 μg/mL ciprofloxacin (1/4 MIC), and the combination of ibuprofen with ciprofloxacin. After 14 h incubation at 37 °C, bacterial cells were harvested and total RNA was extracted using the TriZol reagent (Das and Dash 2014). A nanodrop spectrophotometer was used to evaluate the quality and quantity of the extracted RNA. The cDNA was synthesized using the Thermo Fisher Scientific kit (USA), according to the manufacturer’s protocol. Quantitative real-time PCR was performed using the SYBR Green master mix and gene-specific primers. The sequence of the primers was presented in supplementary Table 1. The rpsl gene was used as an internal control to normalize the expression of target genes.
Biofilm formation
The antibiofilm effect of ibuprofen, alone and in combination with ciprofloxacin, was examined in 96-well microtiter plates (Das and Dash 2014). In brief, bacterial cells were treated with different concentrations of ibuprofen or in the combination with ciprofloxacin and were incubated at 37 °C for 48 h without shaking. After incubation, the plates were washed with distilled water to remove the medium and unattached cells and stained with crystal violet 1% for 15 min. After washing the wells, acid acetic 30% was used to solubilize the dye and finally, the optical absorbance was measured at 570 nm.
Motility assays
To evaluate the swarming motility, a nutrient agar medium containing 5% agar and either drug (ibuprofen 512 µg/mL or ciprofloxacin 8 µg/mL) or both drugs were prepared in 6 cm Petri dishes. Then, 2 µL of bacterial suspension was placed on the surface of the medium. After incubation at 37 °C for 24 h, the swarming zone diameter was measured and compared with the control (Rashid and Kornberg 2000).
In addition, the effect of the drugs on twitching motility was assessed. In brief, a nutrient agar medium (1.5% agar) containing ibuprofen and ciprofloxacin alone or in combination was prepared. The bacterial suspension was inoculated at the bottom of the plates with a sterile toothpick. After 24 h incubation at 37 °C, the agar medium was removed and the plates were stained with crystal violet 0.5% for 20 min. The twitching motility zone of P. aeruginosa on the bottom of the plate was measured and compared with the control (Rashid and Kornberg 2000).
Hemolysis assay
The effect of ibuprofen and ciprofloxacin, alone or in combination, on the hemolytic activity of P. aeruginosa strains, was evaluated using the method described, previously (Lee et al. 2012). Bacterial cells were treated with the drugs at the sub-inhibitory concentration for 16 h at 37 °C. Then, 100 µL of bacterial supernatant was added to the washed red blood cells. The suspension was incubated at 37 °C with shaking for 2 h. Then, the supernatant was collected and the optical density was measured at 430 nm.
Statistical analyses
The assays were conducted in triplicates and data are presented as mean ± SD. The significant difference between the control and treatment groups was assessed by one-way analysis of variance (ANOVA) and Tukey′s post hoc test using the Graphpad Prism 8 software. The differences were regarded as statistically significant when p < 0.05.
Results
Bacterial identification
The P. aeruginosa strains were identified as non-fermentative strains that are H2S, CO2, indol, MR, and urea negative, and also citrate, catalase, and oxidase-positive, and able to grow on cetrimide agar. Also, detection of the bond at 196 bp, corresponding to the rpsL gene, using the specific primer confirmed the identity of the isolated strains.
Growth inhibitory concentrations of the drugs
The MIC of ibuprofen and ciprofloxacin against the clinical P. aeruginosa strains were determined 2048 and 32 µg/mL, respectively. The MIC of ibuprofen for the P. aeruginosa ATCC 85327 was similar to the pathogenic strains, while the MIC of ciprofloxacin was determined 16 µg/mL. The MBC assay revealed that ibuprofen was a bacteriostatic agent, able to inhibit bacterial growth without killing bacterial cells. In contrast, the MBC values of ciprofloxacin for the pathogenic and ATCC strains were recorded 64 and 32 µg/mL, respectively. Based on the results, the ¼ MIC of the drugs was selected as a sub-inhibitory concentration for subsequent experiments.
Synergism of ibuprofen and ciprofloxacin
Based on the results, the synergistic effect of ibuprofen and ciprofloxacin against P. aeruginosa was evaluated by the checkerboard titration assay. The FIC index was calculated 0.4, suggesting that ibuprofen (at 64 µg/mL) and ciprofloxacin (at 12 µg/mL) had synergistic antibacterial effects on P. aeruginosa strains.
Expression of alginate synthesis and efflux pump genes
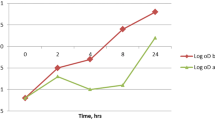
The effect of ibuprofen and ciprofloxacin alone and in combination on the expression of alginate synthesis, alg44 and algT/U, and efflux pump, mexB and oprM genes, was investigated. Our results revealed that the expression of both alginate genes, after treatment with ibuprofen, alone and combined with ciprofloxacin, was significantly reduced. The relative expression of alg44 in ibuprofen and ibuprofen + ciprofloxacin treated cells was decreased (p < 0.05) by 22 and 25%, respectively, compared with the control cells (Fig. 1a). In addition, the expression of algT/U, in ibuprofen and ibuprofen + ciprofloxacin treated cells was decreased by 26 and 37%, respectively (Fig. 1b).
In addition, the expression of mexB and oprM was assessed in ibuprofen and ciprofloxacin treatments. The relative expression of mexB and oprM in ibuprofen, ciprofloxacin, and ibuprofen + ciprofloxacin treated cells was increased, compared with the control. According to the results, the exposure to ibuprofen and ciprofloxacin resulted in an increased expression of mexB gene by 1.6 and 2.1 folds, respectively, while the exposure to the ibuprofen + ciprofloxacin increased the mexB gene by 1.8 folds (Fig. 1c). In addition, the mRNA level of oprM gene was increased following the exposure of the cells to ibuprofen, ciprofloxacin, and ibuprofen + ciprofloxacin by 1.36, 1.4, 1.2, and 1.91 folds, respectively (Fig. 1d).
Effect of ibuprofen and ciprofloxacin on biofilm formation
The antibiofilm effect of the sub-inhibitory concentration of ibuprofen and ibuprofen + ciprofloxacin on P. aeruginosa biofilm was determined by a semi-quantitative plate assay. Based on the results, ibuprofen alone had considerable antibiofilm potential which reduced biofilm formation by 51%, compared with the control. Also, simultaneous treating the cells with ibuprofen and ciprofloxacin reduced biofilm formation by 65%, suggesting the synergic antibiofilm activity of the agents. The results were presented in Fig. 2.
Effect of ibuprofen and ciprofloxacin on bacterial motility
To assess the effect of ibuprofen and ciprofloxacin on swarming and twitching motilities of P. aeruginosa, the bacteria were grown in the presence of a sub-inhibitory concentration of the drugs. The results showed that the combination of ibuprofen and ciprofloxacin has a stronger inhibitory effect on both swarming and twitching motilities of P. aeruginosa compared with either drug alone. The swarming zone in the absence of ibuprofen and ciprofloxacin was 14.0 mm; whereas, in the presence of ibuprofen (1024 μg/mL), ciprofloxacin (16 μg/mL), and ibuprofen + ciprofloxacin the swarming zone reduced to 8.0, 6.0, and 5.0 mm, respectively (Fig. 3). Similarly, the twitching motility zone was 14.0 mm in the control group. The twitching zone in ibuprofen, ciprofloxacin, and ibuprofen + ciprofloxacin treated cells was 14.0, 13.0, and 9.0 mm, respectively (Fig. 4).
Effect of ibuprofen and ciprofloxacin on bacterial hemolytic activity
The effect of ibuprofen and ciprofloxacin alone and in combination, on the hemolytic activity of P. aeruginosa strains was determined. According to the results, the combination of ibuprofen and ciprofloxacin considerably reduced the hemolytic activity of bacterial cells, compared with other treatment groups. Treating P. aeruginosa with ibuprofen and ciprofloxacin alone reduced bacterial hemolysis by 8.97% (OD430 = 0.345) and 17.67% (OD430 = 0.312), respectively, compared with the control (OD430 = 0.379), while simultaneous exposure to ibuprofen and ciprofloxacin reduced the hemolytic activity by 84.96% (OD430 = 0.057). Figure 5 displays the hemolysis inhibition assay for different treatment groups.
Effect of ibuprofen and ciprofloxacin on hemolytic activity of P. aeruginosa. A negative control, B ibuprofen + ciprofloxacin, C ciprofloxacin, D ibuprofen, E positive control. Simultaneous exposure to ibuprofen and ciprofloxacin significantly reduced the hemolytic activity of P. aeruginosa (p < 0.05)
Discussion
Drug-resistant bacterial strains employ several resistance mechanisms which enable them to develop resistance to a wide range of antibiotics. Therefore, treating drug-resistant infections has become a global health challenge. Combination therapy using several antibacterial agents is a novel approach to combat drug-resistance bacteria. In other words, the use of antibiotics combined with non-antibiotics, such as NSAIDs, could be considered an alternative strategy for treating bacterial infections (Chen and Wen 2011; She et al. 2018).
It was reported that NSAIDs, including ibuprofen, are potent antibacterial agents (Elvers and Wright 1995; Obad et al. 2015; Dai et al. 2019). The MIC of ibuprofen for P. aeruginosa, Escherichia coli, Klebsiella pneumoniae, and Proteus vulgaris was reported ≤ 3200 µg/L and has been stated that the MIC value could be reduced by four folds in the presence of some efflux inhibitors (Laudy et al. 2016). According to our result, ibuprofen at 2048 µg/mL could inhibit P. aeruginosa growth but has no bactericidal activity. Our results are similar to the previous studies, however, some contradictory results were observed that could be related to the differences in experimental protocols such as the method used to determine antimicrobial activity, ibuprofen formulation, bacterial strains, and growth conditions.
Ciprofloxacin is a broad-spectrum antibiotic, that inhibits bacterial DNA gyrase II and also inhibits nucleic acid repair and replication (Hooper et al. 1987). The results of the checkerboard titration assay showed that the combination of ibuprofen and ciprofloxacin may have synergy against P. aeruginosa strains. Two main reasons could be hypothesized for this finding, including structural similarity between ibuprofen and fluoroquinolones and competitive inhibition of bacterial efflux systems. Some structural similarities between ibuprofen and fluoroquinolone antibiotics such as ciprofloxacin, levofloxacin, and nalidixic acid have been reported. Moreover, it was found that ibuprofen and other NSAIDs can bind to bacterial DNA gyrase, so inhibiting bacterial nucleic acid replication and repair (Kahlous et al. 2017). In a study, del Prado et al. observed that the animals that received amoxicillin with ibuprofen had fewer bacteria than those received the antibiotic alone (del Prado et al. 2010). Therefore, the synergism of ibuprofen with ciprofloxacin could be associated with the synergistic inhibition of the bacterial topoisomerase enzyme. Also, it was found that ibuprofen can act as a substrate of the MexAB-OprM efflux pump of P. aeruginosa. Thus, the synergism of ibuprofen with ciprofloxacin could also be associated with the competitive inhibition of the bacterial efflux system, which reduces the extrusion of the antibiotic from the cytoplasm.
The bacterial QS system plays a vital role in bacterial physiology regulation and virulence. Molecular docking studies showed that some NSAIDs could interact with the lux-R homolog molecules of pathogenic bacteria and interrupt their QS- systems (Soheili et al. 2015; de Almeida et al. 2018). In P. aeruginosa, the LasR and PqsE proteins are involved with the activation of QS signaling systems, biofilm maturation, and production of virulence factors (Soheili et al. 2015). It was found that many NSAIDs have anti-QS potential against P. aeruginosa strains, mainly via the interference with the LasR and Pqs QS systems. A docking study on the possible interaction of NSAIDs with QS molecules revealed that several NSAIDs could interact with the active site of the LasR and PqsE proteins (Soheili et al. 2015) which could result in the disruption of bacterial QS systems. Moreover, Dai et al. (2019) found that ibuprofen has an inhibitory effect on the level of acyl homoserine lactone, the inducer of the QS system, and has a high binding score for LasI and LasR proteins. These findings reveal that ibuprofen could have an inhibitory effect on virulence factors and QS-dependent traits of P. aeruginosa.
Biofilm formation is one of the major strategies that help P. aeruginosa to survive in different environmental conditions. Previous studies revealed that ibuprofen has a significant inhibitory effect on the biofilm formation of P. aeruginosa (Dai et al. 2019). In this study, the highest biofilm inhibition of ibuprofen was recorded at 1024 μg/mL. It was reported that some NSAIDs, including diclofenac, ibuprofen, and salicylic acid affect bacterial adhesion to abiotic surfaces (Demirag et al. 2007). Also, the exposure of E. coli to ibuprofen reduced the adhesion of bacteria to epithelial cells by inhibiting bacterial fimbriae through the changes in bacterial hydrophobicity and hemolysin production and thus, reduced bacterial biofilm formation (Naves et al. 2010). Therefore, a similar antibiofilm effect of ibuprofen on P. aeruginosa could be hypothesized through the modification of the bacterial surface components. There are more evidence suggesting that NSAIDs, including ibuprofen, may affect biofilm formation through the interruption of P. aeruginosa QS system (Dai et al. 2019). In this study, we found that the combination of ibuprofen and ciprofloxacin has a synergic inhibitory effect on the biofilm formation of P. aeruginosa strains. Previous studies showed that lonazolac, as an NSAID, is an appropriate candidate for inhibiting the QS system and biofilm formation in Salmonella spp. Also, it was suggested that the anti-QS and anti-biofilm potential of NSAIDs could be strengthened in combination with antibiotics (de Almeida et al. 2018). She et al. reported that meloxicam, as an NSAID, combined with some antibiotics may affect the biofilm formation of P. aeruginosa (She et al. 2018). In agreement with previous results, the current work demonstrated the increased antibiofilm activity of ibuprofen with ciprofloxacin against clinical P. aeruginosa.
Alginate production is a major determinant in the biofilm maturation of P. aeruginosa and affects biofilm properties, including cell-to-cell interaction, bio-volume, cell density, surface attachment, and viscoelasticity (Moradali et al. 2015). Previous works showed that some NSAIDs could attenuate the genes associated with biofilm formation in P. aeruginosa. It was found that meloxicam, at 45.62 µg/mL, could significantly inhibit the expression of pslA, pelA, and alg44 genes, the genes responsible for the production of bacterial extracellular polymeric substances (EPS). These results suggest that the inhibition of EPS plays an important role in the antibiofilm activity of meloxicam on P. aeruginosa (She et al. 2018), and thus, could increase their antibiotic susceptibility. According to our results, ibuprofen alone and in combination with an antibiotic can significantly reduce the expression of the genes involved in biofilm formation, alg44, and algT/U. The ibuprofen + ciprofloxacin treated cells showed a further decrease in the expression of the alg genes than the ibuprofen group, However, the difference was not significant, indicating a weak synergism effect between ibuprofen and ciprofloxacin in reducing the expression of the studied genes. There is not enough information available about the effect of ibuprofen and ciprofloxacin on the expression of alg44 and algT/U. In our opinion, the reduction of alginate synthesis genes could be associated with the reduction of the bacterial QS system. The production of bacterial biofilm and several virulence factors are key determinants in bacterial pathogenesis and mainly require the participation of a community of bacteria. The virulence factors of P. aeruginosa are mainly controlled by the QS system, which controls the social behavior of bacteria through multiple interconnected signaling pathways (Karatuna and Yagci 2010).
Our results showed that the expression of the mexB and oprM genes was increased in all treatment groups, compared with the control. However, the relative expression of the mexB gene was significantly lower in ibuprofen + ciprofloxacin-treated cells, compared with the ciprofloxacin alone. Ciprofloxacin is the natural substrate of the MexAB-OprM efflux system. Also, it was reported that some NSAIDs, including ibuprofen, are considered the substrate of this efflux pump (Laudy et al. 2016). Thus, the increased expression of the mentioned genes in all treatment groups could be expected. The MexB protein is a major component of the MexAB-OprM efflux system and is the inner part of the efflux system, which has an important role in the extrusion of the antimicrobials from bacterial cytoplasm to periplasmic space. The reduced expression of the mexB gene in the cells treated with ibuprofen + ciprofloxacin (compared with the ciprofloxacin alone) suggests that the use of ibuprofen could reduce the extrusion of the antibiotic via MexAB-OprM system and could be involved with the increased susceptibility of bacterial cells to the antibiotic. However, to elucidate the exact effect of ibuprofen on the bacterial efflux system, further experiments are required.
Previous studies showed that 3-oxo-C12 HSL, the autoinducer of the QS system, is a substrate of the MexAB-OprM pump in P. aeruginosa and thus, the QS system of P. aeruginosa could be affected by the efflux system. Also, the regulation of the genes regulated by the 3-oxo-C12 HSL and LasR QS system is likely to be affected by the MexAB-OprM pump (Pearson et al. 1999). As mentioned above, ibuprofen can be considered a substrate of RND efflux pumps in P. aeruginosa (Laudy et al. 2016). It could be hypothesized that ibuprofen not only can compete with the extrusion of antibiotics from cells but also can compete with the QS autoinducers and exert inhibitory effects on bacterial QS-dependent traits. Since several virulence factors of P. aeruginosa are controlled by the QS system, we suggest that the use of ibuprofen can reduce the pathogenicity of the bacterium by inhibiting the QS system and also increasing the efficiency of antibiotics.
There are several reports on the antibiofilm and anti-QS activity of NSAIDs and also the reduction of QS-dependent virulence factors in P. aeruginosa (Laudy et al. 2016; Dai et al. 2019). Bacterial motility plays an important role in the development of infection to new sites. The swarming and twitching motility of P. aeruginosa are regulated by the QS system. It was found that NSAIDs, including Ketoprofen and diclofenac, can significantly reduce the swarming of P. aeruginosa (Ulusoy and Bosgelmez-Tinaz 2013). Our work showed that ibuprofen and ciprofloxacin alone and in combination had a significant effect on reducing the swarming of P. aeruginosa. Surprisingly, it was observed that ibuprofen and ciprofloxacin alone had no significant effect on twitching motility but in ibuprofen + ciprofloxacin treated cells the twitching of P. aeruginosa was considerably decreased. Owing to the anti-QS activity of ibuprofen, it could be suggested that ibuprofen in combination with antibiotics could decrease bacterial motility and reduce their colonization on new target surfaces.
The hemolytic activity of P. aeruginosa, another QS-related phenotype, was studied in this work. It was observed that ibuprofen and ciprofloxacin alone considerably reduced the hemolytic activity of P. aeruginosa. However, the combination of the drugs showed the strongest inhibition of bacterial hemolysins. Previous studies reported that treatment of P. aeruginosa with tenoxicam decreased the production of hemolysins which can be due to the anti-QS property of NSAIDs (Askoura et al. 2020). Also, it was reported that diclofenac could decrease hemolysin production by P. aeruginosa and S. aureus by 84 and 66%, respectively (Abbas et al. 2020; Queiroz 2021). In this regard, the synergic effect of ibuprofen and ciprofloxacin in reducing bacterial hemolytic activity could be considered an important anti-virulence feature of the combination therapy.
Many infective diseases, including bacterial infections, cause abnormal accumulation of a variety of inflammatory cells, cytokines, and destructive enzymes which result in local tissue damage and infection development (Čulić et al 2001). Neutrophile dominated inflammatory response is the most common cause of tissue damage in bacterial infections (Čulić et al. 2001). Therefore, host immune responses play both anti-infective and pro-inflammatory roles during bacterial infection. It has been reported that anti-inflammatory treatment could be considered an adjuvent chemotherapy approach for P. aeruginosa infections, especially in cystic fibrosis patients (Chmiel et al. 2013). Owing to the specific activity against neutrophiles, ibuprofen has received considerable attention in the treatment of P. aeruginosa infection, including in cystic fibrosis patients that the infection could be converted to a mucoid and persistence phenotype (Chmiel et al. 2013). Therefore, adjuvant chemotherapy with anti-inflammatory drugs, including ibuprofen could increase the chance of successful treatment of the infection. However, an efficient and therapeutic dose of ibuprofen needs to be established for each patient to gain an efficient therapeutic regime and avoid undesirable effects of the drug (Konstan 2008).
Conclusion
We evaluate the effect of ibuprofen as a widely used NSAID alone and in combination with ciprofloxacin on the expression of alginate synthesis and efflux pump genes of P. aeruginosa. Also, the synergism of ibuprofen and ciprofloxacin on some virulence traits, including biofilm formation, swimming and swarming, and hemolysis of P. aeruginosa was investigated. Our results showed that the combination of ibuprofen and ciprofloxacin significantly reduced bacterial biofilm formation, hemolysin production, swarming and twitching motility. Also, the expression of alginate synthesis genes was significantly reduced, while the mexB-oprM genes were upregulated. Due to the structural similarity to the fluoroquinolones and anti-QS feature of ibuprofen, it could be considered a suitable candidate for the adjuvant chemotherapy of P. aeruginosa infection with ciprofloxacin. However, further in-vivo and ex-vivo experiments are still required.
Data availability
The all related data were included in the manuscript.
References
Abbas HA, Atallah H, El-Sayed MA, El-Ganiny AM (2020) Diclofenac mitigates virulence of multidrug-resistant Staphylococcus aureus. Arch Microbiol 202:2751–2760. https://doi.org/10.1007/s00203-020-01992-y
Askoura M, Mottawea W, Abujamel T, Taher I (2011) Efflux pump inhibitors (EPIs) as new antimicrobial agents against Pseudomonas aeruginosa. Libyan J Med 6:5870
Askoura M, Saleh M, Abbas H (2020) An innovative role for tenoxicam as a quorum sensing inhibitor in Pseudomonas aeruginosa. Arch Microbiol 202:555–565. https://doi.org/10.1007/s00203-019-01771-4
Bajaksouzian S, Visalli MA, Jacobs MR, Appelbaum PC (1997) Activities of levofloxacin, ofloxacin, and ciprofloxacin, alone and in combination with amikacin, against acinetobacters as determined by checkerboard and time-kill studies. Antimicrob Agent Chemother 41:1073–1076. https://doi.org/10.1128/AAC.41.5.1073
Bassetti M, Vena A, Croxatto A, Righi E, Guery B (2018) How to manage Pseudomonas aeruginosa infections. Drugs Context 7:18
Chen L, Wen Y (2011) The role of bacterial biofilm in persistent infections and control strategies. Int J Oral Sci 3:66–73. https://doi.org/10.4248/IJOS11022
Chmiel JF, Konstan MW, Elborn JS (2013) Antibiotic and anti-inflammatory therapies for cystic fibrosis. Cold Spring Harb Perspect Med 3(10):a009779. https://doi.org/10.1101/cshperspect.a009779
CLSI (2021) Performance standards for antimicrobial susceptibility testing; CLSI supplement M100, 31st edn. Clinical and Laboratory Standards Institute, Malvern
Čulić O, Eraković V, Parnham MJ (2001) Anti-inflammatory effects of macrolide antibiotics. Eur J Pharmacol 429(1–3):209–229. https://doi.org/10.1016/S0014-2999(01)01321-8
Dai L, Wu T-q, Xiong Y-s, Ni H-b, Ding Y, Zhang W-c, Chu S-p, S-q Ju, Yu J (2019) Ibuprofen-mediated potential inhibition of biofilm development and quorum sensing in Pseudomonas aeruginosa. Life Sci 237:116947. https://doi.org/10.1016/j.lfs.2019.116947
Das S, Dash HR (2014) Microbial biotechnology—a laboratory manual for bacterial systems. Springer, New Delhi
de Almeida FA, Vargas ELG, Carneiro DG, Pinto UM, Vanetti MCD (2018) Virtual screening of plant compounds and nonsteroidal anti-inflammatory drugs for inhibition of quorum sensing and biofilm formation in Salmonella. Microb Pathog 121:369–388. https://doi.org/10.1016/j.micpath.2018.05.014
del Prado G, Ruiz V, Naves P, Rodríguez-Cerrato V, Soriano F, del Carmen PM (2010) Biofilm formation by Streptococcus pneumoniae strains and effects of human serum albumin, ibuprofen, N-acetyl-l-cysteine, amoxicillin, erythromycin, and levofloxacin. Diagn Microbiol Infect Dis 67:311–318. https://doi.org/10.1016/j.diagmicrobio.2010.03.016
Demirag MK, Esen S, Zivalioglu M, Leblebicioglu H, Keceligil HT (2007) The effect of aspirin on adherence of slime-producing, coagulase-negative staphylococci to vascular grafts. Ann Vasc Surg 21:464–467. https://doi.org/10.1016/j.avsg.2006.06.006
Elvers K, Wright S (1995) Antibacterial activity of the anti-inflammatory compound ibuprofen. Lett Appl Microbiol 20:82–84
Hooper D, Wolfson J, Ng E, Swartz M (1987) Mechanisms of action of and resistance to ciprofloxacin. Am J Med 82:12–20
Kahlous NA, Bawarish MAM, Sarhan MA, Küpper M, Hasaba A, Rajab M (2017) Using chemoinformatics, bioinformatics, and bioassay to predict and explain the antibacterial activity of nonantibiotic Food and Drug Administration drugs. Assay Drug Dev Technol 15:89–105
Karatuna O, Yagci A (2010) Analysis of quorum sensing-dependent virulence factor production and its relationship with antimicrobial susceptibility in Pseudomonas aeruginosa respiratory isolates. Clin Microbiol Infect 16:1770–1775. https://doi.org/10.1111/j.1469-0691.2010.03177.x
Konstan MW (2008) Ibuprofen therapy for cystic fibrosis lung disease: revisited. Curr Opin Pulm Med 14(6):567–573. https://doi.org/10.1097/MCP.0b013e32831311e8
Laudy AE, Mrowka A, Krajewska J, Tyski S (2016) The influence of efflux pump inhibitors on the activity of non-antibiotic NSAIDS against gram-negative rods. PLoS ONE 11:e0147131. https://doi.org/10.1371/journal.pone.0147131
LeBel M (1988) Ciprofloxacin: chemistry, mechanism of action, resistance, antimicrobial spectrum, pharmacokinetics, clinical trials, and adverse reactions. Pharmacotherapy 8:3–30
Lee J-H, Kim Y-G, Cho MH, Kim J-A, Lee J (2012) 7-fluoroindole as an antivirulence compound against Pseudomonas aeruginosa. FEMS Microbiol Lett 329:36–44. https://doi.org/10.1111/j.1574-6968.2012.02500.x
Moradali MF, Rehm BH (2019) The role of alginate in bacterial biofilm formation. Extracellular sugar-based biopolymers matrices. Springer, Cham, pp 517–537
Moradali MF, Donati I, Sims IM, Ghods S, Rehm BH (2015) Alginate polymerization and modification are linked in Pseudomonas aeruginosa. Mbio 6:e00453-e515. https://doi.org/10.1128/mBio.00453-15
Naves P, Del Prado G, Huelves L, Rodriguez-Cerrato V, Ruiz V, Ponte M, Soriano F (2010) Effects of human serum albumin, ibuprofen and N-acetyl-L-cysteine against biofilm formation by pathogenic Escherichia coli strains. J Hosp Infect 76:165–170. https://doi.org/10.1016/j.jhin.2010.05.011
Obad J, Šušković J, Kos B (2015) Antimicrobial activity of ibuprofen: new perspectives on an “Old” non-antibiotic drug. Eur J Pharmaceutic Sci 71:93–98. https://doi.org/10.1016/j.ejps.2015.02.011
Pearson JP, Van Delden C, Iglewski BH (1999) Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J Bacteriol 181:1203–1210. https://doi.org/10.1128/JB.181.4.1203-1210.1999
Queiroz HA (2021) Avaliação in vitro da atividade antibacteriana do diclofenaco sódico frente às cepas de Staphylococcus aureus resistente a meticilina (SARM)
Rasamiravaka T, Labtani Q, Duez P, El Jaziri M (2015) The formation of biofilms by Pseudomonas aeruginosa: a review of the natural and synthetic compounds interfering with control mechanisms. BioMed Res Int. https://doi.org/10.1155/2015/759348
Rashid MH, Kornberg A (2000) Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc Natl Acad Sci USA 97:4885–4890
Rehman A, Patrick WM, Lamont IL (2019) Mechanisms of ciprofloxacin resistance in Pseudomonas aeruginosa: new approaches to an old problem. J Med Microbiol 68(1):1–10. https://doi.org/10.1099/jmm.0.000873
She P, Wang Y, Luo Z, Chen L, Tan R, Wang Y, Wu Y (2018) Meloxicam inhibits biofilm formation and enhances antimicrobial agents efficacy by Pseudomonas aeruginosa. MicrobiologyOpen 7:e00545. https://doi.org/10.1002/mbo3.545
Soheili V, Bazzaz BSF, Abdollahpour N, Hadizadeh F (2015) Investigation of Pseudomonas aeruginosa quorum-sensing signaling system for identifying multiple inhibitors using molecular docking and structural analysis methodology. Microb Pathog 89:73–78. https://doi.org/10.1016/j.micpath.2015.08.017
Ulusoy S, Bosgelmez-Tinaz G (2013) Nonsteroidal anti-inflammatory drugs reduce the production of quorum sensing regulated virulence factors and swarming motility in human pathogen Pseudomonas aeruginosa. Drug Res 63:409–413. https://doi.org/10.1055/s-0033-1343430
Wood LF, Ohman DE (2015) Cell wall stress activates expression of a novel stress response facilitator (SrfA) under σ22 (AlgT/U) control in Pseudomonas aeruginosa. Microbiol 161:30. https://doi.org/10.1099/mic.0.081182-0
Acknowledgements
The author would like to thank the University of Guilan for provision of the facilities to carry out this work.
Funding
No funding was received for this study.
Author information
Authors and Affiliations
Contributions
Sample collection and experiments: SK, FG. Experiment design: HZ. Data analyses: HZ, SK, FG. Manuscript drafting: HZ, SK.Manuscript revision: HZ.
Corresponding author
Ethics declarations
Conflict of interest
The authors of the manuscript have no conflict of interest and competing interests to declare.
Ethical approval
Not applicable.
Research involving human and animal participants
This article does not contain any studies with human participants or animal performed by any of the authors.
Consent to publish
Not applicable.
Consent to participate
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Khodaparast, S., Ghanbari, F. & Zamani, H. Evaluation of the effect of ibuprofen in combination with ciprofloxacin on the virulence-associated traits, and efflux pump genes of Pseudomonas aeruginosa. World J Microbiol Biotechnol 38, 125 (2022). https://doi.org/10.1007/s11274-022-03316-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-022-03316-2