Abstract
Alginates are natural exopolysaccharides produced by seaweeds and bacteria belonging to the genera Pseudomonas and Azotobacter. These natural polymers are important polymeric substances contributing to the formation and development of biofilm matrixes of numerous bacteria enhancing persistence under environmental stresses, antibiotic treatment, and the immune system. Studying bacterial alginates have gained substantial attentions not only for their importance in bacterial pathogenesis but also regarding their biotechnological production for various industrial purposes. The biosynthesis of alginate is unique and has been extensively studied in the opportunistic human pathogen P. aeruginosa. This chapter will present updated data about bacterial production of alginate, its biological function, biosynthesis pathway, and regulation.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
1 An Overview of Bacterial Alginate Discovery
Alginates were first discovered and extracted from seaweeds by E.C.C. Stanford, an English chemist, in 1883 (Stanford 1883). Since this discovery, algal alginates have been extensively characterized for understanding their chemistry and properties, so they have been harnessed greatly for various industrial applications for more than a century. For the first time, Sonnenschein in 1927 reported encapsulated strains of P. aeruginosa (Sonnenschein 1927). Many years later in 1964, alginate production by encapsulated strains of P. aeruginosa was reported by Linker and Jones (Linker and Jones 1964). By analysis of sputum of cystic fibrosis (CF) patients, they found that Pseudomonas species formed unusually large mucoid colonies by producing polysaccharides, which chemically resembled alginic acids (Linker and Jones 1964). Later in 1966, they reported that alginate from these isolates was acetylated contrary to algal alginates, while acetyl groups were removed during alkaline extraction (Linker and Jones 1966). Then, Gorin and Spencer reported that alginic acid was also produced by another bacterial species Azotobacter vinelandii (Gorin and Spencer 1966). In 1973, the slimy polysaccharide produced by P. aeruginosa was reported as consisting of β-1,4-linked D-mannuronic acid residues and variable amounts of its 5-epimer L-guluronic acid (Evans and Linker 1973). Between 1967 and 1976, multiple reports considered strong association of mucoid strains of P. aeruginosa with chronic pulmonary infections in CF patients (Reynolds et al. 1976; Wood et al. 1976; Elston and Hoffman 1967; Doggett 1969). In 1981, other Pseudomonas species including P. fluorescens, P. putida, and P. mendocina were introduced as other alginate-producing bacteria (Govan et al. 1981; Müller and Monte Alegre 2007). The first report of immunogenic properties of alginate from P. aeruginosa was reported in 1983 by Pier et al. (Pier et al. 1983). Later in 1984, Sherbrock-Cox et al. reported the chemical characterization of P. aeruginosa alginate (Sherbrock-Cox et al. 1984). According to this report, alginate produced by P. aeruginosa mainly consisted of random or poly(D-mannuronic acid) block structures while being highly acetylated (Sherbrock-Cox et al. 1984). Between 1985 and 1987, bacterial alginate was profoundly studied in the context of bacterial pathogenesis. In this regard, serial investigations demonstrated alginate binding to the surface of epithelial cells, and structural diversity in P. aeruginosa alginates was thought to be important for this binding (McEachran and Irvin 1985; Ramphal and Pier 1985; Doig et al. 1987).
Studies on the genetic background of alginate production and mucoid strains date back to 1966, when Doggett et al. understood that the mucoid phenotype of P. aeruginosa associated with the CF lung emerges from nonmucoid forms and becomes the predominant population during infection over time (Doggett et al. 1966). Later, others showed that mucoid and nonmucoid strains are the variants of the same strain (Diaz et al. 1970; Williams and Govan 1973). Between 1975 and 1978, various explanations were provided for genetic background of alginate production, and some suggested that the ability of alginate production is transferrable between strains of P. aeruginosa via conjugation (Govan 1976; Fyfe and Govan 1978). In 1980, Fyfe and Govan reported that at least one chromosomal locus is involved in alginate production by bacteria (Fyfe and Govan 1980) followed by introducing chromosomally clustered Alg loci (Ohman and Chakrabarty 1981).
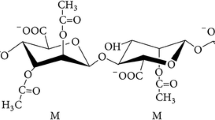
2 Chemical Structure and Physiochemical Properties of the Alginates
Alginates are unbranched anionic polysaccharides produced by brown seaweeds and bacteria belonging to Pseudomonas and Azotobacter genera. Uronic acid residues including β-D-mannuronic acid (M) and its C5 epimer α-L-guluronic acid (G) build up alginate polymer via 1, 4-glycosidic bonds (Fig. 13.1). In nature, alginates are usually found in heteropolymeric forms containing variable numbers and lengths of M blocks, G blocks, MG blocks, and non-acetylated and acetylated residues (Rehm and Valla 1997; Douthit et al. 2005; Remminghorst and Rehm 2006b; Moradali et al. 2015) (Fig. 13.1). Algal alginates are non-acetylated chains, while bacterial alginates have been found highly acetylated (Skjåk-Bræk et al. 1989). The occurrence of G blocks in algal alginates is common, while Pseudomonas alginates lack G blocks contrary to Azotobacter alginates (Gimmestad et al. 2009; Hay et al. 2014; Peteiro 2018; Moradali et al. 2018).
Chemical composition and natural occurrence of various alginates produced by seaweeds and bacteria. Alginate chains efficiently and selectively bind different divalent cations resulting in cross-linked polymeric scaffold and hydrogel formation (Haug and Smidsrod 1970)
Like other polymeric substances, physiochemical properties of the alginates are determined by their composition and molecular mass (Rehm 2010). Alginates were found to exhibit viscoelastic properties (Webber and Shull 2004; Moresi et al. 2004; Moradali et al. 2015). Furthermore, the most important feature of alginates is their ability to efficiently and selectively bind divalent cations, leading to the formation of hydrogels and cross-linked polymeric scaffolds (Mørch et al. 2006) (Fig. 13.1). The intrinsic flexibility of alginates depends on the frequency of constituting blocks as flexibility decreases in the order MG block > MM block > GG block (Meng and Liu 2013; Moradali et al. 2018). The affinity of alginates toward different divalent ions was found to increase in the order Mg2+ <<Mn2+ <Ca2+ <Sr2+ <Ba2+ <Cu2+ <Pb2+; however, the strength, dimension, stability, and mechanical properties of resulting hydrogels differ based on the type of interacting cation, the G content, and the variability of G blocks in polymers (Haug and Smidsrod 1967, 1970; Ouwerx et al. 1998; Sikorski et al. 2007).
The acetylation of alginates occurring as O-acetyl groups notably changes the properties of the alginates by impacting polymer conformation, chain expansion, solubility, water-binding capacity, viscoelasticity, and molecular mass (Moradali et al. 2015). An acetylated alginate absorbs more water due to the better interaction of side chains with water molecules, leading to chain expansion and better solubility (Delben et al. 1982; Skjåk-Bræk et al. 1989; Straatmann et al. 2004).
3 Alginates in Bacterial Biofilms
Biofilms are defined as cellular aggregations surrounded by self-produced polymeric substances and exist as both monospecies and polyspecies cellular communities in a variety of habitats (Costerton et al. 1987; Høiby 2017) (Fig. 13.2). Indeed, adopting the biofilm lifestyle versus planktonic (freestyle) mode provides a survival advantage by which the microorganism can thrive in harsh conditions, stresses, antibiotic treatments, and the immune system (Costerton 1999; Mah and O’Toole 2001; Stewart and William Costerton 2001). Therefore, eradicating biofilms remains a challenge, and a much higher concentration of a killing agent is required to kill biofilm cells compared to planktonic cells (Lewis 2001; Mah and O’Toole 2001; Davies 2003).
Schematic process of biofilm formation by P. aeruginosa with temporal and spatial variation in production of Psl, Pel, and alginate exopolysaccharides (Ghafoor et al. 2011; Colvin et al. 2012; Jennings et al. 2015). To keep the figure simple, each polysaccharide is attributed to distinct cell population
P. aeruginosa is a well-known biofilm former as well as a well-studied alginate producer. It is an opportunistic human pathogen affecting immunocompromised patients and the leading cause of morbidity and mortality in CF patients (Moradali et al. 2017a). Extracellular polymeric substances including polysaccharides, proteins, extracellular DNA (eDNA), and lipids entangle within the biofilm matrix, which is a niche rendering bacteria for intense cell-cell interaction and communication, protecting cells from unfavorable conditions, as well as a reservoir of metabolic substances, nutrients, and energy for fostering growth (Flemming and Wingender 2010; Strempel et al. 2013; Moradali et al. 2017a).
The exopolysaccharides Psl, Pel, and alginate are major constituents of the P. aeruginosa biofilm matrix (Fig. 13.2). They are interacting with other small molecules and macromolecules such as rhamnolipid surfactants and eDNA, determining surface adhesion, biofilm initiation, maturation, and architecture (Davey et al. 2003; Ghafoor et al. 2011; Gellatly and Hancock 2013) (Fig. 13.2). While Pel and Psl are the primary structural polysaccharides in the biofilm of both nonmucoid and mucoid P. aeruginosa strains, alginate overproduction is responsible for the emerging excessive slimy or mucoid biofilm and predominantly occupies the bio-volume of biofilm matrix (Hentzer et al. 2001; Colvin et al. 2011, 2012; Ghafoor et al. 2011) (Fig. 13.2). The formation of mucoid biofilm by P. aeruginosa is the hallmark of chronic infections, and it is indicative of disease progression and long-term persistence (Boucher et al. 1997; Moradali et al. 2017a).
Indeed, the overproduction of alginate confers a highly structured architecture to the biofilms, which comprise microcolonies and so-called mushroom-like structures (Fig. 13.2), while it provides an effective protective layer against opsonophagocytosis, free radicals released from immune cells, and antibiotics used for treatment (Hentzer et al. 2001; Hay et al. 2009a; Strempel et al. 2013).
Likewise, P. putida colonizing the rhizosphere of a variety of plants harnesses alginate in combination with other exopolysaccharides in the structure of biofilm. It was demonstrated that alginate production by this species contributes to the formation of highly structured and developed biofilm mediating stress tolerance particularly under water-limiting conditions (Chang et al. 2007). Chang et al. suggested that alginate is produced by pseudomonads in response to water-limiting conditions to reduce the extent of water loss from biofilm residents (Chang et al. 2007). Alginate shields biofilm cells and maintains a hydrated microenvironment protecting cells from desiccation stress (Chang et al. 2007). Another study showed that alginate production by fluorescent pseudomonads was stimulated in response to high osmolarity and dehydration (Singh et al. 1992).
Recently, a study on the soil bacterium P. alkylphenolia demonstrated that an alginate-like exopolysaccharide biosynthesis gene cluster is necessary for the formation of biofilm aerial and multicellular structures (Lee et al. 2014). This species has the ability to grow in the presence of linear alkylphenols (C1–C5) (Mulet et al. 2015).
Alginate-like exopolysaccharides were also detected in the gel matrix of aerobic granular sludge and normal aerobic flocculent sludge which are complex microbial consortia. Lin et al. noticed that the granular or granular structures of these sludge particles were impacted by different compositions of the alginate-like exopolysaccharides (Lin et al. 2013). They found that aerobic granules contained significantly higher levels of G blocks and less MG blocks, while those derived from aerobic flocculent sludge had equal amounts of G and MG blocks (Lin et al. 2010, 2013). These sources of hydrogels and alginate-like materials have largely attracted researchers and companies to explore on how to recover and utilize these biopolymers from wastewater sludge (van der Hoek et al. 2016).
Our analysis and others showed that gene clusters homologous to alg gene cluster exist in the genomes of a few marine species of the genera Alcanivorax and Marinobacter , but if they are capable of producing alginates remains controversial (Manilla-Pérez et al. 2010; Lee et al. 2014).
4 Biosynthesis of Alginate
Our understanding of alginate biosynthesis is mainly based on the model organism P. aeruginosa and to less extend on A. vinelandii . For many years, understanding the biosynthesis of alginates has been of great importance for scientific community with regard to drug development for treatment of P. aeruginosa infections exacerbated by alginate overproduction as well as establishing the production of bacterial and tailor-made alginates.
Generally, alginate-producing bacteria have the same genetic elements in common for the biosynthesis of the alginates (Fig. 13.3), but they are mainly different at epimerization level and more likely controlling regulatory mechanisms. Genes required for alginate production are mainly clustered in bacterial genome. Except for algC, the genetic elements including algD, alg8 , alg44 , algK, algE (algJ), algG, algX, algL, algI, algJ (algV), algF, and algA are clustered within the alginate (alg) operon (Fig. 12.3).
Proposed model of alginate biosynthesis in bacteria. (a) Operonic organization of the genes encoding different proteins mediating different steps of alginate biosynthesis (Chitnis and Ohman 1993; Hay et al. 2014). (b) Sequential enzymatic events mediating the production of alginate precursor, GDP-mannuronic acid, from fructose-6-phosphate originating from TCA cycle (Narbad et al. 1988; Narbad A. et al. 1990; Rehm and Valla 1997). (c) Proposed model for the alginate biosynthesis/modification/secretion machinery complex and experimentally demonstrated protein-protein interactions (marked with white triangles) (Hay et al. 2012; Rehman et al. 2013; Moradali et al. 2015). This model shows alginate production is positively regulated by c-di-GMP binding to Alg44 which targets the catalytic site of Alg8 polymerase for activating the polymerization of alginate. MucR is proposed to specifically provide c-di-GMP pool in proximity of Alg44. Translocation of alginate across the periplasmic scaffold (AlgG/AlgX/AlgF/AlgI/AlgJ/AlgK) is coupled with modification events (i.e., acetylation and epimerization) (Jain et al. 2003; Robles-Price et al. 2004; Moradali et al. 2015). The alginate lyase, AlgL, is responsible for degrading misguided alginate accumulating in the periplasm (Bakkevig et al. 2005; Wang et al. 2016). Secretion of modified alginate is mediated by two interacting proteins AlgK and AlgE (Hay et al. 2010; Rehman et al. 2013). MucD protein links the complex with the posttranslational alginate regulatory network via an interaction with AlgX (Hay et al. 2012). OM outer membrane, CM cytoplasmic membrane, P(alg) promoter, TCA cycle the tricarboxylic acid cycle, MPI mannose-6-phosphate isomerase, PMM phosphomannomutase, MPG mannose-1-phosphate guanylyltransferase, GMD GDP-mannose 6-dehydrogenase
5 Biosynthesis of Alginate Precursor
The best understood part of alginate biosynthesis is the formation of the activated precursor GDP-mannuronic acid. A series of cytosolic enzymatic steps catalyze the formation of precursor (Fig. 13.3). Synthesis starts with the entry of six carbon substrates to the Entner-Doudoroff pathway (KDPG pathway), resulting in pyruvate, which is channeled toward the tricarboxylic acid (TCA) cycle, while oxaloacetate from the TCA cycle can be converted to fructose-6-phosphate via gluconeogenesis. Three alginate-specific enzymes including AlgA (phosphomannose isomerase/GDP-mannose) , AlgC (phosphomannomutase), and AlgD (GDP-mannose dehydrogenase) catalyze the four biosynthesis steps to convert fructose-6-phosphate to GDP-mannuronic acid (Chitnis and Ohman 1993; Rehm and Valla 1997; Hay et al. 2014). The final step catalyzed by AlgD is a key step in the control of the alginate pathway because GDP-D-mannose is channeled into this pathway and it is irreversibly converted to the precursor GDP-mannuronic acid, a step before alginate polymerization (Roychoudhury et al. 1989; Tavares et al. 1999) (Fig. 13.3).
6 Alginate Polymerization, Modification, and Secretion
In bacteria, alginate polymerization, modifications, and secretion are mediated by a membrane-spanning multiprotein complex in P. aeruginosa (Fig. 13.3). Briefly, this multiprotein complex involves (1) alginate-polymerizing unit (Alg8-Alg44); (2) a proposed periplasmic protein scaffold (Alg44-AlgG-AlgX-AlgK) protecting nascent alginate (polymannuronate) against lyase activity of periplasmic AlgL while translocating polymer across the periplasm aligned with modifications (i.e., epimerization (AlgG)/acetylation (AlgX)); and (3) secretion unit (AlgK-AlgE) responsible for secreting modified alginate across the outer membrane of bacteria (Hay et al. 2012; Rehman et al. 2013; Moradali et al. 2015, 2018). Other subunits including AlgI, AlgJ, and AlgF have been proposed as taking part in the integrity of periplasmic scaffold, while they were found necessary for the acetylation of alginate probably by providing acetylation precursor for terminal acetyltransferase AlgX (Franklin et al. 2004) (Fig. 13.3).
6.1 Polymerization of Alginate
Two interacting membrane-anchored proteins Alg8 (polymerase) and Alg44 (co-polymerase) form the alginate synthase complex catalyzing the formation of polymannuronate chain from activated precursor GDP-mannuronic acid (Fig.13.3). Alg8 is a glycosyltransferase belonging to the glycosyltransferase family 2 that catalyzes the transfer of a sugar molecule from an activated donor, i.e., GDP-mannuronic acid, to an acceptor molecule which is a growing carbohydrate chain. Polymerization mechanism was understood as requiring conformational change of Alg44 upon binding to the second messenger bis-(3′, 5′)-cyclic dimeric guanosine monophosphate (c-di-GMP). In other words, the activation of alginate polymerization is essentially regulated through sensing c-di-GMP at posttranslational level (Remminghorst and Rehm 2006a; Merighi et al. 2007; Oglesby et al. 2008; Remminghorst et al. 2009). Alg44 consists of a cytoplasmic c-di-GMP-sensing PilZ domain, a transmembrane region extending into the periplasm of the bacteria while interacting with other periplasmic subunits (Remminghorst and Rehm 2006a; Oglesby et al. 2008) (Fig. 13.3). Studies based on protein-protein interaction, protein purification, and crystallization proposed that Alg44 is a dimer binding dimeric c-di-GMP toward activating polymerization (Whitney et al. 2015; Moradali et al. 2017b). Our attempt to understand the activation mechanism of alginate polymerization showed that the c-di-GMP binding to Alg44 targets the catalytic sites of Alg8 probably by inducing a conformational change via involving the engagement of some highly conserved amino acid residues of Alg8 at two predicted loops surrounding the catalytic site of Alg8 (Moradali et al. 2017b).
6.2 Modification of Alginate
Modification of polymers by producers aims at gaining physicochemical properties corresponding to their biological function in a given environment. Modification of alginate in bacteria comprises epimerization and acetylation of the polymannuronate chain. While enzymatic modifications occur, both AlgG and AlgX as well as likely AlgF/I/J constitute the proposed periplasmic scaffold, which translocates and guides alginate across the periplasm (Fig. 12.3). Upon epimerization of nascent alginate specifically mediated by the periplasmic AlgG epimerase, M residues are converted to G residues, leading to the formation of polyMG alginate (Jain et al. 2003; Gimmestad et al. 2003). Acetylation is mediated by terminal acetyltransferase AlgX by which O-acetyl ester linkages are added at the C2 or C3 position of M residues, resulting in acetylated alginates (Franklin and Ohman 2002; Baker et al. 2014) (Figs. 13.1 and 13.3).
Unmodified polymannuronate alginates are fairly a stiff polymer which is probably unfavorable for required flexibility in emerging matrix (Smidsrød et al. 1973). This is due to the nature of di-equatorial linkages of M blocks, while equatorial-axial bond of MG block increases the flexibility of the chain (Smidsrød et al. 1973). On the other hand, alginate produced by Pseudomonas species was found lacking G blocks as AlgG cannot generate two consecutive G residues. The presence of G blocks increases cross-linked chains and gelling properties of alginates in the presence of divalent cations such as Ca2+ (Fig. 13.1). The formation of G blocks in A. vinelandii alginate is mediated by other types of mannuronan C-5-epimerases of the so-called Ca2+-dependent AlgE-type, which act extracellularly in a Ca2+-dependent manner (Haug and Larsen 1969; Campa et al. 2004; Gimmestad et al. 2006). The G blocks have been commonly found in algal alginates living in the oceans and coastal areas and also within the coat of dormant cysts of A. vinelandii known as a nitrogen-fixing and soil bacterium (Clementi 1997; Peteiro 2018). This indicates that this type of modification contributes to tolerating physical and mechanical stresses. The acetylation of bacterial alginate increases the interaction of polymers with water and causes chain expansion (Skjåk-Bræk et al. 1989; Moradali et al. 2015).
Furthermore, we found that alginate polymerization and modifications are linked processes. Using various mutants of P. aeruginosa complemented with different combinations of catalytically active and inactive variants of alginate-polymerizing and alginate-modifying genes (i.e., alg8 , alg44 , algG, and algX), we uncovered that AlgG and AlgX had mutually auxiliary role within the multiprotein complex and Alg44 regulates acetylation event beside its association with polymerization step (Moradali et al. 2015, 2017b). In addition, we noticed that the degree of alginate polymerization reflecting the molecular mass is determined by modification events, where the degree of polymerization showed a negative correlation with the degree of epimerization and a positive correlation with the degree of acetylation. Also, we noticed that the epimerization process did interfere with the processivity of alginate polymerization, leading to the formation of shorter chains, while acetylation did not (Moradali et al. 2015). This may explain why alginates produced by P. aeruginosa possess much higher molecular mass compared with algal alginates. Hence, this study resulted in the production of various alginates including those chemically characterized as acetylated polyMG, non-acetylated polyMG, acetylated polyM, and non-acetylated polyM.
The impact of resulting various alginates on the development of biofilms was also examined. We found that the biofilm architecture of P. aeruginosa and viscoelastic property of alginates were remarkably affected by the various alginates. Generally, viscoelastic property is a key physicochemical parameter to assess the behavior of materials under given condition, and it is determined by measuring solid-like elastic modulus (G’) and the liquid-like viscous modulus (G”) . Our analysis showed that all resulting viscoelastic alginates displayed the solid-like elastic modulus (G’) greater than the liquid-like viscous modulus (G”), indicating they had greater elasticity than viscosity. We found out that while molecular mass is a key factor to determine viscoelastic property, the influence of the modification of alginates was also a striking determinant. Here, the presence of acetyl groups lowered viscoelasticity by possibly interfering with intermolecular alginate chain interactions. However increasing molar fractions of M blocks and resulting higher molecular masses increased viscoelasticity of the alginates (Moradali et al. 2015). Regarding biofilm examination, genetically modified P. aeruginosa mutants producing merely acetylated alginates formed well-developed biofilms with highly organized heterogeneous architectures through promoted cell aggregations, when compared with those biofilms formed by the mutant producing non-acetylated alginate. As acetylated alginate displayed much lower viscoelasticity than non-acetylated alginate, we concluded that in the course of biofilm formation, acetyl groups of alginates may function as a signal for cell aggregation regardless of low viscoelastic property (Moradali et al. 2015).
Also, resultant alginates without G residues and acetyl groups (i.e., non-acetylated polyM) caused the formation of undeveloped and narrow microcolonies, which were supported by specific long trails or strips of cells emerging from stigmergic self-organization behavior (Moradali et al. 2015).
6.2.1 Alginate Lyases
The periplasmic alginate lyase AlgL is encoded within the alginate operon in P. aeruginosa (Jain and Ohman 2005) (Fig.13.3). This enzyme belongs to the family of alginate lyases or alginate degrading enzymes which act as either mannuronate or guluronate lyases, but due to their structural diversity, they have not been classified yet (Wong et al. 2000; Ertesvåg 2015). Interestingly, these alginate-modifying enzymes have a wide natural occurrence including in algae (but not brown algae), marine invertebrates, and terrestrial microorganisms (Wong et al. 2000; Ertesvåg 2015). Wide natural distribution of alginate lyases implicate their possible role in digesting alginates for utilizing them as carbon source, while no bacterial alginate producer has been found to be an alginate utilizer. Presumably, in bacteria, alginate lyases play other biological roles. Bacterial AlgL was demonstrated to be essential for degrading misguided alginate trapped in the periplasm, leading to releasing free uronic acid oligomers to avoid the lethal effect of accumulated alginate on cells (Bakkevig et al. 2005) (Fig. 13.3). We showed that AlgL in P. aeruginosa is functionally associated with existing alginate biosynthesis/modification/secretion multiprotein complex (Wang et al. 2016). Such a critical role in the biosynthesis of bacterial alginate is well-understood within various bacterial mutants defective in the production of proposed scaffold forming subunits, i.e., AlgK, AlgX, or AlgG, that destabilize the multiprotein complex. Subsequently, the translocation of alginate across bacterial envelope was supposed to be misguided into the periplasm, leading to AlgL-mediated alginate degradation and the production of unsaturated oligouronides (Jain and Ohman 1998; Jain et al. 2003; Gimmestad et al. 2003; Robles-Price et al. 2004) (Fig. 13.3). We demonstrated that chromosomal expression of algL in P. aeruginosa enhanced alginate O-acetylation and both attachment and dispersal stages of the bacterial biofilm lifecycle were sensitive to the level of O-acetylation (Wang et al. 2016). Alginate lyases have been attractive and promising to study for developing therapeutics in combination with antibiotics in order to eradicate alginate-based biofilms (Alkawash et al. 2006; Lamppa and Griswold 2013).
6.3 Secretion of Alginate
The mechanism of alginate secretion is well-understood in P. aeruginosa . Two interacting proteins AlgK and AlgE, respectively, localized in the periplasm and the outer membrane, mediate alginate secretion across the outer membrane (Whitney et al. 2011; Rehman et al. 2013) (Fig. 13.3). AlgE acts selectively for the secretion of the negatively charged alginate polymer upon possessing a highly electropositive pore constriction formed by an arginine-rich channel (Rehm et al. 1994; Hay et al. 2010; Whitney et al. 2011). On the other hand, the lipoprotein AlgK facilitates proper localization of AlgE at the outer membrane (Keiski et al. 2010). This protein possesses a tetratricopeptide repeat (TPR) protein-protein interaction motif possibly mediating the interaction of AlgK with other subunits of the multiprotein complex (Rehman et al. 2013; Moradali et al. 2015) (Fig. 13.3).
7 Multitier Regulation of Alginate Biosynthesis
The biosynthesis of bacterial alginate is under the control of a complex regulatory network acting at transcriptional, posttranscriptional, and posttranslational levels (Hay et al. 2014) (Fig. 13.4). At transcriptional level, the alternate sigma factor AlgU (previously called AlgT or σ22) plays as the master regulator of alginate biosynthesis, which positively acts on algD promoter. AlgU belongs to the extracytoplasmic function (ECF) family of sigma factors, which are known to be involved in conferring resistance to a wide range of envelope stresses. Recently, it is understood that stimulation of AlgU for cell envelope homeostasis overlaps with controlling pathways for biofilm formation (Wood and Ohman 2012). Under uninduced condition, the anti-sigma factor MucA protein binds to AlgU at the inner membrane and sequesters its activity (Fig. 13.4). The genes encoding AlgU and MucA are clustered with other important chromosomal genes including mucB, mucC, and mucD, and this operon is known as the switch locus for alginate biosynthesis. Multiple studies showed that mutations in this operon are frequent among clinical strains of P. aeruginosa leading to the inactivation of MucA. Consequently, AlgU activates the expression of the alg operon (Fig. 13.4) and its own operon (Boucher et al. 1997; Firoved et al. 2002). It also activates the expression of several other genes involved in alginate biosynthesis and regulation such as algR, algB, algD, algC, and amrZ (Boucher et al. 1997; Mathee et al. 1999; Firoved et al. 2002; Wozniak et al. 2003; Muhammadi and Ahmed 2007).
Schematic major regulatory networks controlling the biosynthesis of alginate and biofilm formation by P. aeruginosa . Key determinants for alginate biosynthesis and overproduction of alginate include elevation of c-di-GMP and releasing AlgU sigma factor as a result of inactivation of MucA (blue rectangle with red cross) by either genetic mutation or proteolysis via the RIP cascade (Boucher et al. 1997; Firoved et al. 2002; Remminghorst and Rehm 2006a; Wood and Ohman 2012; Moradali et al. 2017b). The RIP cascade is thought to be negatively regulated by MucD which also interacts with the alginate biosynthesis multiprotein complex (Damron and Yu 2011). Various proteins localized in the envelope of the cells where these proteins form two-component systems (brown/green rectangles), chemoreceptor-like system (orange complex) and other protein networks sense environmental cues (Moradali et al. 2017a). Either triggered as phosphorylation cascades (small red circle labelled with “P”) or protein-protein interactions, the signals induce diguanylate cyclases (containing GGDEF motif) such as WspR and MucR to synthesize c-di-GMP from two molecules of GTP (guanosine-5′-triphosphate) (Güvener and Harwood 2007; Hay et al. 2009b). Consequently, c-di-GMP-sensing proteins such as Alg44 act as receptor/effector for specific outputs such as induction of alginate and Pel polymerization, inhibition of motility and derepression of psl/pel expression via FleQ protein, induction of attachment, and biofilm formation/maturation (Lee et al. 2007; Baraquet et al. 2012; Li et al. 2012; Moradali et al. 2017b). Gray arrows indicate inductive effect of proteins or molecules, and red blunt headed arrows indicate suppressive effects. The two-component systems are interconnected, and the LadS/RetS/GacS/GacA/RsmA regulatory network (green rectangles) plays a key role in phenotypic switch from motility to biofilm (Brencic et al. 2009; Chambonnier et al. 2016). RIP regulated intramembrane proteolysis, OM outer membrane, CM cytoplasmic membrane
On the other hand, MucA is protected by the periplasmic protein MucB from proteolysis triggered via regulated intramembrane proteolysis (RIP). The RIP signaling cascade involves several proteases and proteolytic events activated by envelope stresses negatively affecting localization or folding of the membrane proteins. The degradation of periplasmic domain of MucA via the RIP cascade and the subsequent release of AlgU represent another regulatory layer of alginate overproduction in response to inducing stresses (Alba et al. 2002; Qiu et al. 2007; Cezairliyan and Sauer 2009; Wood and Ohman 2009) (Fig. 13.4).
It was shown that the alginate biosynthesis multiprotein complex links to the RIP cascade via a periplasmic serine protease and chaperone-like protein called MucD which forms a complex with AlgX (Hay et al. 2012). MucD was proposed to negatively regulate the RIP cascade by chaperoning and/or degrading misfolded membrane proteins that would otherwise activate the RIP cascade (Fig. 13.4). One study proposed that MucD may be sequestered by stable alginate biosynthesis multiprotein complex, while induced destabilization in the protein complex results in releasing MucD from it (Yorgey et al. 2001; Qiu et al. 2007; Damron and Yu 2011; Hay et al. 2012).
Studying clinical isolates of P. aeruginosa with mucoid phenotype unraveled another regulatory pathway for the overproduction of alginate via mutations in the mucA gene which seems frequently observed in these strains. Resulting defective MucA is readily prone to proteolytic degradation as its defective periplasmic domain cannot interact with MucB for being protected, leading to AlgU release (Reiling et al. 2005; Hay et al. 2014). The mutation of mucA is one of the key adaptive mutations in regulatory genes which occur during proposed adaptive radiation of P. aeruginosa over the course of persistent infections in CF patients. By definition, during adaptive radiation initial infecting strains diversify into a variety of genotypes and phenotypes over time until the most favorable and adapted descendants are selected for long-term persistence. Hence, biofilm formation, alginate overproduction, and mucoid phenotype are part of this phenotypic adaptation process (Higgins et al. 2003; Hogardt and Heesemann 2010, 2013; Rau et al. 2010; Winstanley et al. 2016).
The regulation of alginate biosynthesis at transcriptional level via AlgU activity and the regulation of algD promoter overlap or link with other regulatory pathways mainly based on other sigma factors. Hay et al. provided a comprehensive overview on these interconnected regulatory pathways at transcriptional level (Hay et al. 2014).
At posttranscriptional level, a complex regulatory system consists of RsmA/Y/Z complex controls alginate biosynthesis (Fig. 13.4). RsmA is considered as a global regulator acting upon binding to mRNAs and negatively controls biofilm formation pathways such as via inhibition of the elevation of c-di-GMP levels (Irie et al. 2010; Jimenez et al. 2012). Two noncoding RNAs, RsmY and RsmZ, bind to RsmA and counteract its translational repression activity, consequently derepressing the translation of the genes such as involved in biofilm formation, increasing c-di-GMP level, and exopolysaccharides production (Fig. 13.4). This signaling pathway is also under the control of the GacS/A two-component system where in response to external stimuli, a phosphorylated GacA directly induces the synthesis of RsmY and RsmZ noncoding RNAs (Bhagirath et al. 2017; Allsopp et al. 2017; Li et al. 2017) (Fig. 12.4). However, the regulation of alginate biosynthesis at posttranscriptional level is not limited to this RsmA/Y/Z and GacS/A pathways as recently it was shown that AlgU and also AlgR from the two-component transcriptional regulator AlgZ/R control RsmA synthesis, alginate biosynthesis, and several other genes (Stacey et al. 2017).
At posttranslational level, c-di-GMP is a key signal in the regulation of biofilm formation by which timely expression of many genes is controlled (Fig. 13.4). This ubiquitous second messenger present in a wide range of bacteria and principally controls motility-biofilm switch (Römling et al. 2013; Moradali et al. 2017a). Indeed, the cellular level of c-di-GMP is the major determinant for this substantial phenotypic alteration, so that its elevation triggers biofilm formation while inhibiting motility. At least 40 enzymes directly synthesize and/or degrade c-di-GMP in P. aeruginosa which controls cellular level of this small molecule in response to perceived stimuli (Ryan et al. 2006). Previously, we demonstrated that alginate polymerization is impacted by cellular level of c-di-GMP via binding to Alg44 (Moradali et al. 2017b). One particular c-di-GMP-synthesizing protein, MucR, was demonstrated to specifically enhance the level of alginate production in P. aeruginosa, presumably by generating a localized c-di-GMP pool in proximity to the alginate polymerase (Hay et al. 2009b; Moradali et al. 2015, 2017b; Wang et al. 2015) (Fig. 12.4). Likewise, there are many other c-di-GMP receptor proteins which act as effectors for enhancing required pathways for biofilm formation including the production of alginate, Pel, and Psl while they inhibit the expression of motility-relevant genes such as those involved in flagella biosynthesis (Baraquet et al. 2012; Moradali et al. 2017a) (Fig. 13.4).
8 Concluding Remarks
Studying biopolymers constituting the biofilm matrix has gained substantial attention for many years. For more than a century, algal alginates have been of particular interests primarily for their unique physicochemical properties and wide industrial applications. However, the discovery of alginates in mucoid strains of P. aeruginosa and its direct impact on the development of chronic respiratory infections have further increased their importance for human affairs. In contrast to algal alginates, many aspects of the biosynthesis of bacterial alginates have now been unraveled. It is now well-known that microbial communities, either medically or environmentally relevant, harness specific material properties of alginates in response to their environment. Although much has been learned about physicochemical properties, applications, and the biosynthesis of alginates over the last few decades, there are still several key questions mainly about cognate multitier regulation and cross talks among various cellular regulators involved in alginate biosynthesis. In addition, persistent alginate-based biofilms in chronic respiratory infections are still a huge challenge for clinical settings and therapeutic development.
Reporting alginate-like polysaccharides surrounding microbial consortia in recent studies indicated that there are still many possibilities for discovering novel alginates and novel producers in different habitats. Studying novel alginate-producing bacteria from different habitats and comparing cognate regulatory networks would shed light on how environmental cues stimulate the production of various alginates by the bacterial community. On the other hand, understanding molecular mechanisms driving alginate biosynthesis in bacteria would establish a foundation for possible biotechnological production of tailor-made alginates as well as develop therapeutics to interfere with the formation of pathogenic alginate-based biofilms.
References
Alba BM, Leeds JA, Onufryk C, Lu CZ, Gross CA (2002) DegS and YaeL participate sequentially in the cleavage of RseA to activate the σ(E)-dependent extracytoplasmic stress response. Genes Dev 16:2156–2168
Alkawash MA, Soothill JS, Schiller NL (2006) Alginate lyase enhances antibiotic killing of mucoid Pseudomonas aeruginosa in biofilms. APMIS 114:131–138
Allsopp LP, Wood TE, Howard SA, Maggiorelli F, Nolan LM, Wettstadt S, Filloux A (2017) RsmA and AmrZ orchestrate the assembly of all three type VI secretion systems in Pseudomonas aeruginosa. Proc Natl Acad Sci USA 114:7707–7712
Baker P, Ricer T, Moynihan PJ, Kitova EN, Walvoort MT, Little DJ, Whitney JC, Dawson K, Weadge JT, Robinson H, Ohman DE, Codée JD, Klassen JS, Clarke AJ, Howell PL (2014) P. aeruginosa SGNH hydrolase-like proteins AlgJ and AlgX have similar topology but separate and distinct roles in alginate acetylation. PLoS Pathog 10(8):e1004334
Bakkevig K, Sletta H, Gimmestad M, Aune R, Ertesvåg H, Degnes K, Christensen BE, Ellingsen TE, Valla S (2005) Role of the Pseudomonas fluorescens alginate lyase (AlgL) in clearing the periplasm of alginates not exported to the extracellular environment. J Bacteriol 187:8375–8384
Baraquet C, Murakami K, Parsek MR, Harwood CS (2012) The FleQ protein from Pseudomonas aeruginosa functions as both a repressor and an activator to control gene expression from the pel operon promoter in response to c-di-GMP. Nucleic Acids Res 40:7207–7218
Bhagirath AY, Somayajula D, Li Y, Duan K (2017) CmpX affects virulence in Pseudomonas aeruginosa through the Gac/Rsm signaling pathway and by modulating c-di-GMP levels. J Membr Biol 251(1):35–49
Boucher JC, Yu H, Mudd MH, Deretic V (1997) Mucoid Pseudomonas aeruginosa in cystic fibrosis: characterization of muc mutations in clinical isolates and analysis of clearance in a mouse model of respiratory infection. Infect Immun 65:3838–3846
Brencic A, McFarland KA, McManus HR, Castang S, Mogno I, Dove SL, Lory S (2009) The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol Microbiol 73:434–445
Campa C, Holtan S, Nilsen N, Bjerkan TM, Stokke BT, Skjåk-Braek G (2004) Biochemical analysis of the processive mechanism for epimerization of alginate by mannuronan C-5 epimerase AlgE4. Biochem J 381:155–164
Cezairliyan BO, Sauer RT (2009) Control of P. aeruginosa AlgW protease cleavage of MucA by peptide signals and MucB. Mol Microbiol 72:368–379
Chambonnier G, Roux L, Redelberger D, Fadel F, Filloux A, Sivaneson M, de Bentzmann S, Bordi C (2016) The hybrid histidine kinase LadS forms a multicomponent signal transduction system with the GacS/GacA two-component system in Pseudomonas aeruginosa. PLoS Genet 12(5):e1006032
Chang W-S, van de Mortel M, Nielsen L, Nino de Guzman G, Li X, Halverson LJ (2007) Alginate production by Pseudomonas putida creates a hydrated microenvironment and contributes to biofilm architecture and stress tolerance under water-limiting conditions. J Bacteriol 189:8290–8299
Chitnis CE, Ohman DE (1993) Genetic analysis of the alginate biosynthetic gene cluster of Pseudomonas aeruginosa shows evidence of an operonic structure. Mol Microbiol 8:583–593
Clementi F (1997) Alginate production by Azotobacter Vinelandii. Crit Rev Biotechnol 17:327–361
Colvin KM, Gordon VD, Murakami K, Borlee BR, Wozniak DJ, Wong GCL, Parsek MR (2011) The Pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PLoS Pathog 7(1):e1001264
Colvin KM, Irie Y, Tart CS, Urbano R, Whitney JC, Ryder C, Howell PL, Wozniak DJ, Parsek MR (2012) The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ Microbiol 14:1913–1928
Costerton JW (1999) Introduction to biofilm. Int J Antimicrob Agents 11:217–221
Costerton JW, Cheng KJ, Geesey GG, Ladd TI, Nickel JC, Dasgupta M, Marrie TJ (1987) Bacterial biofilms in nature and disease. Annu Rev Microbiol 41:435–464
Damron FH, Yu HD (2011) Pseudomonas aeruginosa MucD regulates the alginate pathway through activation of MucA degradation via MucP proteolytic activity. J Bacteriol 193:286–291
Davey ME, Caiazza NC, O’Toole GA (2003) Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J Bacteriol 185:1027–1036
Davies D (2003) Understanding biofilm resistance to antibacterial agents. Nat Rev Drug Discov 2:114
Delben F, Cesaro À, Paoletti S, Crescenzi V (1982) Monomer composition and acetyl content as main determinants of the ionization behavior of alginates. Carbohydr Res 100:C46–C50
Diaz E, Mosovich LL, Neter E (1970) Serogroups of Pseudomonas aeruginosa and the immune response of patients with cystic fibrosis. J Infect Dis 121:269–274
Doggett RG (1969) Incidence of mucoid Pseudomonas aeruginosa from clinical sources. Appl Microbiol 18:936–937
Doggett RG, Harrison GM, Stillwell NR, Wallis ES (1966) An atypical Pseudomonas aeruginosa associated with cystic fibrosis of the pancreas. J Pediatr 68:215–221
Doig P, Smith NR, Todd T, Irvin RT (1987) Characterization of the binding of Pseudomonas aeruginosa alginate to human epithelial cells. Infect Immun 55:1517–1522
Douthit SA, Dlakic M, Ohman DE, Franklin MJ (2005) Epimerase active domain of Pseudomonas aeruginosa AlgG, a protein that contains a right-handed beta-helix. J Bacteriol 187:4573–4583
Elston HR, Hoffman KC (1967) Increasing incidence of encapsulated Pseudomonas aeruginosa strains. Am J Clin Pathol 48:519–523
Ertesvåg H (2015) Alginate-modifying enzymes: biological roles and biotechnological uses. Front Microbiol 6:523. https://doi.org/10.3389/fmicb.2015.00523
Evans LR, Linker A (1973) Production and characterization of the slime polysaccharide of Pseudomonas aeruginosa. J Bacteriol 116:915–924
Firoved AM, Boucher JC, Deretic V (2002) Global genomic analysis of AlgU (ςsgr;(E))-dependent promoters (sigmulon) in Pseudomonas aeruginosa and implications for inflammatory processes in cystic fibrosis. J Bacteriol 184:1057–1064
Flemming H-C, Wingender J (2010) The biofilm matrix. Nat Rev Microbiol 8:623–633
Franklin MJ, Ohman DE (2002) Mutant analysis and cellular localization of the AlgI, AlgJ, and AlgF proteins required for O acetylation of alginate in Pseudomonas aeruginosa. J Bacteriol 184:3000–3007
Franklin MJ, Douthit SA, McClure MA (2004) Evidence that the algI/algJ gene cassette, required for O acetylation of Pseudomonas aeruginosa alginate, evolved by lateral gene transfer. J Bacteriol 186:4759–4773
Fyfe JAM, Govan JRW (1978) A genetic approach to the study of mucoid Pseudomonas aeruginosa. Proc Soc Gen Microbiol 5:54
Fyfe JA, Govan JR (1980) Alginate synthesis in mucoid Pseudomonas aeruginosa: a chromosomal locus involved in control. J Gen Microbiol 119:443–450
Gellatly SL, Hancock REW (2013) Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathog Dis 67:159–173
Ghafoor A, Hay ID, Rehm BHA (2011) Role of exopolysaccharides in Pseudomonas aeruginosa biofilm formation and architecture. Appl Environ Microbiol 77:5238–5246
Gimmestad M, Sletta H, Ertesvåg H, Bakkevig K, Jain S, Suh S-j, Skjåk-Bræk G, Ellingsen TE, Ohman DE, Valla S (2003) The Pseudomonas fluorescens AlgG protein, but not its mannuronan C-5-epimerase activity, is needed for alginate polymer formation. J Bacteriol 185:3515–3523
Gimmestad M, Steigedal M, Ertesvåg H, Moreno S, Christensen BE, Espín G, Valla S (2006) Identification and characterization of an Azotobacter vinelandii type I secretion system responsible for export of the AlgE-type mannuronan C-5-epimerases. J Bacteriol 188:5551–5560
Gimmestad M, Ertesvåg H, Heggeset TM, Aarstad O, Svanem BI, Valla S (2009) Characterization of three new Azotobacter vinelandii alginate lyases, one of which is involved in cyst germination. J Bacteriol 191:4845–4853
Gorin P, Spencer J (1966) Exocellular alginic acid from Azotobacter vinelandii. Can J Chem 44:993–998
Govan JRW (1976) Genetic studies on mucoid Pseudomonas aeruginosa. Proc Soc Gen Microbiol 3:187
Govan JR, Fyfe JA, Jarman TR (1981) Isolation of alginate-producing mutants of Pseudomonas fluorescens, Pseudomonas putida and Pseudomonas mendocina. J Gen Microbiol 125:217–220
Güvener ZT, Harwood CS (2007) Subcellular location characteristics of the Pseudomonas aeruginosa GGDEF protein, WspR, indicate that it produces cyclic-di-GMP in response to growth on surfaces. Mol Microbiol 66:1459–1473
Haug A, Smidsrod O (1967) Strontium–calcium selectivity of alginates. Nature 215:757
Haug A, Larsen B (1969) Biosynthesis of alginate. Epimerisation of D-mannuronic to L-guluronic acid residues in the polymer chain. Biochim Biophys Acta 192:557–559
Haug A, Smidsrod O (1970) Selectivity of some anionic polymers for divalent metal ions. Acta Chem Scand 24:843–854
Hay ID, Gatland K, Campisano A, Jordens JZ, Rehm BHA (2009a) Impact of alginate overproduction on attachment and biofilm architecture of a supermucoid Pseudomonas aeruginosa strain. Appl Environ Microbiol 75:6022–6025
Hay ID, Remminghorst U, Rehm BH (2009b) MucR, a novel membrane-associated regulator of alginate biosynthesis in Pseudomonas aeruginosa. Appl Environ Microbiol 75:1110–1120
Hay ID, Rehman ZU, Rehm BH (2010) Membrane topology of outer membrane protein AlgE, which is required for alginate production in Pseudomonas aeruginosa. Appl Environ Microbiol 76:1806–1812
Hay ID, Schmidt O, Filitcheva J, Rehm BH (2012) Identification of a periplasmic AlgK-AlgX-MucD multiprotein complex in Pseudomonas aeruginosa involved in biosynthesis and regulation of alginate. Appl Microbiol Biotechnol 93:215–227
Hay ID, Wang Y, Moradali MF, Rehman ZU, Rehm BH (2014) Genetics and regulation of bacterial alginate production. Environ Microbiol 16:2997–3011
Hentzer M, Teitzel GM, Balzer GJ, Heydorn A, Molin S, Givskov M, Parsek MR (2001) Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J Bacteriol 183:5395–5401
Higgins PG, Fluit AC, Milatovic D, Verhoef J, Schmitz FJ (2003) Mutations in GyrA, ParC, MexR and NfxB in clinical isolates of Pseudomonas aeruginosa. Int J Antimicrob Agents 21:409–413
Hogardt M, Heesemann J (2010) Adaptation of Pseudomonas aeruginosa during persistence in the cystic fibrosis lung. Int J Med Microbiol 300:557–562
Hogardt M, Heesemann J (2013) Microevolution of Pseudomonas aeruginosa to a chronic pathogen of the cystic fibrosis lung. Curr Top Microbiol Immunol 358:91–118
Høiby N (2017) A short history of microbial biofilms and biofilm infections. APMIS 125:272–275
Irie Y, Starkey M, Edwards AN, Wozniak DJ, Romeo T, Parsek MR (2010) Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and post-transcriptionally by RsmA. Mol Microbiol 78:158–172
Jain S, Ohman DE (1998) Deletion of algK in mucoid Pseudomonas aeruginosa blocks alginate polymer formation and results in uronic acid secretion. J Bacteriol 180:634–641
Jain S, Ohman DE (2005) Role of an alginate lyase for alginate transport in mucoid Pseudomonas aeruginosa. Infect Immun 73:6429–6436
Jain S, Franklin MJ, Ertesvåg H, Valla S, Ohman DE (2003) The dual roles of AlgG in C-5-epimerization and secretion of alginate polymers in Pseudomonas aeruginosa. Mol Microbiol 47:1123–1133
Jennings LK, Storek KM, Ledvina HE, Coulon C, Marmont LS, Sadovskaya I, Secor PR, Tseng BS, Scian M, Filloux A, Wozniak DJ, Howell PL, Parsek MR (2015) Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc Natl Acad Sci USA 112:11353–11358
Jimenez PN, Koch G, Thompson JA, Xavier KB, Cool RH, Quax WJ (2012) The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol Mol Biol Rev 76:46–65
Keiski CL, Harwich M, Jain S, Neculai AM, Yip P, Robinson H, Whitney JC, Riley L, Burrows LL, Ohman DE, Howell PL (2010) AlgK is a TPR-containing protein and the periplasmic component of a novel exopolysaccharide secretin. Structure 18:265–273
Lamppa JW, Griswold KE (2013) Alginate lyase exhibits catalysis-independent biofilm dispersion and antibiotic synergy. Antimicrob Agents Chemother 57:137–145
Lee VT, Matewish JM, Kessler JL, Hyodo M, Hayakawa Y, Lory S (2007) A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol Microbiol 65:1474–1484
Lee K, Lim EJ, Kim KS, Huang S-L, Veeranagouda Y, Rehm BHA (2014) An alginate-like exopolysaccharide biosynthesis gene cluster involved in biofilm aerial structure formation by Pseudomonas alkylphenolia. Appl Microbiol Biotechnol 98:4137–4148
Lewis K (2001) Riddle of biofilm resistance. Antimicrob Agents Chemother 45:999–1007
Li K, Yang G, Debru AB, Li P, Zong L, Xu T, Wu W, Jin S, Bao Q (2017) SuhB regulates the motile-sessile switch in Pseudomonas aeruginosa through the Gac/Rsm pathway and c-di-GMP signaling. Front Microbiol 8:1045. https://doi.org/10.3389/fmicb.2017.01045
Li Z, Chen JH, Hao Y, Nair SK (2012) Structures of the PelD cyclic diguanylate effector involved in pellicle formation in Pseudomonas aeruginosa PAO1. J Biol Chem 287:30191–30204
Lin Y, de Kreuk M, van Loosdrecht MCM, Adin A (2010) Characterization of alginate-like exopolysaccharides isolated from aerobic granular sludge in pilot-plant. Water Res 44:3355–3364
Lin YM, Sharma PK, van Loosdrecht MCM (2013) The chemical and mechanical differences between alginate-like exopolysaccharides isolated from aerobic flocculent sludge and aerobic granular sludge. Water Res 47:57–65
Linker A, Jones RS (1964) A polysaccharide resembling alginic acid from a Pseudomonas microorganism. Nature 204:187–188
Linker A, Jones RS (1966) A new polysaccharide resembling alginic acid isolated from pseudomonads. J Biol Chem 241:3845–3851
Mah T-FC, O’Toole GA (2001) Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol 9:34–39
Manilla-Pérez E, Reers C, Baumgart M, Hetzler S, Reichelt R, Malkus U, Kalscheuer R, Wältermann M, Steinbüchel A (2010) Analysis of lipid export in hydrocarbonoclastic bacteria of the genus Alcanivorax: identification of lipid export-negative mutants of Alcanivorax borkumensis SK2 and Alcanivorax jadensis T9. J Bacteriol 192:643–656
Mathee K, Ciofu O, Sternberg C, Lindum PW, Campbell JI, Jensen P, Johnsen AH, Givskov M, Ohman DE, Molin S, Høiby N, Kharazmi A (1999) Mucoid conversion of Pseudomonas aeruginosa by hydrogen peroxide: a mechanism for virulence activation in the cystic fibrosis lung. Microbiology 145:1349–1357
McEachran DW, Irvin RT (1985) Adhesion of Pseudomonas aeruginosa to human buccal epithelial cells: evidence for two classes of receptors. Can J Microbiol 31:563–569
Meng S, Liu Y (2013) Alginate block fractions and their effects on membrane fouling. Water Res 47:6618–6627
Merighi MT, Lee V, Hyodo M, Hayakawa Y, Lory S (2007) The second messenger bis-(3′-5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol Microbiol 65:876–895
Moradali FM, Donati I, Sims IM, Ghods S, Rehm BH (2015) Alginate polymerization and modification are linked in Pseudomonas aeruginosa. MBio 6(3):e00453–e00415
Moradali MF, Ghods S, Rehm BH (2017a) Pseudomonas aeruginosa lifestyle: a paradigm for adaptation, survival, and persistence. Front Cell Infect Microbiol 7:39. https://doi.org/10.3389
Moradali MF, Ghods S, Rehm BHA (2017b) Activation mechanism and cellular localization of membrane-anchored alginate polymerase in Pseudomonas aeruginosa. Appl Environ Microbiol 83(9):e03499–e03416
Moradali MF, Ghods S, Rehm BHA (2018) Alginate biosynthesis and biotechnological production. In: Rehm BHA, Moradali MF (eds) Alginates and their biomedical applications. Springer series in biomaterials science and engineering, vol 11. Springer, Singapore
Moresi M, Bruno M, Parente E (2004) Viscoelastic properties of microbial alginate gels by oscillatory dynamic tests. J Food Eng 64:179–186
Muhammadi, Ahmed N (2007) Genetics of bacterial alginate: alginate genes distribution, organization and biosynthesis in bacteria. Curr Genomics 8:191–202
Mulet M, Sánchez D, Lalucat J, Lee K, García-Valdés E (2015) Pseudomonas alkylphenolica sp. Nov., a bacterial species able to form special aerial structures when grown on p-cresol. Int J Syst Evol Microbiol 65:4013–4018
Mørch ÝA, Donati I, Strand BL (2006) Effect of Ca2+, Ba2+, and Sr2+ on alginate microbeads. Biomacromolecules 7:1471–1480
Müller JM, Monte Alegre R (2007) Alginate production by Pseudomonas mendocina in a stirred draft fermenter. World J Microbiol Biotechnol 23:691–695
Narbad A, Russell NJ, Gacesa P (1988) Radiolabelling patterns in alginate of Pseudomonas aeruginosa synthesized from specifically-labelled 14C-monosaccharide precursors. Microbios 54:171–179
Narbad A, Gacesa P, Russell NJ (1990) Biosynthesis of alginate. In: Gacesa P, Russell NJ (eds) Pseudomonas infection and alginates. Springer, Dordrecht
Oglesby LL, Jain S, Ohman DE (2008) Membrane topology and roles of Pseudomonas aeruginosa Alg8 and Alg44 in alginate polymerization. Microbiology 154:1605–1615
Ohman DE, Chakrabarty AM (1981) Genetic mapping of chromosomal determinants for the production of the exopolysaccharide alginate in a Pseudomonas aeruginosa cystic fibrosis isolate. Infect Immun 33:142–148
Ouwerx C, Velings N, Mestdagh MM, Axelos MAV (1998) Physico-chemical properties and rheology of alginate gel beads formed with various divalent cations. Polym Gels Netw 6:393–408
Peteiro C (2018) Alginate production from marine macroalgae, with emphasis on kelp farming. In: Rehm BHA, Moradali MF (eds) Alginates and their biomedical applications. Springer series in biomaterials science and engineering, vol 11. Springer, Singapore
Pier GB, Matthews WJ, Eardley DD (1983) Immunochemical characterization of the mucoid exopolysaccharide of Pseudomonas aeruginosa. J Infect Dis 147:494–503
Qiu D, Eisinger VM, Rowen DW, Yu HD (2007) Regulated proteolysis controls mucoid conversion in Pseudomonas aeruginosa. Proc Natl Acad Sci USA 104:8107–8112
Ramphal R, Pier GB (1985) Role of Pseudomonas aeruginosa mucoid exopolysaccharide in adherence to tracheal cells. Infect Immun 47:1–4
Rau MH, Hansen SK, Johansen HK, Thomsen LE, Workman CT, Nielsen KF, Jelsbak L, Høiby N, Yang L, Molin S (2010) Early adaptive developments of Pseudomonas aeruginosa after the transition from life in the environment to persistent colonization in the airways of human cystic fibrosis hosts. Environ Microbiol 12:1643–1658
Rehm BH (2010) Bacterial polymers: biosynthesis, modifications and applications. Nat Rev Microbiol 8:578–592
Rehm BH, Valla S (1997) Bacterial alginates: biosynthesis and applications. Appl Microbiol Biotechnol 48:281–288
Rehm BH, Boheim G, Tommassen J, Winkler UK (1994) Overexpression of algE in Escherichia coli: subcellular localization, purification, and ion channel properties. J Bacteriol 176:5639–5647
Rehman ZU, Wang Y, Moradali MF, Hay ID, Rehm BH (2013) Insights into the assembly of the alginate biosynthesis machinery in Pseudomonas aeruginosa. Appl Environ Microbiol 79:3264–3272
Reiling SA, Jansen JA, Henley BJ, Singh S, Chattin C, Chandler M, Rowen DW (2005) Prc protease promotes mucoidy in mucA mutants of Pseudomonas aeruginosa. Microbiology 151:2251–2261
Remminghorst U, Rehm BH (2006a) Alg44, a unique protein required for alginate biosynthesis in Pseudomonas aeruginosa. FEBS Lett 580:3883–3888
Remminghorst U, Rehm BH (2006b) Bacterial alginates: from biosynthesis to applications. Biotechnol Lett 28:1701–1712
Remminghorst U, Hay ID, Rehm BH (2009) Molecular characterization of Alg8, a putative glycosyltransferase, involved in alginate polymerisation. J Biotechnol 140:176–183
Reynolds HY, Di Sant’Agnese PA, Zierdt CH (1976) Mucoid Pseudomonas aeruginosa. A sign of cystic fibrosis in young adults with chronic pulmonary disease? JAMA 236:2190–2192
Robles-Price A, Wong TY, Sletta H, Valla S, Schiller NL (2004) AlgX is a periplasmic protein required for alginate biosynthesis in Pseudomonas aeruginosa. J Bacteriol 186:7369–7377
Roychoudhury S, May TB, Gill JF, Singh SK, Feingold DS, Chakrabarty AM (1989) Purification and characterization of guanosine diphospho-D-mannose dehydrogenase. A key enzyme in the biosynthesis of alginate by Pseudomonas aeruginosa. J Biol Chem 264:9380–9385
Ryan RP, Fouhy Y, Lucey JF, Dow JM (2006) Cyclic di-GMP signaling in bacteria: recent advances and new puzzles. J Bacteriol 188:8327–8334
Römling U, Galperin MY, Gomelsky M (2013) Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77:1–52
Sherbrock-Cox V, Russell NJ, Gacesa P (1984) The purification and chemical characterisation of the alginate present in extracellular material produced by mucoid strains of Pseudomonas aeruginosa. Carbohydr Res 135:147–154
Sikorski P, Mo F, Skjåk-Bræk G, Stokke BT (2007) Evidence for egg-box-compatible interactions in calcium−alginate gels from fiber X-ray diffraction. Biomacromolecules 8:2098–2103
Singh S, Koehler B, Fett WF (1992) Effect of osmolarity and dehydration on alginate production by fluorescent pseudomonads. Curr Microbiol 25:335–339
Skjåk-Bræk G, Paoletti S, Gianferrara T (1989) Selective acetylation of mannuronic acid residues in calcium alginate gels. Carbohydr Res 185:119–129
Smidsrød O, Glover RM, Whittington SG (1973) The relative extension of alginates having different chemical composition. Carbohydr Res 27:107–118
Sonnenschein C (1927) Die mucosus form des Pyocyaneus-Bakteriums, bacterium pyocyaneum mucosum. Zentralblatt für Bakteriologie [Naturwiss] 104:365–373
Stacey SD, Williams DA, Pritchett CL (2017) The Pseudomonas aeruginosa two-component regulator AlgR directly activates rsmA expression in a phosphorylation independent manner. J Bacteriol 199(18):e00048–e00017
Stanford E (1883) On align: a new substance obtained from some of the commoner species of marine algae. Chem News 47:254–257
Stewart PS, William Costerton J (2001) Antibiotic resistance of bacteria in biofilms. Lancet 358:135–138
Straatmann A, Windhues T, Borchard W (2004) Effects of acetylation on thermodynamic properties of seaweed alginate in sodium chloride solutions. In: Lechner MD, Börger L (eds) Analytical ultracentrifugation VII. Springer, Berlin/Heidelberg, pp 26–30
Strempel N, Neidig A, Nusser M, Geffers R, Vieillard J, Lesouhaitier O, Brenner-Weiss G, Overhage J (2013) Human host defense peptide LL-37 stimulates virulence factor production and adaptive resistance in Pseudomonas aeruginosa. PLoS One 8(12):e82240
Tavares IM, Leitão JH, Fialho AM, Sá-Correia I (1999) Pattern of changes in the activity of enzymes of GDP-D-mannuronic acid synthesis and in the level of transcription of algA, algC and algD genes accompanying the loss and emergence of mucoidy in Pseudomonas aeruginosa. Res Microbiol 150:105–116
van der Hoek JP, de Fooij H, Struker A (2016) Wastewater as a resource: strategies to recover resources from Amsterdam’s wastewater. Resour Conserv Recycl 113(Suppl C):53–64
Wang Y, Hay ID, Rehman ZU, Rehm BH (2015) Membrane-anchored MucR mediates nitrate-dependent regulation of alginate production in Pseudomonas aeruginosa. Appl Microbiol Biotechnol 99:7253–7265
Wang Y, Moradali MF, Goudarztalejerdi A, Sims IM, Rehm BH (2016) Biological function of a polysaccharide degrading enzyme in the periplasm. Sci Rep 6:31249. https://doi.org/10.1038/srep31249
Webber RE, Shull KR (2004) Strain dependence of the viscoelastic properties of alginate hydrogels. Macromolecules 37:6153–6160
Whitney JC, Hay ID, Li C, Eckford PD, Robinson H, Amaya MF, Wood LF, Ohman DE, Bear CE, Rehm BH, Howell PL (2011) Structural basis for alginate secretion across the bacterial outer membrane. Proc Natl Acad Sci USA 108:13083–13088
Whitney JC, Whitfield GB, Marmont LS, Yip P, Neculai AM, Lobsanov YD, Robinson H, Ohman DE, Howell PL (2015) Dimeric c-di-GMP is required for post-translational regulation of alginate production in Pseudomonas aeruginosa. J Biol Chem 290:12451–12462
Williams RJ, Govan JR (1973) Pyocine typing of mucoid strains of Pseudomonas aeruginosa isolated from children with cystic fibrosis. J Med Microbiol 6:409–412
Winstanley C, O’Brien S, Brockhurst MA (2016) Pseudomonas aeruginosa evolutionary adaptation and diversification in cystic fibrosis chronic lung infections. Trends Microbiol 24:327–337
Wong TY, Preston LA, Schiller NL (2000) Alginate lyase: review of major sources and enzyme characteristics, structure-function analysis, biological roles, and applications. Annu Rev Microbiol 54:289–340
Wood LF, Ohman DE (2009) Use of cell wall stress to characterize sigma 22 (AlgT/U) activation by regulated proteolysis and its regulon in Pseudomonas aeruginosa. Mol Microbiol 72:183–201
Wood LF, Ohman DE (2012) Identification of genes in the σ(22) regulon of Pseudomonas aeruginosa required for cell envelope homeostasis in either the planktonic or the sessile mode of growth. MBio 3(3):e00094–e00012
Wood RE, Boat TF, Doershuk CF (1976) Cystic fibrosis. Am Rev Respir Dis 113:833–878
Wozniak DJ, Sprinkle AB, Baynham PJ (2003) Control of Pseudomonas aeruginosa algZ expression by the alternative sigma factor AlgT. J Bacteriol 185:7297–7300
Yorgey P, Rahme L, Tan MW, Ausubel F (2001) The roles of mucD and alginate in the virulence of Pseudomonas aeruginosa in plants, nematodes and mice. Mol Microbiol 41:1063–1076
Acknowledgments
The authors are grateful to the current and former member of the Rehm research group for their invaluable contributions providing insights into alginate biosynthesis in bacteria.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Moradali, M.F., Rehm, B.H.A. (2019). The Role of Alginate in Bacterial Biofilm Formation. In: Cohen, E., Merzendorfer, H. (eds) Extracellular Sugar-Based Biopolymers Matrices. Biologically-Inspired Systems, vol 12. Springer, Cham. https://doi.org/10.1007/978-3-030-12919-4_13
Download citation
DOI: https://doi.org/10.1007/978-3-030-12919-4_13
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-12918-7
Online ISBN: 978-3-030-12919-4
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)