Abstract
Saccharomyces cerevisiae has been widely used for wine fermentation and bio-fuels production. A S. cerevisiae strain Sc131 isolated from tropical fruit shows good fermentation properties and ethanol tolerance, exhibiting significant potential in Chinese bayberry wine fermentation. In this study, RNA-sequence and RT-qPCR was used to investigate the transcriptome profile of Sc131 in response to ethanol stress. Scanning Electron Microscopy were carried out to observe surface morphology of yeast cells. Totally, 937 genes were identified differential expressed, including 587 up-regulated and 350 down-regulated genes, after 4-h ethanol stress (10% v/v). Transcriptomic analysis revealed that, most genes involved in regulating filamentous growth or pseudohyphal growth were significantly up-regulated in response to ethanol stress. The complex protein quality control machineries, Hsp90/Hsp70 and Hsp104/Hsp70/Hsp40 based chaperone system combining with ubiquitin–proteasome proteolytic pathway were both activated to recognize and degrade misfolding proteins. Genes related to biosynthesis and metabolism of two well-known stress-responsive substances trehalose and ergosterol were generally up-regulated, while genes associated with amino acids biosynthesis and metabolism processes were differentially expressed. Moreover, thiamine was also important in response to ethanol stress. This research may promote the potential applications of Sc131 in the fermentation of Chinese bayberry wine.
Graphical Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Saccharomyces cerevisiae, an efficient ethanol-producing microorganism, has been widely used for wine fermentation and bio-fuels production. Whereas during fermentation processes cells growth of S. cerevisiae and some enzymes activity were inhibited, and fermentation productivity and ethanol yield were limited (Kasavi et al. 2016). As the main product of fermentation, ethanol is considered to be the primary factor limiting the fermentation process (Lam et al. 2014) and ethanol at a concentration over 9% will affect the viability of yeast cells (Fujita et al. 2004). In response to ethanol stress, cell cycle and growth, together with amino acid metabolism, lipid metabolism, mitochondrial function and trehalose biosynthesis were remarkably altered as shown in several studies (Doğan et al. 2014; Snoek et al. 2015; Ranganathan et al. 2015). Moreover, high levels of ethanol destroy the ion balance and perturb protein conformation causing protein aggregation and dysfunction (Teixeira et al. 2009; Ma and Liu 2010).
Over the past decades, genetic engineering, DNA microarray and genome shuffling were applied to investigate the genetic and molecular mechanism of ethanol tolerance in S. cerevisiae (Fraser 2011; Benjaphokee et al. 2012; Müller et al. 2014). Even though, the underlying mechanism of variety of S. cerevisiae strains tolerant to ethanol remains unclear (Kasavi et al. 2016). Recently, high-throughput sequencing technologies are now widely used in genomes and transcriptome research. Sequencing cDNA library from a sample using next generation sequencing (NGS), known as RNA-seq, provides a more precise and comprehensive measurement of levels of transcripts than other methods (Schuierer et al. 2017).
Chinese bayberry wine is a fermented alcoholic beverage made from Chinese bayberry (Myrica rubra Sieb. and Zucc.), which is rich in essential micronutrients and bioactive constituents (Du et al. 2016; Chen et al. 2016). Nevertheless, only a few special yeast strains are used for brewing bayberry wine (Du et al. 2016). In our previous study (data not shown), a S. cerevisiae strain Sc131 isolated from tropical fruit, has been shown to have good fermentation properties and exhibits significant potential in Chinese bayberry wine fermentation. Sc131 can normally grow at pH 3.30–5.50 and possesses high ethanol productivity, high ethanol tolerance, and well-balanced productivity of flavors, in contrast to other yeast strains. However, few works were focus on RNA-seq of Sc131 under ethanol stress to our knowledge.
In this work, RNA-seq was used to investigate gene expression profiles of Sc131 under short-term ethanol stress during alcohol fermentation process. This research shed light on the molecular response to ethanol in Sc131, which may be helpful to guide the fermentation of Chinese bayberry wine.
Materials and methods
Strain, culture conditions and experimental design
Sc131, a diploid S. cerevisiae strain, was isolated from tropical fruit, and preserved in food biotechnology laboratory of Ningbo University. Before culture for the RNA-seq experiments, the yeast cells were activated and pre-cultured. Briefly, a loopful of cells from single colony on the YPD-Agar (1% yeast extract, 2% peptone, 2% glucose, and 2% agar) plate was inoculated in 50 mL YPD medium (1% yeast extract, 2% peptone and 2% glucose) and incubated at 30 °C in an orbital shaker at 150 rpm for 12 h. Then 0.5 mL pre-cultured Sc131 was inoculated in 50 mL of high-sugar YPD medium (1% yeast extract, 2% peptone and 10% glucose) and incubated at 30 °C in an orbital shaker at 150 rpm till the mid-exponential phase.
Ethanol was added into the experimental cultures to a final ethanol concentration of 10% (v/v). Three biological replicates were performed for RNA-seq analysis. T1–T3 were the ethanol treated group and T4–T6 as the control group. The Sc131 cells were cultured in orbital shaker and monitored by measuring the optical density of the culture medium at 600 nm (OD600) with a spectrophotometer, UVmini-1240 (Shimadzu Corporation, Japan). Cells for RNA-seq analysis were collected about 4 h after the addition of ethanol. All samples were stored in − 80 °C refrigerator prior to use.
Scanning electron microscopy (SEM)
After treatment with ethanol, yeast cells from each culture were collected by centrifugation at 5000×g, 4 °C for 10 min and washed three times with physiological saline. Then the cells were resuspended in 2.5% glutaraldehyde for 4 h at 4 °C and washed three times with 0.1 M PBS (pH 7.4) for 15 min each time. Subsequently, the cells were eluted with a gradient concentration of ethanol solutions (30, 50, 70, 80, 90, 95 and 100%) for 10 min in each ethanol solution, and with a gradient of tert-butanol and anhydrous ethanol mix (ratio 1:3, 1:1, 3:1, 3:0) for 10 min in each mix. Finally, yeast cells were dried and coated with a gold/palladium alloy (40:60) to a thickness of 10–20 nm and observed with a Hitachi S3400N scanning electron microscopy system.
RNA preparation, library construction and sequencing
The total RNA was isolated from each sample, using the TRIZOL (Aidlab Biotech, Beijing, China). The purity and integrity of RNA samples were assessed by NanoPhotometer® spectrophotometer (IMPLEN, CA, USA) and the RNA Nano 6000 Assay Kit, respectively. And RNA concentration was detected by Qubit® RNA Assay Kit in Qubit® 2.0. Then samples of 2.0 µg high-quality RNA from each sample were mixed into a single large pool in order to maximize the diversity of transcriptional units.
The construction of the libraries and the RNA-seq were performed by Beijing BioMarker Technologies (Beijing, China). In brief, mRNAs were broken into small pieces in fragmentation buffer. The RNA fragments were copied into first strand cDNA using random primers subsequently the second strand cDNA synthesis was performed. A single ‘A’ baset was ligated with the resulting cDNAs and the sequencing adapters. Fragments (200 ± 25 bp) were then separated by agarose gel electrophoresis and selected for PCR amplification as sequencing templates. Finally, sequencing was performed with the Illumina HiSeqTM2500 sequencing platform.
Reads mapping, annotation and analysis
To get high-quality data, the raw reads were filtered by trimming adapters and removing low-quality sequencing. Besides the minimum acceptance length was 60 bp in order to exclude the effect of sequencing artifacts.
The sequence reads were initially annotated based on the S. cerevisiae reference genome and gene names of different sequences were determined according to the best hit. Then each matched gene was compared against the National Center for Biotechnology Information (NCBI) non-redundant protein (nr) database, Swiss-Prot, Protein Research Foundation (PRF), Protein Information Resource (PIR) and Protein Data Bank (PDB) with an E-value ≤ 10−5. The Blast2 GO program was used to assign gene ontology (GO) terms with an E-value ≤ 10−5 (Han et al. 2013). Annotations of Sc131 unigenes were then used to predict biochemical pathways using the Pathway Tools (Guo et al. 2010). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were retrieved from KEGG web server (http://www.genome.jp/kegg/) (Kanehisa et al. 2004).
Defining differentially expressed genes
Gene expression levels were calculated based on the obtained reads from six libraries using Reads Per Kilobases per Million mapped reads (RPKM) values. DEGs under ethanol stress were detected based on the combined criteria of fold change (FC) ≥ 2 and a Chi square test (P values < 0.05, false discovery rate (FDR) < 0.001).
Quantitative real-time PCR validation
Ten representative genes were analyzed using real-time PCR method to evaluate the validity of Illumina analysis. Total RNA of Sc131 was reverse-transcribed into cDNA using Fastquant RT Kit (with gDNase) (TIANGEN BIOTECH). Then PCR reactions were carried out using a LightCycler96® (Roche Diagnostics GmbH, Mannheim, Germany) thermocycler with the following programs: 94 °C for 3 min, followed by 40 cycles of 95 °C for 3 s, 60 °C for 25 s, and a final extension of 72 °C for 2 min. The relative expression level of target genes was measured with the 2−ΔΔCT method and ALG9 was used as the reference gene (Teste et al. 2009). All tests were performed at least three times. The primers are listed in Table 1.
Results
Comparison of growth and physiological phenotypes of ethanol-treated and control samples
The growth of Sc131 cells was strongly inhibited by ethanol addition at the midexponential phase (Fig. 1). The electron micrographs from SEM observations showed serious morphological damage due to ethanol treatment, and many interesting phenomena were found (Fig. 2). Figure 2b, d showed that the surface of many cells are significantly deformed and cracks appeared in the cell walls of some cells under ethanol stress. However, Fig. 2a, c showed that cell surfaces of the control group were smooth and most of yeast cells were budding.
Illumina sequence data
Six libraries representing transcriptome of three control samples (T1–T3) and three ethanol-treated samples (T4–T6) were constructed respectively for RNA-sEq. Totally, 28.89 Gb clean data were obtained, and the average quality value was ≥ 30 for more than 85% reads. The GC content of these samples was consistently about 42%. Substantial proportion of reads of each library (74.82% in ethanol-treated library and 74.09% in control library) were mapped to S. cerevisiae genome, suggesting good quality of library construction and RNA sequencing.
Defining DEGs
DEGs with P values < 0.05 and FDR < 0.001 were considered statistically significant and P values < 0.01 considered highly significant. Totally, 937 DEGs were identified, including 587 up-regulated genes and 350 down-regulated genes. As shown in Fig. 3, DEGs between control group (T1–T3) and ethanol-treated group were screened out. The Volcano plot (Fig. 3a) plotted FDR against FC and MA plot (Fig. 3b) plotted the M (log FC) against A (log RPKM/expression level) for all DEGs were generated to represent expression level difference and statistically significant degree between two groups in overall. Each point represents a gene, the green spots represent the differentially expressed genes and the red represent a gene that does not show significant difference. For a global perspective on gene expression during ethanol pressure, a heat map representing the transcription levels of all DEGs was generated (Fig. 3c) to display the expression profiles of two groups.
Screening and Expression profilings of DEGs in ethanol stressed Sc131. a Volcano plot for all DEGs, which plotted FDR against FC and b MA plot which plotted the M (log FC) against A (log RPKM/expression level) for all DEGs were generated to represent expression level difference and statistically significant degree between two groups in overall. Each point represents a gene, the green dot represents a significantly differentially expressed gene and the red represents a gene that does not show significant difference. c Heatmap for DEGs using hierarchical clustering analysis. The signal ratios are shown in a red-green color scale, where red represents up-regulation and green represents down-regulation. Each column (T1, T2, T3, T4, T5 and T6) represents the expression value of the RNA-Seq and each row represents a DEG. T1–T3: three biological replicates of the control group; T4–T6: three biological replicates of the ethanol-treated group
Functional annotation and categorization of DEGs
The DEGs were annotated by blasting in diverse protein databases including the NCBI nr database, KEGG, Swiss-Prot, GO and pfam databases. 937 DEGs were successfully mapped in at least one database. 937 DEGs were annotated in the nr (937), KEGG (488), Swiss-Prot (937), GO (675) and COG (368) databases.
GO classification analysis
To gain insight into the functional categories of the DEGs, 675 DEGs were categorized into 48 functional sub-groups GO categories, Fig. 4 showed the distributions of the GO terms in biological process, molecular function, and cellular component. The shallow bars show the number and percentage of DEGs in each category, while the dark bars show those of all annotated genes in each category. In the ontology of cellular components, the DEGs were mainly distributed in the term ‘cell part’, ‘cell’ and ‘organelle’. ‘Binding’ and ‘catalytic activities’ were the main categories in molecular function ontology. For biological processes, most DEGs were gathered at ‘cellular process’, ‘metabolic process’ and ‘single-organism process’. The GO classification was obviously shown that ethanol stress affected enzyme activities, organelle functions and major biological processes. With the help of GO functional classification, we could understand the distribution of gene function at the macro level and predict the potential molecular role of each gene.
KEGG pathway analysis of ethanol responsive genes
Figure 5 showed the major distribution of metabolic/biological pathways that contained more than 3 DEGs. Notably, 57 DEGs were annotated to ‘ribosome’, 19 DEGs were annotated ‘ribosome biogenesis in eukaryotes’ and 14 DEGs were annotated to protein processing in endoplasmic reticulum (ER). It’s obvious that ribosome and ER play critical role in response to ethanol stress. Expression of genes related to meiosis-yeast was also significantly changed. It is possible that Sc131 entered sexual reproduction (meiosis) underwent sporulation, and produced ascus and ascospore in response to ethanol pressure. Regarding carbohydrate metabolism, ‘citrate cycle (TCA cycle)’ and ‘glycolysis/gluconeogenesis’ were significantly changed. Additionally, the gene expression related to signal transduction pathway and RNA transport were changed to response to ethanol stress.
Protein–protein interaction network in response to stress and stimulus
The protein–protein interactions in response to stress and stimulus were assessed by STRING 10.5 (Fig. 6). In protein–protein interaction network, a cluster of proteins with complex interactions with each other usually perform similar functions together. Colored lines between the protein nodes indicate the various types of interactions. Proteins that interact closely with other proteins are important in the interaction network. The FUS3 and vicinity proteins were functional related to Mitogen-Activated Protein Kinase pathway (MAPK) in eukaryotes, which are involved in directing cellular responses to a diverse array of stimuli, such as mitogens, osmotic stress and heat shock (Good et al. 2009; Patterson et al. 2010). Hsp82, Hsc82, SSE2, SSE1, KAR2, SSA2, SSB1, SSB2, SIS1 had pivotal position in the interaction network, which are HSP proteins or chaperones belonging to heat shock protein 70 or 90 family. TPS1 and TPS2 encode critical proteins in biosynthesis of trehalose, which is star molecule in stress-related cell response.
The interaction network of important proteins in response to stress and stimulates. The network nodes represent proteins, and edges represent protein–protein associations. Small nodes: proteins of unknown three-dimensional structure, large nodes: some three-dimensional structure is known or predicted
RNA-seq expression validation by quantitative real-time PCR
In order to quantitatively determine the reliability of the transcriptome results, we detected the expression of ten candidate DEGs using Reverse Transcription quantitative PCR (RT-qPCR). These candidates included six up- and four down-regulated genes. RT-qPCR results showed that, expression of heat shock protein family genes Hsp40, Hsp82, Hsp104, KAR2 was found up-regulated. Two genes ERG3 and ERG24 involved in ergosterol biosynthesis were up-regulated. PRO1, PIR3, FUS1 and NRG2 were down-regulated. A high consistency was displayed between the RNA-seq and RT-qPCR data (Fig. 7), proving the validity of RNA-seq data for genes with distinct transcript abundance.
Discussion
Filamentous growth
Transcriptome data showed that genes involved in regulating filamentous growth were significantly differentially expressed in response to ethanol stress. Most of DEGs involved in filamentous growth such as OPI1, MIT1, GLK1, UBC11, UBC4 and RAD6 were up-regulated, among which MIT1 is considered as a key transcriptional regulator of pseudohyphal growth (Cain et al. 2012). Current study simulating wine brewing is a fermenting process with a high level of initial sugar and ethanol is the major metabolite. In the presence of fermentable carbon sources, relatively nitrogen starvation may induce pseudohyphal growth (Tesnière et al. 2015) and accumulation of ethanol could induce S. cerevisiae filamentous formation (Lorenz et al. 2000). One possible explanation is that, a high surface to volume ratio of filaments can promote transport of nutrients (Garny et al. 2009). Moreover, the genes regulated meiosis and sporulation were up-regulated. Schröder et al. (2000) had proved that meiosis, pseudohyphal growth and sporulation were the responses of yeast to nutrition starvation especially nitrogen starvation. Based on the above results, 10% ethanol might have caused nitrogen starvation which was consist with the hypothesis that ethanol can induce a common nitrogen starvation-like cell response in S. cerevisiae (Kasavi et al. 2016; Stanley et al. 2010). The results indicated that Sc131 form filament through stretching the cell membrane to deal with ethanol stress.
Mitochondrial function and energy metabolism
This RNA-seq analysis revealed that most of genes related to ribosome and ribosome biogenesis in eukaryotes pathways were down-regulated under ethanol stress. However, the transcript levels of genes related to mitochondrial ribosome biosynthesis didn’t seem to change. It meant that ethanol inhibited ribosome biosynthesis in Sc131 cells. We also guess that Sc131 might use mitochondrial ribosomes or change the ribosome components to synthesize additional proteins to resist ethanol stress.
Furthermore, mitochondrion as the energy center drives lots of energy requiring processes in the cell (Mourier and Larsson 2011). Up-regulated genes COX1, COX2 and COX3 were not only related to mitochondrial respiratory chain complex III, IV, mitochondrial electron transport and oxidative phosphorylation, but also encoding cytochrome-c oxidase (COX) which catalyzes both electron transfer from cytochrome c to molecular oxygen and the concomitant proton pumping across the inner mitochondrial membrane, generate large amounts of ATP (Valnot et al. 2000; Dimauro et al. 2012). It indicated that Sc131 cells response to ethanol stress by regulating a series of large enzyme complexes activity that couples the transfer of electrons to the creation of a concentration gradient of protons across the inner mitochondrial membrane to promote ATP synthesis.
Heat shock proteins and protein processing in ER
Endoplasmic reticulum serves many general functions, including the folding and the transport of synthesized proteins, and is the center of various of chaperones and enzymes in eukaryotic, play an important role in response to ethanol stress. As shown in Fig. 5, protein–protein interaction network analysis illustrated that heat shock proteins Hsp70, Hsp90 and Hsp104, as well as genes encoding HSP homolog proteins and their co-chaperones such as SIS1, SSE1 and SSE2 had a complex interaction and were notably clustered. As shown in Fig. 8, genes involved in protein recognition by luminal chaperones, ER-associated degradation (ERAD) and ubiquitin ligase complex in cytoplasm, such as HSP40, Hsp70, Hsp90, BiP and sHFS, were significantly up-regulated. For example, genes encoding members of conserved Hsp70 family, such as SSE1, SSE2 and BiP, were up-regulated. SSA2 encoding ATP binding protein related to protein folding and vacuolar import of proteins were up-regulated more than threefold. Gene encoding Hsp70 family ATPase KAR2, a chaperone to regulate protein folding in the ER and may be important for soluble proteins export in ER (Kimura et al. 2005) were up-regulated. Genes involved in ERAD machinery, encoding proteins that degrade the permanently misfolded proteins. Amm et al. (2014) reported that the ERAD avoids the cell from smothering in its secretory protein waste.
Hsp90s are conserved and abundant molecular chaperone that promotes the maturation, structural maintenance and regulation of some target proteins related to lots of essential cellular processes such as cell cycle control, signal transduction and response to cellular stress (Li et al. 2012). In yeast, Hsp82 and Hsc82 are the cytoplasmic forms of Hsp90 family. Hsp70s are important molecular chaperones that can promote protein folding, prevent unfolded or misfolded protein aggregates (Behnke and Hendershot 2014; Jin et al. 2017). Hsp90 and Hsp70 work together to form a multi chaperone complex and are involved in the folding and maturation of key regulatory proteins and the dealing of the aggregation of misfolding proteins. Hsp104 is constantly expressed at low levels in unstressed cells but significantly up-regulated 12-fold during ethanol stress. Reports have showed that in concert with Ydj1p (Hsp40) and Ssa1p (Hsp70), Hsp104 can refold and reactivate previously denatured, aggregated proteins after ethanol or other environmental stresses (Glover and Lindquist 1998; Kempf et al. 2017), forming another chaperone system that rescues previously aggregated proteins.
It could be speculated that, during ethanol pressure the Hsp70s firstly bind and prevent the aggregation of misfolding proteins, then work together with Hsp90 and a multitude of the co-chaperones to solubilize protein aggregates and relieve ER stress. Simultaneously, Hsp104 in concert with Hsp40 and Hsp70, can reactivate proteins that have been denatured and allowed to aggregate.
Moreover, the DEGs involved in ubiquitination were all notably up-regulated. Protein ubiquitination is performed by continuously enzymatic reactions, among them E3 ubiquitin-protein ligase enzymes are important since they determine the specificity of substrates (Lee et al. 2014) and often worked together with HSP40, Hsp70 and Hsp90 chaperone to recognize misfolding proteins then achieve protein quality control (Berndsen and Wolberger 2014). Our results showed that, UBR1, encoding E3 ubiquitin-protein ligase, plays an important role in cytoplasmic protein quality control in yeast (Nillegoda et al. 2010) were up-regulated, and the up-regulated gene Hsp70 and SSE1 are essential for the ubiquitination of UBR1 substrates. HSP40, Hsp70 and UbcH5 encoding ubiquitin ligase complex in cytoplasm were up-regulated. The ubiquitin–proteasome system can degrade old proteins that exceed their functional lifetime and conduct the destruction of most unfolded and misfolded proteins, especially secretory and integral membrane proteins (Amm et al. 2014).
It demonstrated that misfolded proteins, probably membrane proteins which affect the integrity and function of plasma membrane, were severely aggregated in ER and the accumulation of improperly folded proteins is certainly toxic to the cell. Thus, assembly system to properly fold and refold proteins, and quality control machinery to recognize and degrade misfolding proteins were both activated to cope with this situation. Hsp90/Hsp70 and Hsp104/Hsp70/Hsp40 based chaperone machinery combining with ubiquitin–proteasome proteolytic pathway may play extensive roles in Sc131 in response to ethanol stress.
Trehalose, ergosterol and amino acids biosynthesis and metabolism
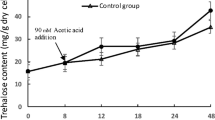
As a well-known stress-responsive substance, the metabolism and biosynthesis of trehalose were identified significantly differentially expressed. TPS1 and TPS2 encoding trehalose-6-phosphate synthase that catalyzes the production of trehalose, were sharply up-regulated. It’s obvious that trehalose was accumulated in Sc131 cells under ethanol stress. Trehalose and the HSPs as two important protective agents worked together to resist ethanol stress. At first trehalose prevents protein denaturation then HSPs deter protein aggregation and finally the disaccharide is degraded.
Manual inspection indicated that the DEGs related to steroid biosynthesis, especially in ergosterol biosynthesis (ERG2, ERG3, ERG24), were all remarkable up-regulated. Ergosterol has the ability to protect cell membrane integrity and enhance membrane fluidity and reports have established that it was involved in stress response (Martínezmontañés et al. 2013; Ren et al. 2014). However, previous research indicated that all the genes related to ergosterol biosynthesis were down-regulation under ethanol stress (Ma and Liu 2010). It meant that 10% ethanol could induce ergosterol production in Sc131 cells.
Amino acids, as the primary metabolites, are involved in many cellular biosynthesis and metabolism processes. Researches had revealed that supplementation of proline and tryptophan will significantly enhance ethanol tolerance of S. cerevisiae (Ma and Liu 2010). Proline strengthens the stability of membranes and proteins, and inhibits protein aggregation (Martos et al. 2007). PRO1 is the key gene to affect proline biosynthesis, and a PRO1-deletion strain is more sensitive to ethanol stress (Yoshikawa et al. 2009). Interestingly, in our results PRO1 involved in proline biosynthetic process was significantly down-regulation, it indicated that high level of ethanol inhibits proline biosynthesis in Sc131 cells. Whereas, the genes encoding tryptophan were remarkable up-regulated under ethanol stress, therefore, tryptophan might be essential for Sc131 in response to ethanol pressure.
Thiamine biosynthesis
Thiamine (vitamin B1) as the cofactor function of thiamine free radicals and subsequent oxidation, is essential for all living organisms (Nagae et al. 2016). Recently reports have established a hypothesis that thiamine can help different organisms to against stress (Wolak et al. 2014). Additionally, thiamine pyrophosphate (TPP) and thiamine diphosphate (TDP) as cofactors for many central metabolic enzymes, is required for all forms of life. In our study, genes THI4, THI5 and THI13 involved in thiamine biosynthetic process and genes PDC1 and PDC5 involved in TPP biosynthetic process were up-regulated. Therefore, thiamine is an important substance that helps the Sc131 to resist ethanol stress.
Conclusions
Transcriptome profile provides an overall view of the transcriptomic changes of Sc131 under ethanol stress. The present comprehensive transcriptional overview provides a complete theory basis for subsequent studies, and has practical significance for application in bayberry wine production. Further study focusing on the ethanol tolerance mechanism of Sc131 and some critical metabolites related to wine product quality will be carried out using proteomics and metabolomics technology.
References
Amm I, Sommer T, Wolf DH (2014) Protein quality control and elimination of protein waste: the role of the ubiquitin-proteasome system. Biochim Biophys Acta 1843:182–196
Behnke J, Hendershot LM (2014) The large hsp70 grp170 binds to unfolded protein substrates in vivo with a regulation distinct from conventional hsp70s. J Biol Chem 289:2899–2907
Benjaphokee S, Hasegawa D, Yokota D, Asvarak T, Auesukaree C, Sugiyama M, Kaneko Y, Boonchird C, Harashima S (2012) Highly efficient bioethanol production by a Saccharomyces cerevisiae strain with multiple stress tolerance to high temperature, acid and ethanol. New Biotechnol 29:379–386
Berndsen CE, Wolberger C (2014) New insights into ubiquitin E3 ligase mechanism. Nat Struct Mol Biol 21:301–307
Cain CW, Lohse MB, Homann OR, Sil A, Johnson AD (2012) A conserved transcriptional regulator governs fungal morphology in widely diverged species. Genetics 190:511–521
Chen YY, Zhang ZH, Zhong CY, Song XM, Lin QH, Huang CM, Huang RH, Chen W (2016) Functional analysis of differentially expressed proteins in Chinese bayberry (Myrica rubra sieb. et Zucc.) fruits during ripening. Food Chem 190:763–770
Dimauro S, Tanji K, Schon EA (2012) The many clinical faces of cytochrome c oxidase deficiency. Adv Exp Med Biol 748:341–357
Doğan A, Demirci S, Aytekin AÖ, Şahin F (2014) Improvements of tolerance to stress conditions by genetic engineering in Saccharomyces cerevisiae, during ethanol production. Appl Biochem Biotechnol 174:28–42
Du J, Han F, Yu P, Li J, Fan L (2016) Optimization of fermentation conditions for Chinese bayberry wine by response surface methodology and its qualities. J Inst Brew 122:763–771
Fraser HB (2011) Genome-wide approaches to the study of adaptive gene expression evolution: systematic studies of evolutionary adaptations involving gene expression will allow many fundamental questions in evolutionary biology to be addressed. Bioessays 33:469–477
Fujita K, Matsuyama A, Kobayashi Y, Iwahashi H (2004) Comprehensive gene expression analysis of the response to straight-chain alcohols in Saccharomyces cerevisiae using cDNA microarray. J Appl Microbiol 97:57–67
Garny K, Neu TR, Horn H (2009) Sloughing and limited substrate conditions trigger filamentous growth in heterotrophic biofilms—measurements in flow-through tube reactor. Chem Eng Sci 64:2723–2732
Glover JR, Lindquist S (1998) Hsp104, hsp70, and hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell 94:73–82
Good M, Tang G, Singleton J, Reményi A, Lim WA (2009) The ste5 scaffold directs mating signaling by catalytically unlocking the fus3 map kinase for activation. Cell 136:1085–1097
Guo S, Zheng Y, Joung JG, Liu SQ, Zhang ZH, Crasta OR, Sobral BW, Xu Y, Huan SW, Fei ZJ (2010) Transcriptome sequencing and comparative analysis of cucumber flowers with different sex types. BMC Genomics 11:384
Han XJ, Wang YD, Chen YC, Lin LY, Wu QK (2013) Transcriptome sequencing and expression analysis of terpenoid biosynthesis genes in litsea cubeba. PLoS ONE 8:e76890
Jin H, Komita M, Aoe T (2017) The role of BiP retrieval by the KDEL receptor in the early secretory pathway and its effect on protein quality control and neurodegeneration. Front Mol Neurosci 10:222
Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori AM (2004) The kegg resource for deciphering the genome. Nucleic Acids Res 32:277–280
Kasavi C, Eraslan S, Oner ET, Kirdar B (2016) An integrative analysis of transcriptomic response of ethanol tolerant strains to ethanol in Saccharomyces cerevisiae. Mol Biosyst 12:464–476
Kempf C, Lengeler K, Wendland J (2017) Differential stress response of saccharomyces hybrids revealed by monitoring hsp104 aggregation and disaggregation. Microbiol Res 200:53–63
Kimura T, Hosoda Y, Sato Y, Kitamura Y, Ikeda T, Horibe T, Kikuchi M (2005) Interactions among yeast protein-disulfide isomerase proteins and endoplasmic reticulum chaperone proteins influence their activities. J Biol Chem 280:31438–31441
Lam FH, Ghaderi A, Fink GR, Stephanopoulos G (2014) Engineering alcohol tolerance in yeast. Science 346:71–75
Lee D, Kim M, Cho KH (2014) A design principle underlying the paradoxical roles of E3 ubiquitin ligases. Sci Rep 4:5573
Li J, Soroka J, Buchner J (2012) The hsp90 chaperone machinery: conformational dynamics and regulation by co-chaperones ☆. Biochim Biophys Acta 1823:624–635
Lorenz MC, Cutler NS, Heitman J (2000) Characterization of alcohol-induced filamentous growth in Saccharomyces cerevisiae. Mol Biol Cell 11:183–199
Ma M, Liu ZL (2010) Mechanisms of ethanol tolerance in Saccharomyces cerevisiae. Appl. Microbiol Biotechnol 87:829–845
Martínezmontañés F, Rienzo A, Povedahuertes D, Pascualahuir A, Proft M (2013) Activator and repressor functions of the Mot3 transcription factor in the osmostress response of Saccharomyces cerevisiae. Eukaryot Cell 12:636–647
Martos GI, Minahk CJ, Valdez GFD, Morero R (2007) Effects of protective agents on membrane fluidity of freeze-dried Lactobacillus delbrueckii ssp. bulgaricus. Lett Appl Microbiol 45:282–288
Mourier A, Larsson N (2011) Tracing the trail of protons through complex I of the mitochondrial respiratory chain. PLoS Biol 9:e1001129
Müller CA, Hawkins M, Retkute R, Malla S, Wilson R, Blythe MJ, Nakato R, Komata M, Shirahige K, Moura APSD, Nieduszynski CA (2014) The dynamics of genome replication using deep sequencing. Nucleic Acids Res 42:e3
Nagae M, Parniske M, Kawaguchi M, Takeda N (2016) The thiamine biosynthesis gene THI1 promotes nodule growth and seed maturation. Plant Physiol 172:2033–2043
Nillegoda NB, Theodoraki MA, Mandal AK, Mayo KJ, Ren HY, Sultana R, Wu K, Johnson J, Cyr DM, Caplan AJ (2010) Ubr1 and Ubr2 function in a quality control pathway for degradation of unfolded cytosolic proteins. Mol Biol Cell 21:2102–2116
Patterson JC, Klimenko ES, Thorner J (2010) Single-cell analysis reveals that insulation maintains signaling specificity between two yeast mapk pathways with common components. Sci Signal 3:625–638
Ranganathan S, Bai G, Lyubetskaya A, Knapp GS, Peterson MW, Gazdik M, Gomes ALC, Galagan JE, McDonough KA (2015) Characterization of a cAMP responsive transcription factor, Cmr (Rv1675c), in TB complex mycobacteria reveals overlap with the DosR (Devr) dormancy regulon. Nucleic Acids Res 44:134–151
Ren B, Dai HQ, Pei G, Tong YJ, Zhuo Y, Yang N, Su MY, Huang P, Yang YZ, Zhang LX (2014) ABC transporters coupled with the elevated ergosterol contents contribute to the azole resistance and amphotericin B susceptibility. Appl Microbiol Biot 98:2609–2616
Schröder M, Chang JS, Kaufman RJ (2000) The unfolded protein response represses nitrogen-starvation induced developmental differentiation in yeast. Gene Dev 14:2962–2975
Schuierer S, Carbone W, Knehr J, Petitjean V, Fernandez A, Sultan M (2017) A comprehensive assessment of RNA-seq protocols for degraded and low-quantity samples. BMC Genomics 18:442
Snoek T, Nicolino MP, Bremt SVD, Mertens S, Saels V, Verplaetse A, Steensels J, Verstrepen KJ (2015) Large-scale robot-assisted genome shuffling yields industrial Saccharomyces cerevisiae yeasts with increased ethanol tolerance. Biotechnol Biofuels 8:32
Stanley D, Bandara A, Fraser S, Chambers PJ, Stanley GA (2010) The ethanol stress response and ethanol tolerance of Saccharomyces cerevisiae. J Appl Microbiol 109:13–24
Teixeira MC, Raposo LR, Mira NP, Lourenço AB, Sá-Correia I (2009) Genome-wide identification of Saccharomyces cerevisiae genes required for maximal tolerance to ethanol. Appl Environ Microbiol 75:5761–5772
Tesnière C, Brice C, Blondin B (2015) Responses of Saccharomyces cerevisiae, to nitrogen starvation in wine alcoholic fermentation. Appl Microbiol Biotechnol 99:7025–7034
Teste MA, Duquenne M, François JM, Parrou JL (2009) Validation of reference genes for quantitative expression analysis by real-time RT-PCR in Saccharomyces cerevisiae. BMC Mol Biol 10:99
Valnot I, Osmond S, Gigarel N (2000) Mutations of the sco1 gene in mitochondrial cytochrome c oxidase deficiency with neonatal-onset hepatic failure and encephalopathy. Am J Hum Genet 67:1104–1109
Wolak N, Kowalska E, Kozik A, Rapala-Kozik M (2014) Thiamine increases the resistance of baker’s yeast Saccharomyces cerevisiae against oxidative, osmotic and thermal stress, through mechanisms partly independent of thiamine diphosphate-bound enzymes. FEMS Yeast Res 14:1249–1262
Yoshikawa K, Tanaka T, Furusawa C, Nagahisa K, Hirasawa T, Shimizu H (2009) Comprehensive phenotypic analysis for identification of genes affecting growth under ethanol stress in Saccharomyces cerevisiae. FEMS Yeast Res 9:32–44
Acknowledgements
Authors gratefully acknowledge the financial support of National Natural Science Foundation of China (NNSF No. 31471709), K. C. Wong Magna Fund in Ningbo University and the Scientific Research Foundation of Graduate School of Ningbo University.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Li, R., Xiong, G., Yuan, S. et al. Investigating the underlying mechanism of Saccharomyces cerevisiae in response to ethanol stress employing RNA-seq analysis. World J Microbiol Biotechnol 33, 206 (2017). https://doi.org/10.1007/s11274-017-2376-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-017-2376-5