Abstract
The innovative CRISPR–Cas based genome editing technology provides some functionality and advantages such as the high efficiency and specificity as well as ease of handling. Both aspects of the CRISPR–Cas9 system including genetic engineering and gene regulation are advantageously applicable to the construction of microbial cell factories. As one of the most extensively used cell factories, E. coli has been engineered to produce various high value-added chemical compounds such as pharmaceuticals, biochemicals, and biofuels. Therefore, to improve the production of valuable metabolites, many investigations have been performed by focusing on CRISPR–Cas- based metabolic engineering of this host. In the current review, the biology underlying CRISPR–Cas9 system was briefly explained and then the applications of CRISPR–Cas9/CRISPRi tools were considered for cell factory construction in E. coli.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Construction of optimal microbial factories engineered for the bio-production of a range of heterologous proteins or the enhanced production of native compounds, often requires knock-out, knock-down, and overexpression of multiple gene targets (Xu et al. 2020; Ziegler and Takors 2020; Zou et al. 2020). Although some useful genome engineering tools have been utilized for the past several decades, strain development is considered as a very time consuming process. Because one or a few genetic engineering steps are not usually applicable on achieving economically efficient cell factories, so tedious multi-step metabolic engineering methods are often required. Therefore, the development of simple and high-throughput methods for the targeted genome editing is of great importance for industrial biotechnology (Adiego-Pérez et al. 2019; Marcellin and Nielsen 2018; Zhan et al. 2019).
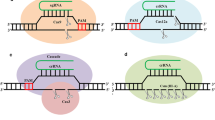
The essential first step in genome editing is the formation of DNA double-stranded break (DSB) at the target genomic region. Also, in early genome editing tools such as transcription activator–like effector nucleases (TALENs) and zinc finger nucleases (ZFNs), the targeted DSBs at specific genomic loci were induced by endonucleases with customized catalytic domains for the specific binding to target loci through DNA. The necessity of a new protein design in each experiment restricts the application of the previous methods. However, these approaches are still used in some experiments (Boch et al. 2009; Carroll 2011; Nandy et al. 2020). In contrast with ZFN and TALEN tools, in recently developed Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)–Cas system, one nuclease is able to target different specific loci in the genome and also to introduce DSBs, by the use of different specific small guide RNAs, which lead to the stimulation of DNA repair pathway by homolog-directed repair (HDR) or by non-homologous end joining (NHEJ) (Abdelaal and Yazdani 2020; Cho et al. 2018b). HDR and NHEJ are two pathways responsible for DSBs repair in most organisms. Although NHEJ is a major mechanism of mammalian cell DNA repair, it is a weak process in some prokaryotes including Mycobacterium, Pseudomonas and Bacillus subtilis and doesn’t exist in Escherichia coli (E. coli). The common rout of DSB repair for prokaryotes is HDR (Ebrahimi and Hashemi 2020; Trimidal et al. 2019). Upon cleavage by Cas9, in the absence of an appropriate repair template, NHEJ is able to efficiently introduce insertion/deletion mutations (indels) with various lengths. In this regard, if occur within a coding exon, indels can lead to gene knock-outs. On the other hand, in the presence of an exogenously supplied DNA, HDR-mediated repair use a donor template to insert the desired sequence at the target locus. In addition, homology arms to the DSB flanking region is necessary for the repair template (Fig. 1) (Pawelczak et al. 2018). Using the HDR pathway, CRISPR–Cas9 system can be exploited in gene knockout/in as well as in the introduction of precise point mutations (Liu et al. 2019). Also, due to the capability of introducing DSBs at multiple sites, several target genes can be modified by CRISPR–Cas system in a single transformation experiment. Therefore, this powerful targeted genome editing platform is known as a remarkably high specific, simple, efficient, and suitable platform for high- throughput gene editing in a wide range ofbiological systems (Koonin and Makarova 2019; Mougiakos et al. 2018; Song 2017).
This is the first study to specifically focus on E. coli as a microbial cell factory and the examples of how CRISPR–Cas-based technologies have been applied for the construction and improvement of this cell factory are reviewed. Finally, a selection of frequently used tools for computer-aided design of guide-RNAs with minimal off-target potentials is listed.
Biology of the CRISPR–Cas system
CRISPR–Cas system, as a three-step antiviral defense mechanism, was initially observed in the E. coli (Hille et al. 2018; Moon et al. 2019). At first, Cas proteins can integrate the short DNA spacers generated by invaders into a CRISPR array in the host genome. Secondly, CRISPR array composed of the adapted spacers and identical palindromic repeats, was transcribed into a long transcript called pre-CRISPR RNA (pre-crRNA). After the processing, mature small CRISPR RNAs (crRNAs) were generated from pre-crRNA and to form a duplex with complementary trans-activating RNAs (tracrRNAs). Thirdly, a crRNA–tracrRNA hybrid complex to guide the Cas9 nuclease to an invading DNA target, as a protospacer, resulted in the DSB formation, and finally to cleavage of foreign DNA. It is believed that, this immune response can destroy the genetic material of the invader through the foreign DNA cleavage and stopping the infection (Fig. 2) (Chen et al. 2020; Hashemi 2018; Singh 2020; Tian et al. 2017; Wright et al. 2016).
The naturally occurring CRISPR–Cas bacterial immunity. Short DNA spacers generated by invaders can be integrated into a CRISPR array in the host genome by naturally occurring CRISPR bacterial immunity. First CRISPR locus is transcribed to yield precursor crRNA (pre-crRNA) which is then processed into mature crRNAs. These mature crRNAs can form a duplex with complementary trans-activating RNAs (tracrRNAs). Finally, the gRNA-guided Cas protein can cleave foreign DNA or RNA
Depending on the comparative genomic, phylogenetic, and protein structural analyses, the CRISPR–Cas system can be divided into two classes (class 1 and class 2) (Shmakov et al. 2015). Accordingly, class 1 has a multisubunit effector complexes composed of multiple Cas proteins for mediating the interference activities against foreign DNA. In contrast with Class 1, interference in Class 2 was performed by a single, large, and multidomain Cas protein. In this regard, these classes can also be further sub divided into several types given the presence of a specific signature protein. In addition, class 2 is divided into various three types (type II, type V, and type VI) and many subtypes. In this Class, the type II CRISPR–Cas9 as well as type V CRISPR–Cas12a (Cpf1) are found as almost exclusively in bacteria, and then, they have been broadly used for genome engineering across microorganisms (Charpentier et al. 2019; Makarova et al. 2015). Notably, the gene clusters encoding the Cas proteins are located near the CRISPR array. Cas9 is known as the effector nuclease for type II system and can also be guided to target sequence in the genome denoted as a protospacer by the crRNA–tracrRNA complex (Van Der Oost et al. 2014). Indeed, in the 5′-end of the crRNA module, there is a 20-nucleotide guide sequence known as spacer that can form a duplex with protospacer. In order to be recognized by Cas9, protospacer should be flanked by protospacer adjacent motif (PAM), which is a specific short DNA motif on the 3′ end of the target. Moreover, Cas9 is stabilized via binding to the 3′ end of tracrRNA and besides, produces a DSB in the target sequence (Bortesi and Fischer 2015; Nishimasu et al. 2014). In this regard, Cas9 is originated from Streptococcus pyogenes, which requires a PAM composed of two guanines and one random nucleotide (NGG) for target selection (Geng et al. 2016). Cas9 derived from Treponema denticola, Neisseria meningitidus, and Streptococcus thermophilus require different PAMs such as 5′-NAAAAN-3′, 5′-NNNNGA/ CTT-3′, and 5′-NNAGAAW-3′, respectively. Also, for further simplification of the genome editing design, two separate functional sequences (crRNA and tracrRNA) can be connected by a linker to form a single synthetic guide RNA (sgRNA) (Hashemi 2018; Terns and Terns 2014).
CRISPR interference (CRISPRi)
In addition to gene deletion or integration, the precise control of gene expression is considered to be an important approach in metabolic engineering (Vigouroux and Bikard 2020). Although siRNA-mediated transcriptional regulation has long been utilized in eukaryotic systems, the dCas9 based CRISPR interference (CRISPRi) has recently provided such a silencing tool for prokaryotes (Ibrahim et al. 2019; Lunge et al. 2020). dCas9 is a catalytically inactive variant of the Cas9 endonuclease, which can only be connected to the location specified in the genome by sgRNA. In this regard, when specifically binding to genomic locus, dCas9 is able to block RNA polymerase (RNAP) sterically for progressing to the downstream gene (Chaikind et al. 2020; Schultenkämper et al. 2020). This simple interference system can, transiently or constitutively, suppress the expression of target genes (Deyell et al. 2019). Moreover, the strength of this repression can be precisely controlled through exploiting an inducible expression system for the expression of the sgRNA module or dCas9. Accordingly, this is more important when a basal expression level of the target gene is needed or when the accumulation of toxic intermediates should be prevented (McCarty et al. 2020; Vigouroux and Bikard 2020).
CRISPR–Cas9 mediated construction of E. coli cell factory
Up to now, the investigators have reported the efficiency of the CRISPR–Cas9 toolkit as an editing approach in diverse bacterial cells like E. coli, which is currently one of the most widely used cell factories for the bio-production of different enzymes, biofuels, pharmaceuticals, and biochemical (Choudhary et al. 2020; Das et al. 2020; Mitsui et al. 2019; Wang et al. 2020). Therefore, to improve the production of the valuable metabolites, many studies have been performed by focusing on the CRISPR–Cas- mediated metabolic engineering of this host (An et al. 2020; Cho et al. 2018b; Satowa et al. 2020) (Table 1). For example, a heterologous pathway for biosynthesis of β-carotene was integrated into the E. coli genome by the use of a combination of CRISPR-based method and λ-RED recombineering. Also, for overproduction of β-carotene, central metabolic and the methylerythritol-phosphate (MEP) pathways were modulated through several deletions as well as the promoter/RBS substitutions. β‐carotene, as an isoprenoid, is a red–orange pigment, which is widely used in nutraceutical and pharmaceutical industries due to having some properties such as antioxidant, anticancer, and anti‐inflammatory activities. β‐carotene has also been shown to have a preventive role against cardiovascular diseases. In this study, for exploring the metabolic landscape, more than 100 genetic variants were constructed. Consequently, 2 g L − 1 of β-carotene was produced in the developed E. coli with 15 mutations using a fed-batch culture. Moreover, Li et al. observed an increased editing efficiency in cells with an intact mismatch repair pathway in this investigation. Also, the great potential of the Cas9-based tools for efficient genome editing was revealed in this extensive study (Fig. 3) (Li et al. 2015). In another study, Liang et al. have reported the capability of the CRISPR-enabled trackable genome engineering (CREATE) tool that was developed in terms of the Cas9-recombineering method to construct 903 various isopropanol producing variants of E. coli. The best producing variant named as PA14, is able to produce 7.1 g/L of isopropanol during 24 h (1.5-fold higher than the initial strain). Moreover, the maximum volumetric productivity of isopropanol in PA14 was also improved markedly compared to the parent strain (0.22 g/L/h higher than the initial strain named as PA07) (Liang et al. 2017). Furthermore, CRISPR–Cas9 based genome editing was shown to be effective on n-butanol overproduction under the microaerobic condition in E. coli. Accordingly, for this purpose, the expression level of the genes involved in n-butanol synthesis has increased in the engineered E. coli. Moreover, gltA gene encoding citrate synthase was downregulated via CRISPR–Cas9 based on the modification of its 5′-untranslated region. In addition, this modification resulted in the effective redirection of carbon flux from acetyl-CoA to citric acid cycle toward acetoacetyl-CoA and mostly to the n-butanol production (Heo et al. 2017). Moreover, broad potential applications of 5-Aminolevulinic acid (ALA) has recently attracted much attention. In this regard, it can be used as a precursor for synthesizing several compounds such as heme, chlorophyl tetrapyrrole, cytochrome, and vitamin B12. Moreover, due to the low-yield of its traditional chemical synthesis methods, many studies have focused on inexpensive sources such as E. coli for its production. For this purpose, C4 pathway was introduced in E. coli based on CRISPR–Cas9 system for the production of ALA from glucose. The titer of ALA was further increased (from 20 to 689 mg/L) when hemA gene derived from Rhodobacter capsulatus was heterologously expressed in E. coli based on the engineering of its ribosome binding site. The most of the improvement in ALA production was observed via modification of coenzyme A, succinyl-CoA, and glycine biosynthesis pathways as well as the downregulation of hemB expression. Notably, the engineered E. coli named AAGSB-1 can produce ALA at a titer of 2.81 g/L in batch-fermentation (Ding et al. 2017). By combining the CRISPR–Cas9 system with λ-Red recombineering, the lipid content of the E. coli was efficiently enhanced in a research conducted by Xia et al. also, the strains with the inactivated phosphoenolpyruvate carboxylase were selected as well as fatty acid regulatory transcription factor (fadD) and fadR, Δ9 acyl-lipid desaturase (Δ9 desaturase), and acetyl-CoA carboxylase (acc) genes were simultaneously integrated into the genome. No change was observed in fatty acid composition between the recombinant strains and wild-type strains. All the recombinant strains had a higher total lipid content (nearly 5.3%) compared to the wild-type strain (Xia et al. 2016). The CRISPR–Cas9 system was also exploited for the production of amino acids in E. coli. The aromatic amino acid, named L-tyrosine (tyrosine) is extensively used in the nutraceutical and pharmaceutical industries. Also, for efficiently performing de novo biosynthesis of L-tyrosine, Wang et al. improved HGXP, as a L-tyrosine producing strain, in which tyrP and aroP genes encoding two distinct permeases that regulate the intracellular transport of L-tyrosine in E. coli were knocked-out by the CRISPR–Cas system. The results of the fermentation experiments revealed that, 3.45 and 3.74 g/L of L-tyrosine were produced by the tyrP and aroP knock-out mutants, respectively. Moreover, the final L-tyrosine yields of tyrP and aroP knock-out mutants obtained from a 3-L fermentor and under the optimized culture conditions, were further increased to 35.1 and 44.5 g/L, respectively (Wang et al. 2019). In another study, the parallel metabolic pathway engineering (PMPE) method was reported by Fujiwara et al. So, using this strategy, shikimate pathway derivatives can be achieved from the glucose–xylose co-substrate, which are known as two major components of lignocellulose. In this study, using the CRISPR–Cas two-plasmid system, a xylose catabolic pathway into E. coli was introduced to recover cell growth. Therefore, based on the PMPE strategy, the target chemical was mainly produced from glucose, whereas the essential metabolites for cell growth were supplied by xylose. They also produced cis, cis-muconic acid (MA) as well as L-tyrosine as the shikimate pathway derivatives to confirm the versatility of PMPE. Consequently, in the PMPE E. coli strain, MA was produced at a titer of 4.09 g/L and L-tyrosine was also achieved with 64% of the theoretical yield. In addition, they revealed that, the bio-production efficiency can be improved by the effective utilization of glucose–xylose co-substrate. Also, other value-added chemicals can be achieved from the lignocellulosic resources using the PMPE method (Fujiwara et al. 2020).
A metabolic engineering strategy for efficient formation of β-carotene in E. coli using CRISPR–Cas9 meditated genome editing. Schematic represents central metabolic pathways, MEP pathway, and β-carotene biosynthetic pathway. Genes tested for overexpression are shown in green and those tested for deletion are depicted in red. G6P, glucose-6-phosphate; FBP, fructose 1,6-bisphosphate; F6P, fructose-6-phosphate; DHAP, Dihydroxyacetone phosphate; 3PG, 3-phosphoglycerate; G3P, glyceraldehyde 3-phosphate; PEP, phosphoenolpyruvate; 6PG, 6-phosphogluconate; Pyr, pyruvate; X5P, xylulose-5-phosphate; E4P, erythrose-4-phosphate; R5P, ribose-5-phosphate; S7P, sedoheptulose-7-phosphate; 2-kg, α-oxoglutarate; KDPG, 2-keto-3-deoxy-6-phosphogluconate; OAA, oxaloacetate; Glu, glutamic acid; MEP, 2C-methyl-D-erythritol-4-phosphate; DXP, 1-deoxy-Dxylulose-5-phosphate; CDP-ME, 4-diphosphocytidyl-2C-methyl-D-erythritol; MEC, 2C-methyl-D-erythritol 2,4-cyclodiphosphate; GGPP, geranylgeranyl diphosphate; CDP-ME-2P, 4-diphosphocytidyl-2Cmethyl-D-erythritol-2-phosphate; HMBDP, 1-hydroxy-2-methyl-2-(E)-butenyl-4-diphosphate; DPP, dimethylallyl diphosphate; IPP, isopentenyl diphosphate; FPP, farnesyl diphosphate
CRISPRi mediated construction of E. coli cell factory
CRISPRi, as a promising tool for transcriptional regulation, is able to modulate the expression of target genes, constitutively or transiently, with no gene disruption. By considering the metabolic pathways, CRISPRi can repress the competing pathway genes, and thus, can direct flux toward the target product synthesis (Dong 2019; Dong et al. 2020; Tian et al. 2020; Wu et al. 2020). CRISPRi mediated metabolic engineering has also been successfully utilized in model organisms like E. coli (Dasgupta et al. 2020; Mougiakos et al. 2018) (Table 1). For example, poly (3-hydroxybutyrate-co-4-hydroxybutyrate) (P (3HB-co-4HB)) polymer was produced in E. coli by Lv et al. as follows. Firstly, they introduced the pathway responsible for the polyhydroxyalkanoate (PHA) biosynthesis from glucose into the E. coli. Also, for increasing the polymer level, using CRISPRi-based tool, they have downregulated the expression of several genes that are involved in TCA cycle and lead to an increase in the level of succinate semialdehyde, which is a precursor for 4HB synthesis (as the main fraction of the [P(3HB-co-4HB)] polymer) (Fig. 4) (Lv et al. 2015). Resveratrol (3, 5, 4′-trihydroxy-trans-stilbene) (RES) is a non-flavonoid polyphenol linked to a wide variety of functions such as anti‐inflammation, anticancer, and antioxidant properties. It has also been recognized that, the grapes, peanuts, and berries are rich in this polyphenol; however, its application has been hampered by an inefficient extraction process (Huminiecki and Horbańczuk 2018; Li et al. 2019b). So, different metabolic engineering approaches have been promoted for the efficient preparation of resveratrol in microbial cell factories. In this regard, Wu et al. have introduced a malonate assimilation pathway from Rhizobium trifolii into E. coli leading to an increase in the supply of malonyl-CoA, which is known as a key precursor for the synthesis of Resveratrol. Moreover, the malonyl-CoA consumption pathway was inactivated by the CRISPRi mediated downregulation of the fatty acid biosynthesis pathway. These genetic modifications also increased the final resveratrol titer to 304.5 mg/L. By the use of the CRISPRi strategy, Wu et al. developed a simple and efficient procedure for preparing resveratrol in a common cell factory (Wu et al. 2017b). Multiplex silencing was utilized via a sgRNA approach by most of these studies, whereas a dual RNA (crRNA/tracrRNA) approach named CRISPathBrick is a rapid CRISPR-array assembly method, which was used by Cress et al. Accordingly, using this strategy, the expression of plasmid-based genes was tuned and chromosomal targets in virulent, E. coli, and probiotic strains were repressed. So, CRISPathBrick provides multiplex CRISPRi-based silencing in organisms in which the genetic toolbox is limited (Cress et al. 2015). In addition, flavonoids have provided a valuable resource to be applied in human health as well as in their nutrition diet. Furthermore, they can be widely used as anti-obesity, antiviral, and anti-cancer agents. Malonyl-coenzyme A (malonyl-CoA) is the initial substrate for the production of the naringenin, which is a common precursor of most flavonoids. The biosynthesis of flavonoids is hampered by the limited amount of malonyl-CoA in E. coli, and using the CRISPRi system, Wu et al. overcame this limitation via fine-tuning of the central metabolic pathways. Accordingly, their results showed that, the intracellular level of malonyl-CoA can increase by over 223%. Moreover, no significant change was observed in the final biomass accumulation due to the efficient tuning of the target genes (less than 10% decrease in the final OD600). Finally, a high yield of naringenin (421.6 mg/L that was increased up to 7.4-fold compared to the parent strain) was also achieved via the CRISPRi based multiple gene repression (Wu et al. 2015). In another study, fine-tuning of a biosynthetic pathway based on the CRISPRi system could successfully direct the carbon flux toward the synthesis of the target products in E. coli engineered to harbor the plant-derived terpenoid synthases as well as the mevalonate (MVA) pathway. In this study, an efficient regulatable CRISPRi system has been developed for the suppression of the transcription of the acetoacetyl-CoA thiolase enzyme, which has been responsible for catalyzing the first step in the biosynthetic MVA pathway. Consequently, the production of lycopene (C40) has also increased as well as (-)-α-bisabolol (C15) as a result of this modulation (Kim et al. 2016). Also, spending on excess biomass formation, carbon and energy can limit the production of proteins or biochemicals in the microbial cell factories. So, for circumventing this problem, Li et al. controlled the growth of E. coli via CRISPRi based modulation of dnaA and oriC leading to the repression of the DNA replication machinery as well as the modulation of pyrF or thyA, which lead to nucleotide synthesis block. In this study, the production of biochemicals was decoupled from the growth and then led to an increase up to 41% in the mevalonate yield (Li et al. 2016). Moreover, in a study conducted by Kim et al., the expression level of endogenous genes was modulated in a multiplex manner by CRISPRi, as a tunable system. In this investigation, pta, ldhA, adhE, and frdA genes responsible for the formation of byproducts including acetate, lactate, ethanol, and succinate, were successfully repressed either individually or in double, triple, or quadruple combination. In addition, the reduced formation of byproducts (acetate, lactate, succinate, and ethanol) via multiplex CRISPRi led to an enrichment in acetyl-CoA supply and directed the carbon flux toward the enhanced n-butanol production. In this engineered cell factory, the productivity and yield of n-butanol have enhanced up to 3.2- and 5.4- fold compared to the parent strain, respectively. These data represent a successful CRISPRi-mediated approach for repressing the endogenous genes in a multiplex manner, which can be used for the rapid evaluation of multiplex interventions to develop some well-organized cell factories (Kim et al. 2017). Furthermore, in several studies, E. coli has been metabolically engineered by utilizing the CRISPR–Cas tool combined with the CRISPRi system. For example, for the production of 1,4-butanediol (1,4-BDO) in E. coli, at first, gltA was point mutated exploiting the CRISPR system,, native lpdA was replaced with heterologous lpdA, sad was knocked-out, and then two large gene cassettes encoding cat1, 4hbd, sucD, bld, cat2, and bdh in the 1,4-BDO biosynthesis pathway were inserted into the genome. This engineered E. coli was able to produce 0.9 g/L of 1,4-BDO in 2 d. The 1,4-BDO titer was further increased up to 1.8 g/L when the expression of the competing genes including gabD, tesB, and ybgC was repressed using the CRISPRi tool. This repression also led to a decrease in the formation of byproducts including succinate and gamma-butyrolactone. In this study, metabolic flux regulation was successfully performed in E. coli by the combined use of the CRISPR–Cas9 system with the CRISPRi tool (Wu et al. 2017c). Pinosylvin (trans-3,5-dihydroxystilbene) is considered as a promising pharmaceutical or nutraceutical due to having cardioprotective, antioxidative, anti-cancer, and anti-inflammatory properties. But unfortunately, it can be synthesized just in a very low amount in genus Pinus. So, a rational modular design approach was developed for the efficient biosynthesis of this medicinally important product in E. coli strain. Accordingly, in this strategy, using the CRISPRi system, the genes including aroFwt, TcPAL, and pheAfbr as well as 4-coumarate: coenzyme A ligase (4CL) and stilbene synthase (STS) involved in module I, were overexpressed. Besides, central metabolism-related genes (fabB/fabF, adhE, eno, fumC, and sucC) involved in module III were also repressed. This engineered E. coli was able to produce pinosylvin to a titer of 281 mg/L from d-glucose. Notably, some efficient microbial cell factories can be developed using this rational modular design approach for the production of several valuable chemicals (Wu et al. 2017a). In another study, Cress et al. introduced a plant anthocyanin synthesis pathway into E. coli to produce peonidin 3-O-glucoside (P3G) from an abundant precursor, named as the flavan-3-ol substrate ( +)-catechin. In this regard, the endogenous metabolites including S-adenosyl-l-methionine (SAM or AdoMet) and UDP-glucose are also required for this pathway. So, they exploited CRISPRi system to deregulate the methionine biosynthetic pathway resulting in the improvement of SAM availability for O-methylation of the biosynthetic precursor of P3G known as cyanidin 3-O-glucoside (C3G). Based on these modulation, P3G was successfully produced at a final titer of 51 mg/L. This valuable strategy can also be used for the production of the other O-methylated anthocyanin pigments from any methylated products (Cress et al. 2017). A systematic metabolic engineering based on the CRISPRi system was also reported for the production of D-pantothenic acid in E. coli W3110 as follows. Firstly, a titer of 0.49 g/L has been achieved by editing in several genes including pantothenate synthetase, acetohydroxy acid synthase II, 3-methyl-2-oxobutanoate, 2-dehydropantoate 2-reductase, hydroxymethyltransferase, and ketol-acid reductoisomerase. Afterward, the yield was further enhanced to 1.48 g/L by the repression of those genes involved in L-valine biosynthesis (which is a competing pathway to the D-pantothenic acid biosynthetic pathway). Pantothenate kinase mutagenesis as well as threonine deaminase deletion could increase the production titer to 1.78 g/L. Also, DPA-9/pTrc99a-panBC (C.G) strain produced D-pantothenic acid at a final titer of 28.45 g/L in the fed-batch fermentations (Zhang et al. 2019).
Application of CRISPRi for metabolic engineering of E. coli. Schematic represents the central metabolic and P(3HB-co-4HB) biosynthetic pathways. The deleted genes are shown in red. phaA, β-ketothiolase; phaB, NADPH-dependent acetoacetyl-CoA reductase; phaC, PHA synthase; sucD, succinate semi-aldehyde dehydrogenase; 4hbD, 4-hydroxybutyrate dehydrogenase; orfZ, CoA transferase; sdhA and sdhB encoding succinate dehydrogenase of E. coli; sucC and sucD, the succinyl-CoA synthetase of E. coli; sad and gabD, succinate semi-aldehyde dehydrogenase of E. coli
Computational tools supporting sgRNA design
Despite previous technologies, the CRISPR–Cas9 system enables an efficient genome editing in diverse cells and organisms. Obviously, the accurate and efficient targeting of the CRISPR–Cas9 tool to the desired location is required for this system. So, the correct identification of the optimal target-site and subsequent design of the complimentary gRNA are two main reasons for the success of an experiment. The on-target activity of an optimal gRNA should be maximum (efficiency) while its potential off-target effects (specificity) should be minimum (Wilson et al. 2018). Currently, efficiency and specificity of sgRNAs can be calculated by several online tools (Sledzinski et al. 2020). On-target efficiency of sgRNA is determined based on the position of the binding site within the gene model and the nucleotide composition of the binding site. In contrast with efficiency, because no specific phenotype can be detected in most of the off-targets, specificity of sgRNAs is more difficult to be predicted and current available data are not sufficient to predict all the off-targets bioinformatically (Alkhnbashi et al. 2020).
Nowadays, there are few available bioinformatics tools for designing single-guide RNAs used in bacteria. The same principles are followed by these tools. Firstly, candidate target regions are identified depending on the application (knock-out, activation or repression), the gene of interest, and the PAM of the utilized CRISPR–Cas9 system. The candidate target regions are then assessed based on low off-target activity as well as high on-target efficiency (Alkhnbashi et al. 2020). For example, CRISPR-ERA supports the sgRNA design in different organisms. So, sgRNA sequences needed for the CRISPRi mediated gene activation or repression can be generated using this program (https://CRISPR-era.stanford.edu) (Chuai et al. 2017; Liu et al. 2015). Furthermore, potential off-target cleavage sites can be predicted using sgRNAcas9 software (Xie et al. 2014). A selection of frequently used tools can be found in Table 2.
Challenges of CRISPR–Cas9 mediated gene editing
Despite its wide-spread use, CRISPR machinery faces major obstacles need to be solved (Ebrahimi and Hashemi 2020). CRISPR system application can be hampered by off-target DNA cleavage by Cas9 nuclease (Herai 2019). This ability is the result of the evolutional combat between bacteria and viruses in which bacteria Cas protein is able to cleave targets with a minor number of mismatches and in return, the viruses try to escape from Cas nuclease attack via mutation in its genetic sequence (Li et al. 2019a). Some approaches have been proposed to limit this unwanted effect including experimental off-target cleavage validation, Cas9-sgRNA delivery modification, computational predication, guide RNA engineering, and high-fidelity Cas9 engineering (Shen et al. 2019; Yin et al. 2018). Moreover, CRISPR machinery is sometimes suppressed and fail to properly edit resulting in escaper colonies survival which contain undesired edits or wild-type sequences. Deactivation of the CRISPR system resulted from mutations in genes related to either Cas9 or gRNA or both is the main mechanism involved in the formation of the escaper colonies (Vento et al. 2019). Optimized expression of gRNA and Cas9 endonuclease can inhibit the formation of these colonies (Guo et al. 2019; Li et al. 2019c; Song et al. 2017). Furthermore, Cas9 endonuclease overexpression can be toxic to E. coli. The DNA cleavage feature as well as the transient PAM recognition and binding across the DNA are suggested to be two main reasons for the enzyme cytotoxicity (Cho et al. 2018a; Vento et al. 2019). Reduced toxicity was reported when an inducible expression system was used for Cas9 endonuclease (Reisch and Prather 2015). Besides, compared to Cas9 endonuclease, the Cas9n variant showed less toxicity since it could cleave only one strand of the DNA (Standage-Beier et al. 2015). Taken together, circumventing these barriers may largely increase the successful rate of CRISPR-mediated gene modification.
Concluding remarks and future perspectives
Adopting those rapidly advancing methodologies like CRISPR–Cas tool, which is a multiplex, fast, and efficient tool can offer great prospects in the development of microbial cell factories. Multiplex editing capability in E. coli, as a model organism, has been improved using the high-throughput tools like crMAGE, which can combine the CRISPR–Cas9 system with the recombineering tools (Fokum et al. 2019; Pan and Reed 2018). Expectedly, the industrial strain will be more rapidly developed by further improvement of these high-throughput tools into the automated platforms. Furthermore, the Cas-based engineering toolbox will be further developed via the recruitment of alternative CRISPR systems. For example, The Cpf1 (Cas12a) RNA-guided DNA endonuclease can be considered as an efficient alternative tool for bacterial genome editing. Interestingly, Cpf1 has an intrinsic RNAse activity leading to the processing of its own precursor crRNA with no need for a tracrRNA (Bhushan 2020). Consequently, in multiplex engineering, the barrier issues when using Cas9 and multiple sgRNAs can be overcomed using Cpf1. In this regard, Zhang et al. have recently reported successful multiplex silencing in E. coli exploiting a DNase-dead Cpf1 (ddCpf1) variant (Zhang et al. 2017).
References
Abdelaal AS, Yazdani SS. (2020). Development and use of CRISPR in industrial applications. In: Genome Engineering via CRISPR-Cas9 System. Elsevier, pp 177–197
Adiego-Pérez B, Randazzo P, Daran JM, Verwaal R, Roubos JA, Daran-Lapujade P, Van Der Oost J. (2019). Multiplex genome editing of microorganisms using CRISPR-Cas FEMS microbiology letters 366:fnz086
Alkhnbashi OS, Meier T, Mitrofanov A, Backofen R, Voß B (2020) CRISPR-Cas Bioinformatics Methods 172:3–11
An J, Zhang W, Jing X, Nie Y, Xu Y. (2020). Reconstitution of TCA cycle involving l-isoleucine dioxygenase for hydroxylation of l-isoleucine in Escherichia coli using CRISPR-Cas9 3 Biotech 10:1–10
Bae S, Park J, Kim J-S. (2014). Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases Bioinformatics 30:1473–1475
Bhushan K. (2020). Evolution and molecular mechanism of CRISPR/Cas9 systems. In: Genome Engineering via CRISPR-Cas9 System. Elsevier, pp 15–25
Boch J et al (2009) Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326:1509–1512
Bortesi L, Fischer R. (2015). The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnology advances, 33: 41–52
Carroll D (2011) Genome engineering with zinc-finger nucleases. Genetics 188:773–782
Chaikind B, Van Rossum HM, Miller A, Perkovich P, Szyjka S, Patel K. (2020). Applications of CRISPRI in high throughput metabolic engineering. Google Patents
Charpentier E, Elsholz A, Marchfelder A. (2019). CRISPR-Cas: more than ten years and still full of mysteries. Taylor & Francis
Chen S, Yao Y, Zhang Y, Fan G. (2020). CRISPR system: Discovery, development and off-target detection Cellular Signalling, 109577
Cho S, Choe D, Lee E, Kim SC, Palsson B, Cho B-K. (2018). High-level dCas9 expression induces abnormal cell morphology in Escherichia coli ACS synthetic biology 7:1085–1094
Cho S, Shin J, Cho B-K (2018) Applications of CRISPR/Cas system to bacterial metabolic engineering. Int J Mol Sci 19:1089
Choudhary M, Joshi S, Singh P, Srivastava N. (2020). Biofuel production from lignocellulosic biomass: Introduction and metabolic engineering for fermentation scale-up. In: Genetic and Metabolic Engineering for Improved Biofuel Production from Lignocellulosic Biomass. Elsevier, pp 1–12
Chuai G-h, Wang Q-L, Liu Q (2017) In silico meets in vivo: towards computational CRISPR-based sgRNA design. Trends Biotechnol 35:12–21
Cress BF, Leitz QD, Kim DC, Amore TD, Suzuki JY, Linhardt RJ, Koffas MA. (2017). CRISPRi-mediated metabolic engineering of E. coli for O-methylated anthocyanin production Microbial cell factories 16:10
Cress BF et al. (2015). CRISPathBrick: modular combinatorial assembly of type II-A CRISPR arrays for dCas9-mediated multiplex transcriptional repression in E. coli. ACS Syn Biol 4:987–1000
Das M, Patra P, Ghosh A (2020) Metabolic engineering for enhancing microbial biosynthesis of advanced biofuels. Renew Sustain Energy Rev 119:109562
Dasgupta A, Chowdhury N, De RK (2020) Metabolic pathway engineering: perspectives and applications. Comput Methods Programs Biomed 192:105436
Deyell M, Ameta S, Nghe P. (2019). Large scale control and programming of gene expression using CRISPR. In: Seminars in cell & developmental biology, Elsevier
Ding W, Weng H, Du G, Chen J, Kang Z. (2017). 5-Aminolevulinic acid production from inexpensive glucose by engineering the C4 pathway in Escherichia coli. J Indus Microbiol Biotechnol 44:1127–1135
Doench JG et al. (2016). Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol, 34:184
Dong C. (2019). Programming bacterial gene expression using synthetic CRISPR-Cas transcriptional regulators.
Dong X et al (2020) CRISPRi-guided multiplexed fine-tuning of metabolic flux for enhanced Lacto-N-neotetraose production in Bacillus subtilis. J Agricul Food Chem 68:2477–2484
Ebrahimi V, Hashemi A. (2020). Challenges of in vitro genome editing with CRISPR/Cas9 and possible solutions: a review Gene, 144813
Fokum E et al. (2019). Metabolic engineering of bacterial strains using CRISPR/Cas9 systems for biosynthesis of value-added products. Food Biosci, 28:125–132
Fujiwara R, Noda S, Tanaka T, Kondo A. (2020). Metabolic engineering of Escherichia coli for shikimate pathway derivative production from glucose–xylose co-substrate Nature Communications, 11:1–12
Geng Y, Deng Z, Sun Y. (2016). An insight into the protospacer adjacent motif of Streptococcus pyogenes Cas9 with artificially stimulated RNA-guided-Cas9 DNA cleavage flexibility. RSC Adv 6:33514–33522
Guo T, Xin Y, Zhang Y, Gu X, Kong J (2019) A rapid and versatile tool for genomic engineering in Lactococcus lactis. Microb Cell Fact 18:22
Hashemi A (2018) CRISPR-Cas system as a genome engineering platform: applications in biomedicine and biotechnology. Curr Gene Ther 18:115–124
Heo M-J, Jung H-M, Um J, Lee S-W, Oh M-K. (2017). Controlling citrate synthase expression by CRISPR/Cas9 genome editing for n-butanol production in Escherichia coli. ACS Synth Biol 6:182–189
Herai RH. (2019). Avoiding the off-target effects of CRISPR/cas9 system is still a challenging accomplishment for genetic transformation. Gene 700:176–178
Hille F, Richter H, Wong SP, Bratovič M, Ressel S, Charpentier E (2018) The biology of CRISPR-Cas: backward and forward. Cell 172:1239–1259
Hsu PD et al. (2013). DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol, 31:827
Huminiecki L, Horbańczuk J (2018) The functional genomic studies of resveratrol in respect to its anti-cancer effects. Biotechnol Adv 36:1699–1708
Ibrahim A, ÖZSÖZ M SZ, Tirah G, Gideon O (2019) Genome engineering using the CRISPR Cas9 system. J Biomed Pharm Sci, 2:2
Kim SK, Han GH, Seong W, Kim H, Kim S-W, Lee D-H, Lee S-G (2016) CRISPR interference-guided balancing of a biosynthetic mevalonate pathway increases terpenoid production. Metab Eng 38:228–240
Kim SK, Seong W, Han GH, Lee D-H, Lee S-G (2017) CRISPR interference-guided multiplex repression of endogenous competing pathway genes for redirecting metabolic flux in Escherichia coli. Microb Cell Fact 16:188
Koonin EV, Makarova KS (2019) Origins and evolution of CRISPR-Cas systems. Philosoph Trans Royal Soc B 374:20180087
Li D, Zhou H, Zeng X. (2019). Battling CRISPR-Cas9 off-target genome editing. Springer
Li Q-S, Li Y, Deora GS, Ruan B-F (2019) Derivatives and analogues of resveratrol: recent advances in structural modification. Mini Rev Med Chem 19:809–825
Li Q, Seys FM, Minton NP, Yang J, Jiang Y, Jiang W, Yang S. (2019) CRISPR–Cas9D10A nickase‐assisted base editing in the solvent producer Clostridium beijerinckii. Biotechnol Bioeng, 116:1475–1483
Li S, Jendresen CB, Grünberger A, Ronda C, Jensen SI, Noack S, Nielsen AT (2016) Enhanced protein and biochemical production using CRISPRi-based growth switches. Metab Eng 38:274–284
Li Y et al. (2015). Metabolic engineering of Escherichia coli using CRISPR–Cas9 meditated genome editing. Metabolic Eng, 31:13–21
Liang L, Liu R, Garst AD, Lee T, Beckham GT, Gill RT (2017) CRISPR EnAbled trackable genome engineering for isopropanol production in Escherichia coli. Metab Eng 41:1–10
Liu H, Wei Z, Dominguez A, Li Y, Wang X, Qi LS. (2015). CRISPR-ERA: a comprehensive design tool for CRISPR-mediated gene editing, repression and activation. Bioinformatics, 31:3676–3678
Liu M, Rehman S, Tang X, Gu K, Fan Q, Chen D, Ma W (2019) Methodologies for improving HDR efficiency. Front Genet 9:691
Lunge A, Choudhary E, Sharma R, Gupta R, Agarwal N. (2020). Functional understanding of CRISPR interference: its advantages and limitations for gene silencing in bacteria. In: Genome Engineering via CRISPR-Cas9 System. Elsevier, pp 199–218
Lv L, Ren Y-L, Chen J-C, Wu Q, Chen G-Q. (2015). Application of CRISPRi for prokaryotic metabolic engineering involving multiple genes, a case study: controllable P (3HB-co-4HB) biosynthesis. Metabol Eng, 29:160–168
Ma M, Ye AY, Zheng W, Kong L. (2013). A guide RNA sequence design platform for the CRISPR/Cas9 system for model organism genomes. BioMed Res Int 2013
Macpherson CR, Scherf A (2015) Flexible guide-RNA design for CRISPR applications using protospacer workbench. Nat Biotechnol 33:805
Makarova KS et al. (2015). An updated evolutionary classification of CRISPR–Cas systems. Nat Rev Microbiol, 13:722–736
Marcellin E, Nielsen LK (2018) Advances in analytical tools for high throughput strain engineering. Curr Opin Biotechnol 54:33–40
McCarty NS, Graham AE, Studená L, Ledesma-Amaro R (2020) Multiplexed CRISPR technologies for gene editing and transcriptional regulation Nature. Communications 11:1–13
Mitsui R, Yamada R, Ogino H (2019) CRISPR system in the yeast Saccharomyces cerevisiae and its application in the bioproduction of useful chemicals. World J Microbiol Biotechnol 35:111
Montague TG, Cruz JM, Gagnon JA, Church GM, Valen E. (2014). CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res, 42:W401-W407
Moon SB, Ko J-H, Kim Y-S (2019) Recent advances in the CRISPR genome editing tool set. Exp Mol Med 51:1–11
Mougiakos I, Bosma EF, Ganguly J, van der Oost J, van Kranenburg R (2018) Hijacking CRISPR-Cas for high-throughput bacterial metabolic engineering: advances and prospects. Curr Opin Biotechnol 50:146–157
Naito Y, Hino K, Bono H, Ui-Tei K. (2015). CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics, 31:1120–1123
Nandy D, Maity A, Mitra AK (2020) Target-specific gene delivery in plant systems and their expression: Insights into recent developments. J Biosci 45:30
Nishimasu H et al. (2014). Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 156:935–949
O’Brien A, Bailey TL. (2014). GT-Scan: identifying unique genomic targets Bioinformatics 30:2673–2675
Pan S, Reed JL (2018) Advances in gap-filling genome-scale metabolic models and model-driven experiments lead to novel metabolic discoveries. Curr Opin Biotechnol 51:103–108
Pawelczak KS, Gavande NS, VanderVere-Carozza PS, Turchi JJ (2018) Modulating DNA repair pathways to improve precision genome engineering. ACS Chem Biol 13:389–396
Prykhozhij SV, Vinothkumar Rajan DG, Berman JN. (2015). CRISPR multitargeter: a web tool to find common and unique CRISPR single guide RNA targets in a set of similar sequences, PloS one, 10
Reisch CR, Prather KL. (2015). The no-SCAR (S carless C as9 A ssisted R ecombineering) system for genome editing in Escherichia coli. Sci Rep, 5:1–12
Satowa D et al. (2020). Metabolic engineering of E. coli for improving mevalonate production to promote NADPH regeneration and enhance acetyl‐CoA supply. Biotechnol Bioeng
Schultenkämper K, Brito LF, Wendisch VF. (2020). Impact of CRISPR interference on strain development in biotechnology. Biotechnol Appl Biochem
Shen C-C, Hsu M-N, Chang C-W, Lin M-W, Hwu J-R, Tu Y, Hu Y-C. (2019). Synthetic switch to minimize CRISPR off-target effects by self-restricting Cas9 transcription and translation. Nucleic Acids Res, 47:e13-e13
Shmakov S et al. (2015). Discovery and functional characterization of diverse class 2 CRISPR-Cas systems, Mol cell, 60:385–397
Singh V. (2020). An introduction to genome editing CRISPR-Cas systems. In: Genome Engineering via CRISPR-Cas9 System. Elsevier, pp 1–13
Sledzinski P, Nowaczyk M, Olejniczak M (2020) Computational tools and resources supporting CRISPR-Cas. Exp Cell 9:1288
Song M. (2017). The CRISPR/Cas9 system: Their delivery, in vivo and ex vivo applications and clinical development by startups. Biotechnol Progress, 33:1035–1045
Song X, Huang H, Xiong Z, Ai L, Yang S. (2017). CRISPR-Cas9D10A nickase-assisted genome editing in Lactobacillus casei. Appl Environ Microbiol, 83:e01259–01217
Standage-Beier K, Zhang Q, Wang X (2015) Targeted large-scale deletion of bacterial genomes using CRISPR-nickases. ACS Synth Biol 4:1217–1225
Stemmer M, Thumberger T, del Sol Keyer M, Wittbrodt J, Mateo JL. (2015). CCTop: an intuitive, flexible and reliable CRISPR/Cas9 target prediction tool, PloS one, 10
Terns RM, Terns MP (2014) CRISPR-based technologies: prokaryotic defense weapons repurposed. Trends Genet 30:111–118
Tian J, Yang G, Gu Y, Sun X, Lu Y, Jiang W. (2020). Developing an endogenous quorum-sensing based CRISPRi circuit for autonomous and tunable dynamic regulation of multiple targets in industrial Streptomyces bioRxiv
Tian P, Wang J, Shen X, Rey JF, Yuan Q, Yan Y (2017) Fundamental CRISPR-Cas9 tools and current applications in microbial systems. Synth Sys Biotechnolo 2:219–225
Trimidal SG et al (2019) Can designer indels be tailored by gene editing? can indels be customized? BioEssays 41:1900126
Van Der Oost J, Westra ER, Jackson RN, Wiedenheft B (2014) Unravelling the structural and mechanistic basis of CRISPR–Cas systems. Nat Rev Microbiol 12:479–492
Vento JM, Crook N, Beisel CL (2019) Barriers to genome editing with CRISPR in bacteria. J Indus Microbiol Biotechnol 46:1327–1341
Vigouroux A, Bikard D. (2020). CRISPR tools to control gene expression in bacteria. Microbiol Mol Biol Rev, 84
Wang B, Guo Y, Xu Z, Tu R, Wang Q. (2020). Genomic, transcriptomic, and metabolic characterizations of Escherichia coli adapted to branched-chain higher alcohol tolerance. Appl Microbiol Biotechnol, 1–14
Wang Q, Zeng W, Zhou J (2019) Effect of gene knock-out of L-tyrosine transport system on L-tyrosine production in Escherichia coli Sheng wu gong cheng xue bao. Chin J Biotechnol 35:1247–1255
Wilson LO, O’Brien AR, Bauer DC (2018) The current state and future of CRISPR-Cas9 gRNA design tools. Front Pharmacol 9:749
Wong N, Liu W, Wang X (2015) WU-CRISPR: characteristics of functional guide RNAs for the CRISPR/Cas9 system. Genome Biol 16:218
Wright AV, Nuñez JK, Doudna JA (2016) Biology and applications of CRISPR systems: harnessing nature’s toolbox for genome engineering. Cell 164:29–44
Wu J, Du G, Chen J, Zhou J (2015) Enhancing flavonoid production by systematically tuning the central metabolic pathways based on a CRISPR interference system in Escherichia coli. Sci Rep 5:13477
Wu J, Zhang X, Zhu Y, Tan Q, He J, Dong M (2017) Rational modular design of metabolic network for efficient production of plant polyphenol pinosylvin. Sci Rep 7:1–15
Wu J, Zhou P, Zhang X, Dong M (2017) efficient de novo synthesis of resveratrol by metabolically engineered Escherichia coli. J Indus Microbiol Biotechnol 44:1083–1095
Wu M-Y, Sung L-Y, Li H, Huang C-H, Hu Y-C (2017) Combining CRISPR and CRISPRi systems for metabolic engineering of E. coli and 1, 4-BDO biosynthesis. ACS Synth Biol 6:2350–2361
Wu Y et al (2020) Design of a programmable biosensor-CRISPRi genetic circuits for dynamic and autonomous dual-control of metabolic flux in Bacillus subtilis. Nucleic Acids Res 48:996–1009
Xia J et al (2016) Expression of Shewanella frigidimarina fatty acid metabolic genes in E. coli by CRISPR/cas9-coupled lambda Red recombineering. Biotechnol let 38:117–122
Xie S, Shen B, Zhang C, Huang X, Zhang Y. (2014). sgRNAcas9: a software package for designing CRISPR sgRNA and evaluating potential off-target cleavage sites. PloS one, 9
Xu H et al (2015) Sequence determinants of improved CRISPR sgRNA design. Genome Res 25:1147–1157
Xu S, Wang Q, Zeng W, Li Y, Shi G, Zhou J. (2020). Construction of a heat-inducible Escherichia coli strain for efficient de novo biosynthesis of L-tyrosine. Process Biochem
Yin H et al (2018) Partial DNA-guided Cas9 enables genome editing with reduced off-target activity. Nat Chemical Biol 14:311
Zhan T, Rindtorff N, Betge J. (2019). Ebert MP, Boutros M CRISPR/Cas9 for cancer research and therapy. In: Seminars in cancer biology, Elsevier, pp 106–119
Zhang B, Zhang X-M, Wang W, Liu Z-Q, Zheng Y-G (2019) Metabolic engineering of Escherichia coli for d-pantothenic acid production. Food Chem 294:267–275
Zhang H, Cheng Q-X, Liu A-M, Zhao G-P, Wang J (2017) A novel and efficient method for bacteria genome editing employing both CRISPR/Cas9 and an antibiotic resistance cassette. Front Microbiol 8:812
Zhu LJ, Holmes BR, Aronin N, Brodsky MH. (2014). CRISPRseek: a bioconductor package to identify target-specific guide RNAs for CRISPR-Cas9 genome-editing systems, PloS one, 9
Ziegler M, Takors R (2020) Reduced and minimal cell factories in bioprocesses: towards a streamlined chassis. Minimal Cells: Design. Construction, Biotechnological Applications. Springer, pp 1–44
Zou X et al (2020) Pathway construction and metabolic engineering for fermentative production of β-alanine in Escherichia coli. Appl Microbiol Biotechnol 104:2545–2559
Acknowledgements
This work was supported by the research deputy of Shahid Beheshti University of Medical Sciences in Tehran, Iran.
Funding
Not applicable.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Hashemi, A. CRISPR–Cas9/CRISPRi tools for cell factory construction in E. coli. World J Microbiol Biotechnol 36, 96 (2020). https://doi.org/10.1007/s11274-020-02872-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-020-02872-9