Abstract
Therapeutic failures against diseases due to resistant Gram-negative bacteria have become a major threat nowadays as confirmed by surveillance reports across the world. One of the methods of development of multidrug resistance in Escherichia coli and Pseudomonas aeruginosa is by means of RND efflux pumps. Inhibition of these pumps might help to combat the antibiotic resistance problem, for which the structure and regulation of the pumps have to be known. Moreover, judicious antibiotic use is needed to control the situation. This paper focuses on the issue of antibiotic resistance as well as the structure, regulation and inhibition of the efflux pumps present in Escherichia coli and Pseudomonas aeruginosa.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The development of antibiotic resistance in Gram-negative bacteria is a worldwide concern. Frequencies of therapeutic failures against bacterial infections have increased due to the propagation of multidrug resistant (MDR) Gram-negative bacteria (Saier et al. 1998; Fernandez-Recio et al. 2004; Moreira et al. 2004; Rice 2006; Pagès and Amaral 2009; Fernández and Hancock 2012; Nikaido and Pagès 2012; Fernando and Kumar 2013). Antibiotic resistant bacteria have been declared as a major health threat by World Health Organization (WHO 2014). Antibiotic resistance can be classified into three types: intrinsic resistance due to inherent properties of the microorganism; acquired resistance due to incorporation of new genetic material or adaptive resistance due to alteration of gene expression under stress conditions (Fernández and Hancock 2012). Resistance in Gram-negative bacteria might be due to acquired genes, which are part of mobile elements known as gene cassettes (Hall and Collins 1998). Acquired resistance can be a result of alteration in expression of those genes encoding for multidrug transporters due to the occurrence of mutation and amplification in them. Mutation in global regulatory genes can also lead to acquired resistance in bacteria (Moreira et al. 2004).
The bacterial MDR phenotype has been found to be the result of decreased influx or increased efflux of antibiotics (Lomovskaya and Watkins 2001; Fernandez-Recio et al. 2004; Poole 2008; Nikaido and Pagès 2012). Gram-negative bacteria such as Escherichia coli and Pseudomonas aeruginosa export antibiotics by means of efflux pumps (Poole et al. 1993; Ma et al. 1995; Nikaido 1998a, b; Eswaran et al. 2004; Fernandez-Recio et al. 2004; Nikaido and Takatsuka 2009; Xu et al. 2012; Fernando and Kumar 2013). Efflux pumps are transport proteins which are involved in extrusion process of toxic substances from the cell (Webber and Piddock 2003). Efflux pumps contribute to intrinsic antibiotic resistance (Hancock 1998; Rosenberg et al. 2000). Efflux system of bacteria can be divided into five classes—Major facilitator superfamily (MFS), resistance-nodulation division (RND) family, ATP binding cassette (ABC) family, small multidrug resistance (SMR) family and multi-drug and toxic compound extrusion (MATE) family (Lomovskaya and Watkins 2001; Ramos et al. 2002; Schweizer 2003; Webber and Piddock 2003; Moreira et al. 2004; Poole 2004; Mahamoud et al. 2007; Stavri et al. 2007; Fernández and Hancock 2012; Fernando and Kumar 2013). Efflux pumps which are concerned only with expulsion of antibiotics are usually encoded on transposons and transmissible plasmids, while others such as the multidrug resistance (MDR) pumps concerned with expulsion of compounds having different antibacterial mode of action are a part of bacterial chromosomes (Lomovskaya and Watkins 2001; Moreira et al. 2004). Susceptibility of bacteria to antibiotics is linked to membrane permeability, which can be modified by decreased production of porin or increased efflux pump expression. In bacterial MDR phenotype, these phenomena are associated with acquisition of additional resistance mechanisms, such as antibiotic target mutation or production of enzymes capable of degradation of antibiotics (Moreira et al. 2004; Poole 2007; Pagès and Amaral 2009; Nikaido 1998a, b; Nikaido and Pagès 2012). RND efflux pumps are common in Gram-negative bacteria and are almost always chromosomally encoded (Nikaido 1998a, b; Poole 2004, 2008). RND efflux pumps monitor the level of metabolite accumulated due to pathway blockages and prevent them from reaching toxic levels (Poole 2008).
Structures of efflux systems, having RND pumps, of Escherichia coli as well as Pseudomonas aeruginosa have been resolved with the help of X-ray crystallography (Yu et al. 2003). Structural studies have revealed that the transporters capture the substrate either from the outer leaflet of cytoplasmic membrane or from the periplasm (Pagès and Amaral 2009; Nikaido and Pagès 2012; Fernando and Kumar 2013). AcrAB–TolC and MexAB–OprM are RND efflux systems present in E. coli and P. aeruginosa and have been found essential for the survival, colonization and virulence in them (Moreira et al. 2004; Poole 2008; Pagès and Amaral 2009; Nikaido and Pagès 2012; Fernando and Kumars 2013). AcrAB in E. coli makes it resistant to the abundant bile salts present in their normal habitat (Nikaido 1998a, b; Poole 2000). The deletion or deactivation of the main efflux pump system leads to its replacement by RND efflux pump systems which are genetically distinct (Poole 2007; Nikaido and Pagès 2012). Efflux pump inhibitors might target inactivation of efflux parameters which impairs antibiotic efflux (Pagès and Amaral 2009).
For the development of new and effective therapy to deal with infections of MDR Gram-negative bacteria, it is essential to understand MDR efflux pump structure and function (Saier et al. 1998; Pagès and Amaral 2009) along with every interaction as well as processes that regulate the pump efficacy. To understand interactions, the identification of amino acid residues of pump and pharmacophoric groups at drug surface has to be done (Nikaido and Pagès 2012).
Efflux and multidrug resistance in Gram-negative bacteria E. coli and P. aeruginosa
A bacterium might be resistant or susceptible to a drug based on the presence or absence of functional RND components (Blair and Piddock 2009). Efflux pump when exposed to one of its substrates can favour its over-expression, leading to cross resistance to other substrates including antimicrobials which are clinically relevant (Moreira et al. 2004). MDR is associated with RND system over-expression (Piddock 2006; Blair and Piddock 2009; Ruggerone et al. 2013). During treatment of Gram negative bacterial infections, the polyspecificity of AcrAB pump triggers spreading of efflux producing bacteria. The minimum inhibitory concentrations (MIC) of antimicrobials agents are affected by overproduction of AcrAB–TolC and it has been observed that the pumps are able to expel out several antimicrobial agents including antibiotics, biocides, etc (Nikaido and Pagès 2012) as shown in Fig. 1. Experiments have confirmed the active export of chloramphenicol from E. coli cells (Moreira et al. 2004). Tetracycline efflux occurs by classical tet efflux pumps (Nikaido 1996) encoded by genes of monophyletic origin and catalyzed by drug:H+ antiport (Aminov et al. 2002). Tetracycline efflux in E. coli occurs in two stages, first stage is of low resistance through efflux while second stage is of high resistance due to accumulation of other resistance mechanism (Fernando and Kumar 2013).
AcrB of E. coli determines substrate specificity and is driven by proton motive force (Vargiu and Nikaido 2012). It can deal with various structurally diverse compounds such as cephalosporins, fluoroquinolones, penicillins, chloramphenicol, etc. Studies have revealed that AcrB of E. coli has a distal binding pocket and a proximal binding pocket. These pockets help in broad spectrum antibiotic resistance as they can accommodate substrates of varying sizes and properties (Vargiu and Nikaido 2012; Blair et al. 2015). Resolved structure of AcrB, suggests that, there is interaction between substrate and hydrophobic interior of the substrate binding site present within AcrB (Nikaido and Takatsuka 2009). World Health Organization (WHO) has revealed high levels of resistance of E. coli to the third generation cephalosporins and fluoroquinolones (FQs). Resistance of E. coli to FQs is mainly due to mutation and resistance to third generation cephalosporins which indicates that they produce ESBL (extended spectrum beta-lactamase). In 2013, it was found that in 17 out of 22 countries, about 85–100% isolates of E. coli were ESBL positive (The state of the world’s antibiotic 2015). Studies have shown the role of efflux pumps in the FQ resistant isolates of E. coli and it has also been reported that FQ treatment contributes towards emergence of efflux producers (Nikaido and Pagès 2012). The FQ efflux system of E. coli is found to be encoded by acrAB–tolC genes. For therapeutic intervention, it is logical to target FQ-MDR efflux pumps, which when inactivated increase bacterial susceptibility to fluoroquinolones and other antibiotics and prevent emergence of resistance to fluoroquinolones (Poole 2000). Another mechanism of FQ resistance found in clinical isolate of E. coli is QepA, which is a plasmid-mediated efflux pump (Yamane et al. 2007).
In case of the opportunistic pathogen Pseudomonas aeruginosa, responsible for severe infections, RND efflux pumps like MexAB–OprM, MexCD–OprJ, MexEF–OprN and MexXY–OprM are involved which contribute strongly to its reduced susceptibility towards antibiotics (Mine et al 1999; Lomovskaya et al. 2001; Poole 2001; Aeschlimann 2003; Schweizer 2003; Moreira et al. 2004; Nikaido and Pagès 2012). P. aeruginosa has 10–12 RND family transporters which regulate independently and share antibiotic substrates (Poole 2008; Fernando and Kumar 2013). The MexAB–OprM system in clinical isolates of P. aeruginosa is highly conserved indicating its role in intrinsic resistance of the bacteria. The MexAB–OprM system pumps out a large number of antimicrobials such as chloramphenicol, tetracycline, streptonigrin, ciprofloxacin and nalidixic acid. It pumps out antibiotics of the class quinolones, Poole 2001; Moreira et al. 2004; Fernando and Kumar 2013). Overproduction of MexAB–OprM effux system due to mutations in nalB repressor gene of P. aeruginosa renders the bacteria more resistant to several agents (Nikaido 1998a, b; Li et al. 2000). In contrast to MexAB–OprM system, the MexCD–OprJ doesn’t pump out the conventional cephalosporins but is concerned with pumping out of cephalosporins having quaternary-nitrogen-containing constituents at the 3-position. There is a possibility that MexCD–OprJ needs the presence of at least one positively charged group at the hydrophilic end of its substrate, while there is no such need in case of MexAB–OprM. MexCD–OprJ cannot eliminate Nikaido 1998). MexCD–OprJ system is responsible for expulsion of various drugs such as quinolones, macrolides, lincomyscin, novobiocin and tetracyclines (Moreira et al. 2004). The MexEF–OprN system in P. aeruginosa increases their resistance to chloramphenicol, quinolones, trimethoprim and imipenem. The MexEF–OprN system provides resistance to fluoroquinolones but sensitivity has been found to cefotaxime, novobiocin, ceftazidime, cefoperazone, cefpirone, carbenicilin, cefozopran, meropenem, andaztreonam (Moreira et al. 2004). MexCD–OprJ and MexEF–OprN are found in nfxB and nfxC mutants (Lomovskaya et al. 1999; Aeschlimann 2003). Both contribute towards acquired resistance in the bacterium (Masuda et al. 2000). P. aeruginosa mutants with deletions for the genes responsible for encoding three best characterized FQ-MDR efflux systems were found hypersusceptible to fluoroquinolones (Poole 2000). The MexXY–OprM system provides resistance to acriflavine, ethidium bromide, fluoroquinolones and erythromycin. It also provides certain amount of resistance to tetracycline, kanamycin, chloramphenicol and some Moreira et al. 2004).
Evolution of efflux pumps in E. coli and P. aeruginosa
Regarding the evolution of multidrug transporters, there are two hypotheses; According to the first hypothesis, bacteria due to the exposure to various toxic substances have undergone evolution and accordingly have developed mechanisms such as multidrug transporters for detoxification and elimination of the toxins (Ramos et al. 2002). According to the second hypothesis, the export of toxic compounds by the multidrug transporters is opportunist and occurs only under exposure of drug in clinic or experimental situations (Moreira et al. 2004). Out of 29 putative efflux pumps identified in E. coli, 5 belong to RND family and the phylogenetic analysis of RND family members showed that these proteins based on their function might be classified into three subfamilies, of which the first subfamily deals with divalent heavy metal ions, second one specific for lipooligosaccharides and the third subfamily catalyses efflux of multiple drugs (Saier et al. 1998). AcrB protein of E. coli and MexB of P. aeruginosa are putative pumps of RND family (George 1996).
Structure of efflux pumps
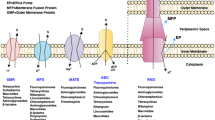
Multidrug efflux pumps have three components- An integral inner membrane protein, which is either an ABC transporter or proton antiporter of RND, a protein of TolC exit duct family anchored in outer membrane and projecting across periplasm and a periplasmic adapter protein anchored to inner membrane by single transmembrane helix (Zgurskaya and Nikaido 2000; Eswaran et al. 2004; Moreira et al. 2004; Poole 2008; Nikaido and Takatsuka 2009). The multidrug efflux system of E. coli is AcrAB system having an RND type transporter ArcB, accessory protein AcrA in periplasm and outer membrane channel TolC as shown in Fig. 2 (Nikaido and Zgurskaya 2001; Ramos et al. 2002; Fernandez-Recio et al. 2004; Nikaido and Takatsuka 2009; Xu et al. 2012). Pathogenic bacteria have tripartite pumps with broad and overlapping substrate specificities (Eswaran et al. 2004). P. aeruginosa has at least four distinct major efflux (Mex) systems, whereas AcrA–ArcB–TolC, the major pump of E.coli determines resistance to antibiotics, detergents, organic solvents, dyes and bile salts (Eswaran et al. 2004; Fernando and Kumar 2013). In E. coli, AcrD transporter is also present which shares similar topology as AcrB and is responsible for aminoglycoside efflux (Rosenberg et al. 2000; Elkins and Nikaido 2002).
AcrB is a trimer having 100 Å diameter and 50 Å long transmembrane domain, comprising 36 α helices and there is a funnel like structure at the top of the periplasmic domain with internal diameter of 30 Å, which has been termed as TolC-docking domain, due to its similarity with the model of TolC entrance in open state (Eswaran et al. 2004). TolC in open state bind more robustly to AcrA and AcrB than TolC in closed state (Fernandez-Recio et al. 2004). TolC is a 140 Å long tapered cylinder, composed of 40 Å long outer membrane β barrel and 100 Å long α helical barrel across periplasm and a mixed α/β structure forms a band around the mid portion of α helical barrel (Eswaran et al. 2004; Nikaido and Takatsuka 2009). In the planar lipid bilayer, TolC forms channels which enable export of antibiotics across outer membrane of E. coli (Poole 2000). TolC can be found in isolated state which suggests that it must be in closed state, otherwise there would be a large open pore in outer membrane. The interaction of TolC with other pump components perhaps helps in its allosteric transition to open state (Fernandez-Recio et al. 2004). Functional TolC is essential for AcrAB efflux pump operation (Fralick 1996). The AcrAB proteins are homologous to Mex proteins of P. aeruginosa, while there is limited homology between TolC and OEPs of P. aeruginosa (Poole 2000). AcrB and MexB have 70% similar amino acid sequences (Tikhonova et al. 2002).
The monomer of MexA has three linearly arranged subdomains- β barrel, lipoyl domain and 47 Å α helical hairpin with a straight C-terminal helix and an N-terminal helix (Eswaran et al. 2004). It has been predicted that coiled region of AcrA is about 14 residues longer than that of MexA (Fernandez-Recio et al. 2004). The length of AcrA is predicted to be 17 nm and it is the best-characterized membrane fusion protein in efflux pumps. Its first 24 amino acids show feature of a typical bacterial lipoprotein signal peptide. The amino terminal cysteine residue, after cleavage of the signal peptide, is acylated with the fatty acids of inner membrane (Ramos et al. 2002).
Docking AcrA model into TolC open state manually, provided eight possible orientations, which when further refined, showed −40.5 kcal/mol scoring energy forming three closed interfacial contacts (Fernandez-Recio et al. 2004). From the crystal structure of AcrB in complex with the drugs, it has been found that for opening of periplasmic pore, there is a need for drug binding, conduction of protons as well as protein–protein interaction with AcrB and TolC (Fernandez-Recio et al. 2004). The structural analysis of multidrug transport proteins has revealed that they possess hydrophobic binding sites. They bind to the substrates by both hydrophobic interactions as well as electrostatic attraction (Moreira et al. 2004).
Regulation of efflux pumps
RND pumps are regulated by both local and global regulators. Overexpression of the pump may be due to the interaction of certain molecules with the local repressor, which causes the derepression of the operon. Another possible reason of overexpression might be the mutation in the repressor coding genes (Webber and Piddock 2003), as seen in case of AcrAB pump of E. coli and the MexAB–OprM pump of P. aeruginosa, resulting from the mutation in their repressor coding genes, acrR and mexR respectively (Nikaido 1996; Fernando and Kumar 2013). MexR is involved in auto-regulation by repression of mexR promoter in addition to control of mexA promoter transcription (Evans et al. 2001; Moreira et al. 2004). The nalB mutation causes overexpression of MexAB–OprM (Nikaido 1996; Lomovskaya et al. 1999). MexCD–OprJ overexpression is controlled by nfxB which is a local repressor gene (Nikaido 1998a, b). In E. coli AcrAB pump, induced by bile salts and fatty acids, overexpression has also been a result of activation of expression by global regulator Rob (Fernando and Kumar 2013). AcrAB is regulated by MarA and its homologs SoxS (Webber and Piddock 2003) and RobA, which increase efflux by increasing transcription of the operon acrAB and also downregulates OmpF porin synthesis, through enhanced production of MicF, an antisense RNA (Okusu et al. 1996; Nikaido 1996, 1998a, b; Poole 2000; Fernando and Kumar 2013).
Inhibition of efflux pumps
An important area of drug discovery is the development of efflux pump inhibitors (EPIs) (Bambeke et al. 2003). EPIs are agents that inhibit efflux (Piddock et al. 2010) as shown in Fig. 3. The increase in accumulation of drug under the presence of an inhibitor suggests that they block the efflux of drugs (Stavri et al. 2007). Efflux pump of Gram-negative bacteria may be inhibited by altering the regulation of their expression, altering functional assembly of its components, blockage of outer membrane channel with the help of plug, collapsing efflux energy, competitive or non-competitive inhibition of affinity sites of efflux pump with a non-antibiotic molecule and changing antibiotic chemical design in order to reduce its affinity towards efflux pump binding sites (Pagès and Amaral 2009).
The repression of expression of genes, using antisense oligonucleotides or molecules and small interfering RNA, was exemplified for inhibition of efflux pump in E. coli, but it can be applied to every pump of known regulatory mechanism (Pagès and Amaral 2009). The functional assembly of components of efflux pumps like typical envelope lipoprotein AcrA might be blocked by globomycin, an inhibitor of signal peptidase II, which is responsible for removal of lipoprotein signal sequence of the exported membrane proteins (Pagès and Amaral 2009). However it has been observed that MexCD–OprJ pump expels cerulenin which is an inhibitor of fatty acid biosynthesis in bacteria (Pagès and Amaral 2009). To obstruct the central cavity of AcrB pump, which allows periport of drugs, periplasmic transit blockers can be used (Pagès et al. 2005). For pore blocker designing, it is essential to know the dynamics of drug flux within the channel (Pagès and Amaral 2009). Efflux energy can be collapsed by several compounds such as potassium cyanide and carbonyl cyanide m-chlorophenylhydrazone (Pagès and Amaral 2009).
EPIs might fulfill certain criteria; they must show activity only in those strains having efflux pump, amplify accumulation and reduce extrusion of substrates of efflux pumps and the proton gradient present across the inner membrane must not be affected (Lomovskaya and Watkins 2001).
Various compounds such as globomycin, carbonyl cyanide m-chlorophenylhydrazone (CCCP), phenylalanyl arginyl Pagès et al. 2005). Among arylpiperazines, 1-(1-naphthylmethyl)-piperazine (NMP) was found to increase accumulation of substrates in E. coli, which are otherwise effluxed by the pumps, reduce MICs of two or more antibiotics in strains having overexpressed efflux pumps and showed no inhibition in growth of the strains deficient of efflux pumps (Bohnert and Kern 2005).
Discovery of novel natural product as EPI is of immense importance for future use in inhibiting MDR pumps (Stavri et al. 2007). Berberine is a common alkaloid in plant species belonging to family Berberidaceae, which shows weak antibiotic properties as it is pumped out (Stermitz et al. 1999). Plant extracts from Berberis fremontii, which alone had no impact on cell growth but showed inhibition in presence of berberine suggests the presence of inhibitor in them, which was found to be 5́-Methoxyhydnocarpin (5́-MHC) (Stermitz et al. 1999). Ciprofloxacin is a substrate common to many efflux pumps (Piddock et al. 2010). Chloroform extract of Berberis aetnensis, when fractioned further into green and yellow sub-fractions, then used in combination with ciprofloxacin, lowered its MIC for strains of E. coli and P. aeruginosa (Stavri et al. 2007). Plant based natural product, p-Coumaric acid showed inhibitory action when used with ciprofloxacin in P. aeruginosa with overexpressed MexAB–OprM efflux pumps (Choudhury et al. 2016).
Efflux pump inhibitor development approach for therapy is quite challenging because of its possible effects on eukaryotic transporters and so inhibitors specific for prokaryotic cells are to be designed (Bambeke et al. 2003). In the process of identification of EPI, it is important to note whether a compound actually inhibits pump or other has other antibacterial properties (Stavri et al. 2007).
Conclusion
Antibiotic resistant bacteria are responsible for health crisis all over the world. Efflux pumps are one of the mechanisms by which the bacteria develop resistance to drugs. EPIs help to inhibit the expulsion of drugs from the cell and reduce the MIC of the drugs. However, till date no EPI has been clinically approved. So it is important to make use of available information about efflux systems and search for non-toxic, more effective and broad spectrum EPIs in natural resources, which would potentiate the already available antibiotics. There is a need to determine the clinical potential of the natural product based EPIs.
References
Aeschlimann JR (2003) The role of multidrug efflux pumps in the antibiotic resistance of Pseudomonas aeruginosa and other Gram-negative bacteria. Pharmacotherapy 23:916–924
Aminov RI, Chee- Stanford JC, Garrigues N, Teferedegne B, Krapac IJ, White BA, Mackie RI (2002) Development, validation, and application of PCR primers for detection of tetracycline efflux genes of Gram-negative bacteria. Appl Environ Microbiol 68:1786–1793
Bambeke FV, Glupczynski Y, Plésiat P, Pechère JC, Tulkens PM (2003) Antibiotic efflux pumps in prokaryotic cells: occurrence, impact on resistance and strategies for the future of antimicrobial therapy. J Antimicrob Chemother 51:1055–1065
Blair JMA, Piddock LJV (2009) Structure function and inhibition of RND efflux pumps in Gram-negative bacteria: an update. Curr Opin Microbiol 12:512–519
Blair JMA, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJV (2015) Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol 13:42–51
Bohnert JA, Kern WV (2005) Selected arylpiperazines are capable of reversing multidrug resistance in Escherichia coli overexpressing RND efflux pumps. Antimicrob Agents Chemother 49:849–852
Choudhury D., Talukdar A.D., Chetia P., Bhattacharjee A. and Choudhary M.D. (2016) Screening of natural products and derivatives for identification of RND efflux pump inhibitors. Comb Chem High Throughput Screen 19:705–713
Elkins CA, Nikaido H (2002) Substrate specificity of the RND-type multidrug efflux pumps AcrB and AcrD of Escherichia coli is determined predominately by two large periplasmic loops. J Bacteriol 184:6490–6498
Eswaran J, Koronakis E, Higgins MK, Hughes C, Koronakis V (2004) Three’s company: component structures bring a closer view of tripartite drug efflux pumps. Curr Opin Struct Biol 14:741–747
Evans E, Adwoye L, Poole K (2001) MexR repressor of the mexAB-oprM multidrug efflux operon of Pseudomonas aeruginosa: identification of MexR binding sites in mexA-mexR intergenic region. J Bacteriol 183:807–812
Fernández L, Hancock Robert EW (2012) Adaptive and mutational resistance: role of porins and efflux pumps in drug resistance. Clin Microbiol Rev 25:661–681
Fernandez-Recio J, Walas F, Federici L, Pratap JV, Bavro VN, Miguel RN, Mizuguchi K, Luisi B (2004) A model of a transmembrane drug-efflux pump from Gram-negative bacteria. FEBS Lett 578:5–9
Fernando DM, Kumar A (2013) Resistance-nodulation-division multidrug efflux pumps in Gram-negative bacteria: role in virulence. Antibiotics 2:163–181
Fralick JA (1996) Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J Bacteriol 178:5803–5805
George AM (1996) Multidrug resistance in enteric and other gram negative bacteria. FEMS Microbiol Lett 139:1–10
Hall RM, Collins CM (1998) Antibiotic resistance in gram-negative bacteria: the role of gene cassettes and integrons. Drug Resist Updat 1:109–119
Hancock REW (1998) Resistance mechanisms in Pseudomonas aeruginosa and other nonfermentative Gram-negative bacteria. CID 27:S93–S99
Li X-Z, Zhang L, Poole K (2000) Interplay between the MexA–MexB–OprM multidrug efflux system and the outer membrane barrier in the multiple antibiotic resistance of Pseudomonas aeruginosa. J Antimicrob Chemother 45:433–436
Lomovskaya O, Watkins W (2001) Inhibition of efflux pumps as a novel approach to combat drug resistance in bacteria. J Mol Microbiol Biotechnol 3:225–236
Lomovskaya O, Lee A, Hoshino K, Ishida H, Mistry A, Warren MS, Boyer E, Chamberland S, Lee VJ (1999) Use of genetic approach to evaluate the consequences of inhibition of efflux pumps Pseudomonas aeruginosa. Antimicrob Agents Chemother 43:1340–1346
Lomovskaya O, Warren MS, Lee A, Galazzo J, Fronko R, Lee M, Blais J, Cho D, Chamberland S, Renau T, Leger R, Hecker S, Watkins W, Hoshino K, Ishida H, Lee VJ (2001) Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob Agents Chemother 45:105–116
Ma D, Cook DN, Alberti M, Pon NG, Nikaido H, Hearst JE (1995) Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol Microbiol 16:45–55
Mahamoud A, Chevalier J, Alibert-Franco S, Kern WV, Pagès J-M (2007) Antibiotic efflux pumps in Gram-negative bacteria: the inhibitor response strategy. J Antimicrob Chemother 59:1223–1229
Masuda N, Sakagawa E, Ohya S, Gotoh N, Tsujimoto H, Nishino T (2000) Substrate specificities of MexAB–OprM, MexCD–OprJ and MexXY–OprM efflux pumps Pseudomonas aeruginosa. Antimicrob Agents Chemother 44:3322–3327
Mine T, Morita Y, Kataoka A, Mizushima T, Tsuchiya T (1999) Expression in Escherichia coli of a new multidrug efflux pump MexXY, from Pseudomonas aeruginosa. Antimicrob Agents Chemother 43:415–417
Moreira MAS, Souza ECD, Moraes CAD (2004) Multidrug efflux systems in Gram-negative bacteria. Braz J Microbiol 35:19–28
Nikaido H (1996) Multidrug efflux pumps of Gram-negative bacteria. J Bacteriol 178:5853–5859
Nikaido H (1998a) Multiple antibiotic resistance and efflux. Curr Opin Microbiol 1:516–523
Nikaido H (1998b) Antibiotic resistance caused by Gram-negative Multidrug efflux pumps. CID 27:S32–S41
Nikaido H, Pagès JM (2012) Broad-specificity efflux pumps and their role in multidrug resistance of Gram-negative bacteria. FEMS Microbiol Rev 36:340–363
Nikaido H, Takatsuka Y (2009) Mechanisms of RND multidrug efflux pumps. Biochim Biophys Acta 1794:769–781
Nikaido H, Zgurskaya HI (2001) AcrAB and related multidrug efflux pumps of Escherichia coli. J Mol Microbiol 3:215–218
Okusu H, Ma D, Nikaido H (1996) AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J Bacteriol 178:306–308
Pagès JM, Amaral L (2009) Mechanisms of drug efflux and strategies to combat them: challenging the efflux pump of Gram-negative bacteria. Biochim Biophys Acta 1794:826–833
Pagès JM, Masi M, Barbe J (2005) Inhibitors of efflux pumps in Gram-negative bacteria. Trends Mol Med 11:382–389
Piddock LJV (2006) Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin Microbiol Rev 19:382–402
Piddock LJV, Garvey MI, Rahman MM, Gibbons S (2010) Natural and synthetic compounds such as trimethoprim behave as inhibitors of efflux in Gram-negative bacteria. J Antimicrob Chemother 65:1215–1223
Poole K (2000) Efflux mediated resistance to fluoroquinolones in Gram-negative bacteria. Antimicrob Agents Chemother 44:2233–2241
Poole K (2001) Multidrug efflux pumps and antimicrobial resistance in Pseudomonas aeruginosa and related organisms. J Mol Microbiol Biotechnol 3:255–264
Poole K (2004) Efflux mediated multi-resistance in Gram-negative bacteria. Clin Microbiol Infect 10:12–26
Poole K (2007) Efflux pumps as antimicrobial resistance mechanisms. Ann Med 39:162–176
Poole K (2008) Bacterial multidrug efflux pumps serve other functions. Microbe 3:179–185
Poole K, Krebes K, McNally C, Neshat S (1993) Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J Bacteriol 175:7363–7372
Ramos JL, Duque E, Gallegos MT, Godoy P, González MIR, Rojas A, Terán W, Segura A (2002) Mechanisms of solvent tolerance in Gram-negative bacteria. Annu Rev Microbiol 56:743–768
Rice LB (2006) Unmet medical needs in antibacterial therapy. Biochem Pharmacol 71:991–995
Rosenberg EY, Ma D, Nikaido H (2000) AcrD of Escherichia coli is an aminoglycoside efflux pump. J Bacteriol 182:1754–1756
Ruggerone P, Vargiu AV, Collu F, Fischer N, Kandt C (2013) Molecular dynamics computer simulations of multidrug RND efflux pumps. Comput Struct Biotechnol J 5:1–11
Saier MH Jr, Paulsen IT, Sliwinski MK, Pao SS, Skurray RA, Nikaido H (1998) Evolutionary origins of multidrug and drug specific efflux pumps in bacteria. FASEB J 12:265–274
Schweizer HP (2003) Efflux as mechanism of resistance to antimicrobials in Pseudomonas aeruginosa and related bacteria: unanswered questions. Genet Mol Res 2:48–62
Stavri M, Piddock LJV, Gibbons S (2007) Bacterial efflux pump inhibitors from natural sources. J Antimicrob Chemother 59:1247–1260
Stermitz FR, Lorenz P, Tawara JN, Zenewicz LA, Lewis K (1999) Synergy in a medicinal plant: antimicrobial action of berberine potentiated by 5́-methoxyhydnocarpin, a multidrug pump inhibitor. PNAS 97:1433–1437
Tikhonova EB, Wang Q, Zgurskaya HI (2002) Chimeric analysis of the multicomponent multidrug efflux transporters from Gram-negative bacteria. J Bacteriol 184:6499–6507
Vargiu AV, Nikaido H (2012) Multidrug binding properties of the AcrB efflux pump characterized by molecular dynamics simulations. PNAS 109:20637–20642
Webber MA, Piddock L.J.V. (2003) The importance of efflux pumps in bacterial antibiotic resistance. J Antimicrob Chemother 51:9–11
WHO (2014) Antimicrobial resistance: global report on surveillance. WHO Press, World Health Organization, Switzerland
Xu Y, Moeller A, Jun S, Le M, Yoon B, Kim J, Lee K, Ha N (2012) Assembly and channel opening of outer membrane protein in tripartite drug efflux pumps of Gram-negative bacteria. J Biol Chem 287:11740–11750
Yamane K, Wachino J, Suzuki S, Kimura K, Shibata N, Kato H, Shibayama K, Konda T, Arakawa Y (2007) New plasmid-mediated fluoroquinolone efflux pump, QepA, found in Escherichia coli clinical isolate. Antimicrob Agents Chemother 51:3354–3360
Yu EW, Aires JR, Nikaido H (2003) AcrB multidrug efflux pump of Escherichia coli: composite structure binding cavity of exceptional flexibility generates its extremely wide substrate specificity. J Bacteriol 185:5657–5664
Zgurskaya HI, Nikaidos H (2000) Cross-linked complex between oligomeric periplasmic lipoprotein AcrA and the innermembrane-associated multidrug efflux pump AcrB from Escherichia coli. J Bacteriol 182:4264–4267
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Puzari, M., Chetia, P. RND efflux pump mediated antibiotic resistance in Gram-negative bacteria Escherichia coli and Pseudomonas aeruginosa: a major issue worldwide. World J Microbiol Biotechnol 33, 24 (2017). https://doi.org/10.1007/s11274-016-2190-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-016-2190-5