Abstract
Lanzhou lily (Liliumdavidii var. unicolor) is the best edible lily as well as a traditional medicinal plant in China. The microbes associated with plant roots play crucial roles in plant growth and health. However, little is known about the differences of rhizosphere microbes between healthy and wilted Lanzhou lily (Lilium davidii var. unicolor) plants. The objective of this study was to compare the rhizosphere microbial community and functional diversity of healthy and wilted plants, and to identify potential biocontrol agents with significant effect. Paired end Illumina Mi-Seq sequencing of 16S rRNA and ITS gene amplicons was employed to study the bacterial and fungal communities in the rhizosphere soil of Lanzhou lily plants. BIOLOG technology was adopted to investigate the microbial functional diversity. Our results indicated that there were major differences in the rhizosphere microbial composition and functional diversity of wilted samples compared with healthy samples. Healthy Lanzhou lily plants exhibited lower rhizosphere-associated bacterial diversity than diseased plants, whereas fungi exhibited the opposite trend. The dominant phyla in both the healthy and wilted samples were Proteobacteria and Ascomycota, i.e., 34.45 and 64.01 %, respectively. The microbial functional diversity was suppressed in wilted soil samples. Besides Fusarium, the higher relative abundances of Rhizoctonia, Verticillium, Penicillium, and Ilyonectria (Neonectria) in the wilted samples suggest they may pathogenetic root rot fungi. The high relative abundances of Bacillus in Firmicutes in healthy samples may have significant roles as biological control agents against soilborne pathogens. This is the first study to find evidence of major differences between the microbial communities in the rhizospheric soil of healthy and wilted Lanzhou lily, which may be linked to the health status of plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The rhizosphere is defined as the narrow region of soil that surrounds plant roots, which is affected directly by the living roots (Hiltner 1904; Kent and Triplett 2002). It is a “hot” spot for microbial interactions because exudates released by plant roots comprise the main food sources for microorganisms, thereby driving their population density and activities (Raaijmakers et al. 2009). Thus, the rhizosphere is a niche where plants, soil, and microorganisms interact dynamically. Vast numbers of highly diverse microbes are associated with plant roots, and the complex microbial community in the rhizosphere has crucial roles in plant growth and health. Various biotic and abiotic factors are assumed to influence the microbial communities in this region. In particular, plant species and the soil type are considered to shape the structure and function of microbial communities in the rhizosphere, and plants can recruit protective microorganisms after attacks by pathogens or insects, while their activities may suppress pathogens in the rhizosphere (Berendsen et al. 2012; Berg and Smalla 2009). It has been demonstrated that a high proportions of rhizosphere- and other soil-inhabiting microorganisms remain unculturable on synthetic media at present (Fitter et al. 2005; Kent and Triplett 2002), but the development and improvement of next-generation sequencing have revolutionized microbial research, thereby overcoming the inherent bias caused by culture-dependent and traditional molecular techniques (Glenn 2011; Gottel et al. 2011; Metzker 2010). Moreover, the lower cost and greater capacity of Illumina sequencing have facilitated more accurate estimation of microbial community diversity, which makes it a more suitable platform compared with 454 pyrosequencing (Degnan and Ochman 2012).
Many microorganisms of the rhizosphere community have neutral influences on plants, whereas others have deleterious or beneficial effects. Some rhizosphere-inhabiting microbes are known to participate in detrimental interactions with plants, which lead to the development of root, stem, and crown rot diseases, as well as vascular wilting, take-all, and damping-off; whereas others are clearly beneficial for plants by stimulating their growth, enhancing nutrient uptake, and helping to control soil-borne diseases (Berg 2009; Garbeva et al. 2004; Keel and Défago 1997). Indeed, plant-friendly or beneficial microorganisms, including nitrogen-fixing bacteria, endo- and ectomycorrhizal fungi, and plant growth-promoting rhizobacteria and fungi, have been employed as biocontrol agents to promote natural disease suppression via mechanisms such as competition, antibiotic production, parasitism, and induced systemic resistance (Prashar et al. 2014; Raaijmakers et al. 2009; Van Loon et al. 1998). However, most studies of biocontrol for soilborne plant pathogens and root diseases have focused on the interactions between a single pair of species and they have not considered the vast amounts of bacterial and fungal diversity that exist within the functional groups found in the rhizosphere. Thus, we have clearly not exploited the full potential of disease-suppressing microbial communities for controlling soilborne diseases. Therefore, more information is needed to understand the roles and significance of different rhizosphere-inhabiting microorganisms in biological control.
Wilt diseases caused mainly by fungal pathogens have been common in cultivated Lanzhou lily (Lilium davidii var. unicolor) plants for many years (Li and Li 1996; Shang et al. 2014). Lanzhou lily is a native variety of China, where it is an important edible bulb crop that is distributed mostly in the middle area of Gansu province (Shi et al. 2012; Wang et al. 2010). It is also renowned for its flaming and flamboyant color provides its ornamental value. Besides, Lanzhou lily scales, which are considered health food due to their abundant nutritional components of proteins, carbohydrates lipids and amino acids are used in Chinese medicine in different forms as fresh bulbs, dried scales, as well as powder to treat heart and lung ailments (Li et al. 2014b). In recent years, due to the prevalence of fungal diseases, there have been significant decreases in some crop yields (Bai et al. 2013). The main pathogens that cause wilt diseases in Lanzhou lily under field conditions belong to the genus Fusarium (Li and Li 1996; Shang et al. 2014). At present, very few efficient biologically-based control strategies have been developed to control the pathogens that affect Lanzhou lily in field conditions. Therefore, a better understanding of the rhizosphere microbial composition and diversity associated with Lanzhou lily plants grown in natural cultivation fields would facilitate the development of efficient biological control strategies.
In the present study, we employed Illumina-based high-throughput sequencing and BIOLOG technology approaches to profile the main microbial rhizosphere taxa and functional diversity associated with Lanzhou lily plants grown in field conditions, thereby determining whether the rhizosphere microbial community composition and functional diversity differs for healthy and wilted plants. We also explored the potentially beneficial microbes that could be applied to Lanzhou lily to facilitate biological control in field conditions.
Materials and methods
Site information and sample collection
Lanzhou lily plants were obtained in June 2014 from two main cultivation fields located in Lanzhou (35°55′N, 103°46′E), where a high incidence of wilt and root rot diseases has been reported (Bai et al. 2013; Shang et al. 2014). The plants had been cultivated continuously for 3 years under field conditions in grey-cinnamon soils. Some plants in certain fields exhibited wilt symptoms associated with infection by Fusarium spp., whereas others were healthy. The plant samples were collected separately from two healthy fields and two fields (each fields is about 5 ha.) where the plants exhibited symptoms of wilt disease [brown vascular and rot root was confirmed, but it was not due to abiotic factors (Shang et al. 2014)] in early July (before florescence). Rhizospheric soil (RS) was obtained from healthy Lanzhou lily fields (CH, YH) and from wilted Lanzhou lily fields (CD, YD) in Caoyuan village and Yuanjiawan village (Lanzhou city) respectively. Five plants were randomly collected from each sampling field. Each rhizosphere sample comprised the intact root system and the sediment that adhered tightly to each individual plant was sealed in an unopened plastic ziplock bag and put into an ice chest before being transported to the lab. Bulk soil samples around the wilted and healthy roots were also collected in triplicate for BIOLOG and potential biocontrol agents- screening analysis. The roots were shaken gently to remove any loosely adhering soil. RS was carefully removed from fine roots by gently scraping the adhered soil with sterilized fine forceps and surgical knife. Samples from each sampling field were pooled and homogenized, and then stored immediately in a freezer at −80 °C until they were used for molecular analysis.
DNA extraction, PCR amplification, and Illumina sequencing analysis
RS genomic DNA was extracted directly using a soil DNA extraction kit (Omega Bio-Tek, GA, USA) according to the manufacturer’s instructions. The concentration and purity of DNA extracts were determined using a NanoDrop ND-2000C spectrophotometer (Thermo Scientific, Wilmington, USA). Qualified total genomic DNA was amplified using a 515F/806R primer set, which amplifies the V4 region of the 16S rDNA gene (Caporaso et al. 2011), and an ITS5/ITS2 primer set, which amplifies the ITS1 region of the internal transcribed spacer (White et al. 1990), to determine the diversity and compositions of the bacterial and fungal communities in each sample. The forward primer contained a unique 6-bp error-correcting barcode for each sample. DNA was amplified according to a previously described protocol (Caporaso et al. 2011; White et al. 1990). High-throughput sequencing of amplicons was conducted using the Illumina MiSeq platform at Novogene Bioinformatics Technology Co. Ltd (Beijing, China). Complete data sets in this study have been deposited in the NCBI Short Read Archive database under accession numbers SRR2085086 (bacterial data) and SRR2085088 (fungal data).
Pairs of reads from the original DNA fragments were merged using FLASH (Magoč and Salzberg 2011). Sequencing reads were assigned to each sample according to the unique individual barcodes. Sequences were analyzed with the QIIME (Caporaso et al. 2010) software package (Quantitative Insights Into Microbial Ecology) and UPARSE pipeline (Edgar 2013), as well as using custom Perl scripts to analyze the alpha (within sample) and beta (between samples) diversity. First, the reads were filtered using QIIME quality filters. The default settings for Illumina processing in QIIME were employed (r = 3, p = 0.75; total read length: q = 3; n = 0). The UPARSE pipeline was then used to detect operational taxonomic units (OTUs) at 97 % similarity. A representative sequence was selected for each OTU and used to assign the taxonomic composition with the RDP classifier (Wang et al. 2007).
Microbial functional diversity
Functional diversity of the microbial communities in the RS of Lanzhou lily roots was estimated by the BIOLOG method (Gomez et al. 2006). For the BIOLOG assay, EcoPlate™ plates (BIOLOG Inc., Hayward, CA, USA) were used to determine the bacterial carbon source utilization profile. 1.0 g of fresh bulk soil from around either diseased or healthy roots was mixed with 99 ml of 0.85 % sterile NaCl solution, shaken for 30 min on a reciprocal shaker, and 150 μl of the solution were inoculated into each well of the EcoPlate with 31 carbon sources and incubated at 25 °C in the dark. Plates were read every 24 h at 590 nm up to 168 h. Microbial activity, expressed as average well color development (AWCD) was determined to assess the functional diversity of microbial communities in the RS of either healthy or wilted Lanzhou lily roots. The patterns of 31 carbon sources utilization by each sample was measured as described previously (Pessi et al. 2012), and principal coordinate analysis (Hackett and Griffiths 1997) was adopted to detect the time-course of substrate utilization in healthy and wilted samples. The substrates are referred to by number, so that substrate 1 = well A2, substrate 4 = well B1, substrate 31 = well H4 and so on.
Screening of potential bacterial biocontrol agents against Lanzhou lily wilt pathogens
We have identified Fusarium tricinctum as Lanzhou Lily wilt pathogen in our previous study (Shang et al. 2014), meanwhile, we also isolated and identified F. oxysporum as Lanzhou lily wilt pathogen that the species has been reported before (Li and Li 1996). The sequences of ITS gene in both isolates were compared with known sequences in the NCBI GenBank using BLAST and were deposited to the GenBank (Accession Nos. KF728675 and KP091292). Both pathogens used in this study were stored in our lab.
To screen for potential biocontrol agents against F. tricinctum and F. oxysporum, RS were collected from healthy Lanzhou lily fields (CH, YH). Briefly, 10 g of soil was suspended in 90 ml of 0.85 % NaCl solution and the mixed solution was incubated for 30 min at room temperature with gentle shaking. Suspensions were then serially diluted with 0.85 % NaCl solution and plated onto LB plates. All isolates were purified twice and then stored at −80 °C in 25 % glycerol for further use. The potential wild bacteria isolates with antagonistic activities towards F. tricinctum and F. oxysporum were measured as follows: One 7 mm disk of pure fungal pathogens mycelia was placed in the centre of a potato dextrose agar (PDA) plate (containing 200 g of potato, 20 g of glucose, 20 g of agar, made up to 1 l). The suspension (2 μl overnight cultured single colony with LB medium at 37 °C) of the wild isolates was placed around the fungal inoculums at a distance of 1.5 cm. The plates were incubated at 25 °C until clear zones of inhibition became visual and zones of inhibition were measured as described previously (Berg et al. 2005). To identify the bacteria with potential biocontrol effect, the 16S rRNA genes of the wild isolates were amplified by PCR with primers 27F and 1492R. The sequences of 16S rRNA gene in wild isolates were deposited to the GenBank (Accession Nos. KU363819 and KU363820).
Statistical analysis
Alpha diversity was calculated according to rarefaction analysis by Chao1 (Chao and Lee 1992), the observed OTUs, and Shannon index metrics. Rarefaction curves were generated based on these three metrics (Schloss et al. 2009). Heatmaps and Venn diagrams were generated using custom R scripts. QIIME was used to calculate the weighted and unweighted UniFrac, and Bray Curtis. Principal coordinates analysis and the unweighted pair group method with arithmetic mean clustering were conducted with the unweighted and weighted UniFrac measures based on a previously published protocol (Kuczynski et al. 2012). The histograms and broken line graph were created using Microsoft Excel 2010 (Microsoft, Redmond, Washington, USA).
Results
Overall diversity of microbial communities in Lanzhou lily RS
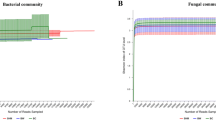
There were differences in the rarefaction curves obtained for the microbial communities derived from healthy and wilted RS, where the bacterial curves had greater richness for the wilted RS whereas the fungal curves exhibited the opposite trend. All of the amplified rarefaction curves increased rapidly from 0 to 2500 sequences, and they tended to approach the saturation plateau for all samples in terms of both bacteria and fungi (Fig. 1). From all of the samples, a total of 386,211 high-quality sequences (170,348 sequences for bacteria and 215,863 for fungi) were obtained after filtering the low-quality reads, chimeras, and attachment sequences. The effective sequence reads comprised 58,127 for bacteria in healthy RS, 112,221 for bacteria in wilted RS, 121,985 for fungi in healthy RS, and for 93,878 fungi in wilted RS (Table 1).
The total number of OTUs detected at 97 % shared sequence similarity was very high in the Lanzhou lily RS in terms of both bacteria and fungi, and the estimated alpha diversity demonstrated that there was abundant microbial diversity in all of the samples (Table 1). For bacteria, the number of different phylogenetic OTUs in all samples ranged from 1783 to 2398, where the wilted RS had higher 16S rRNA gene diversity than the healthy RS. Sample YD had the highest number of OTUs and bacterial diversity among the four samples, whereas CH had the lowest number of OTUs and richness. For fungi, the number of different phylogenetic OTUs in all samples ranged from 361 to 382, and the healthy RS exhibited higher diversity than the wilted RS. Sample YH had the highest number of OTUs and Shannon index among all of the samples, whereas CD and YD had the lowest OTU number, although CD had the highest richness according to Chao1 (Table 1).
Microbial community composition of Lanzhou lily RS
All valid reads were classified from phylum to genus by QIIME using the default settings. The overall microbial compositions of the samples were similar, but some groups from the bacterial and fungal communities in the healthy and wilted RS samples contained different proportions at the phylum level (Fig. 2). Based on the bacterial phylum assignment results (Fig. 2a), we found that Illumina high-throughput sequencing produced few reads that were unclassified and, on average, 99.15 % of the 16S rRNA sequence reads were identified as bacteria belonging to 35 phyla. Proteobacteria was the most abundant phylum in all samples, accounting for 26.96–40.68 % of the total valid reads in all samples, with an average relative abundance of 34.45 %. Acidobacteria was the second most abundant phylum in all samples with an average relative abundance of 12.09 %. The other dominant phyla were Actinobacteria (average of 11.96 %), Bacteroidetes (11.76 %), Crenarchaeota (5.68 %), Gemmatimonadetes (5.15 %), Planctomycetes (4.76 %), Chloroflexi (3.88 %), Verrucomicrobia (3.55 %), and Firmicutes (2.73 %) (a detailed comparison of these phyla is shown in Table S1). At the class level, based on the average relative abundances, the four most frequent classes were Sphingobacteriia, Alphaproteobacteria, Betaproteobacteria, and Gammaproteobacteria, ranging from 11.29 to 8.43 %. In addition, compared with the wilted samples (CD and YD) from the same sites, the healthy samples (CH and YH) contained relative higher percentages of Bacteroidetes (11.74 and 12.41 %, respectively), Crenarchaeota (5.53 and 4.96 %), Verrucomicrobia (2.80 and 1.29 %), Firmicutes (4.21 and 1.89 %), and Sphingobacteriia (11.47 and 11.99 %), but relatively lower percentages of Proteobacteria (12.58 and 10.07 %, respectively), Gemmatimonadetes (7.50 and 9.72 %), Planctomycetes (4.11 and 2.63 %), Alphaproteobacteria (8.58 and 6.75 %), Betaproteobacteria (2.16 and 3.71 %), and Gammaproteobacteria (3.66 and 1.22 %).
The fungal classification results showed that the dominant phylum was Ascomycota in all samples, accounting for 54.84–74.37 % of the total valid reads, with an average relative abundance of 64.01 %. The next most dominant phyla were Basidiomycota and Zygomycota with 12.70 and 9.74 %, respectively (Fig. 2b). At the class level, the most abundant class was Sordariomycetes (45.97 %), followed by Pezizomycetes (5.40 %), Agaricomycetes (5.04 %), and Eurotiomycetes (3.11 %). Compared with the wilted samples (CD and YD) from the same sites, the healthy samples (CH and YH) contained relatively higher percentages of Zygomycota (13.70 and 5.03 %, respective), but relatively lower percentages of Agaricomycetes (5.22 and 1.52 %).
Differences in the microbial communities of healthy and wilted RS samples
The total number of unique bacterial OTUs (data not shown) detected in the four pooled samples was 3128, where 159 OTUs were associated only with the healthy RS and 477 OTUs were associated only with the wilted RS, while 1047 were shared by the healthy and wilted Lanzhou lily RS samples (Fig. 3a). OTUs classified as Candidatus nitrososphaera, OD1, OP11, OP3, Leptospiraceae, Asteroleplasma, TM6, and TM7 comprised 10.7 % of the 159 OTUs detected only in the healthy RS, while Armatimonadetes, Chlamydiales, Fusobacteria, Gemmatimonadetes, and NKB19 accounted for 19.5 % of the 477 OTUs detected only in the wilted RS. In total, 508 different fungal OTUs were identified in this study, 21 of which were associated only with the healthy RS, 18 OTUs were associated only with the wilted RS, and 228 were shared by all samples (Fig. 3b). Among the fungi, OTUs classified as Gymnascella aurantiaca and Leucoagaricus leucothites comprised 14.3 % of the 21 OTUs detected only in the healthy RS, while Pezizales and Mrakia accounted for 11.1 % of the 18 OTUs detected only in the wilted RS.
Heatmaps and the hierarchical clustering analysis results for the 35 dominant genera from the different samples are shown in Fig. 4. Hierarchical clustering analysis detected two major groups that separated healthy or wilted RS samples in terms of bacteria (Fig. 4a), but the same clusters were not obtained for fungi (Fig. 4b). Among bacteria, the differences between healthy and wilted RS were attributable mostly to Flavisolibacter, Opitutus, Adhaeribacter, Steroidobacter, Bacillus, and Lactococcus, which were relatively more abundant in healthy RS (the relative abundances were 0.70, 0.25, 0.27, 1.11, 0.74, and 0.58 % higher, respectively), whereas Skermanella, Planctomyces, Thermomonas, and Phyllobacterium were relatively less abundant (the relative abundances were 0.24, 0.65, 0.56, and 0.24 % lower, respectively). For fungi, hierarchical clustering did not separate the healthy RS from the wilted RS, but some genera were still associated with healthy or wilted RS at higher relative abundances, such as Thielaviopsis and Chaetomium in healthy RS (the relative abundances were 0.05 and 0.11 % higher, respectively), and Geomyces and Rhizoctonia in wilted RS (the relative abundances were 0.03 and 3.34 % higher, respectively). Moreover, compared with the healthy sample (YH), YD contained relatively higher abundances of Wardomyces, Verticillium, Metarhizium, and Alternaria (the relative abundances were 0.13, 0.06, 1.15, and 0.04 % higher, respectively), while CD contained relatively higher abundances of Cephalotrichum, Exophiala, Penicillium, Stilbella, Ilyonectria, Neonectria, Volutella, Scytalidium, and Plectosphaerella (the relative abundances were 0.10, 0.26, 9.07, 1.57, 0.72, 0.84, 0.43, 0.52, and 14.39 % higher, respectively) compared with the healthy sample (CH).
Principal coordinates analysis was also performed using the OTUs to compare the variations in the microbial communities of the healthy and wilted samples. The first principal component obtained by the unweighted Unifrac algorithm separated the CH and YH bacterial communities from those of CD and YD (65.12 % of contribution rate) (Fig. 5a), although the second principal component obtained by the Bray-Curtis algorithm separated the CH and YH fungal communities from those of CD and YD (33.03 % of contribution rate) (Fig. 5b), CH and YH were similar in the fungal community composition at the phylum level (Fig. S1), thus the distinct clusters indicated high variation in the microbial community structures of the healthy RS and wilted RS.
Microbial functional diversity of healthy and wilted RS samples
Average AWCD values were used to calculate functional diversity indices of the microbial communities in healthy and wilted RS. The AWCD for samples from Caoyuan village (CH, CD) and Yuanjiawan village (YH, YD) increased rapidly after incubation for 24 h and generally followed the sigmoidal curve with incubation time, but reached a maximum at different incubation times. Both diseased soils (CD, YD) showed a delayed onset of AWCD increase, a reduced rate of AWCD increase and a reduced AWCD maximum when compared with healthy soils (CH, YH) at each time point respectively. Pairwise comparisons between diseased and healthy soils from the same field indicated a significant reduction in AWCD, ranging from 7.27 to 60.75 % (Caoyuan village) and 13.03 to 85.25 % (Yuanjiawan village) at different time points during the incubation period (Fig. 6).
The degree of sole carbon substrate utilization at lag phase by each sample was shown in Fig. 7. All carbon sources could be metabolized by communities from healthy samples CH and YH; however, in wilted samples CD and YD, except for the most carbon substrates, there are few substrates such as D, L-α-Glycerol Phosphate (29) and 2-Hydroxy Benzoic Acid utilized (10) by the communities at low level. As for the principal coordinates analysis of each carbon substrate utilized over time shown in Fig. 8, communities from healthy and wilted RS were similar in the patterns of most carbon substrates utilization in general. The first principal coordinate represents the degree and rate of colour development: low scores on the axis correspond to a rapid and high level of colour development (e.g. substrate 7: l-Asparagine) while high scores correspond to a gradual and low level increase in colour in the well (e.g. substrate 29: D, L-α-Glycerol Phosphate). Combined the analysis of Fig. 7 with Fig. 8, it is obvious that in both of the healthy and wilted samples, substrates l-Asparagine, l-Serine, D-Galacturonic Acid, pyruvic acid methyl ester, 4-Hydroxy Benzoic Acid and D-Cellobiose (7, 15, 6, 4, 14 and 24) could be metabolized highly and rapidly, while substrates D, L-α-Glycerol Phosphate, α-Ketobutyric Acid, γ-Hydroxybutyric Acid, l-Threonine, Glycyl-l-Glutamic Acid and i-Erythritol (29, 26, 18, 19, 23 and 9) were utilized lowly and gradually. The apparent differences of carbon substrate utilization patterns between healthy and wilted RS communities should be 2-Hydroxy Benzoic Acid and l-Phenylalanine (10 and 11) utilized highly in healthy sample but lowly in wilted sample. Of these, l-Asparagine, l-Serine, 4-Hydroxy Benzoic Acid, l-Threonine, 2-Hydroxy Benzoic Acid and l-Phenylalanine (7, 15, 14, 19, 10 and 11) have been reported as constituents of root exudates (Campbell et al. 1997).
Carbon source utilization by samples CH, YH, CD and YD at 168 h. Data represent OD590 divided by the AWCD. 1 β-methyl-D-glucoside; 2 D-galactonic acid γ-lactone; 3 l-arginine; 4 pyruvic acid methyl ester; 5 D-xylose; 6 D-galacturonic acid; 7 l-asparagine; 8 tween 40; 9 i-erythritol; 10 2-hydroxy benzoic acid; 11 l-phenylalanine; 12 tween 80; 13 D-mannitol; 14 4-hydroxy benzoic acid; 15 l-serine; 16 α-cyclodextrin; 17 N-acetyl-D-glucosamine; 18 γ-hydroxybutyric acid; 19 l-threonine; 20 glycogen; 21 D-glucosaminic acid; 22 itaconic acid; 23 glycyl-l-glutamic acid; 24 D-cellobiose; 25 glucose-1-phosphate; 26 α-ketobutyric acid; 27 phenylethyl-amine; 28 α-d-lactose; 29 D,L-α-glycerol phosphate; 30 D-malic acid; 31 putrescine
Principal coordinates analysis of each carbon substrate utilized over time by microbial communities from rhizosphere soil of healthy (a) and wilted (b) Lanzhou lily plants. The substrates are referred to by number the same as listed in Fig. 7
Wild bacterial isolates with antagonistic abilities against Lanzhou lily wilt pathogens
With F. tricinctum and F. oxysporum as the indicator strains of Lanzhou lily wilt pathogens, two potential biocontrol strains S15B1 and S15B5 with strong antagonistic effects towards both pathogens were screened from the RS of healthy Lanzhou lily. By sequencing and comparing their 16S rRNA gene with reference sequences in the NCBI GenBank, S15B1 was identified as Bacillus subtilis and S15B5 was identified as Bacillus amyloliquefaciens.
The average radius of inhibition zones of S15B1 against F. tricinctum and F. oxysporum were 8.0 mm and 4.6 mm, with inhibitory effects of 69.3 and 72.2 % respectively; and the average radius of inhibition zones of S15B5 against F. tricinctum and F. oxysporum were 12.8 mm and 5.3 mm, with inhibitory effects of 61.3 and 64.4 % respectively. Both of the Bacilli strains could form biofilm on the medium (Fig. 9).
Discussion
In this study, we used next generation sequencing and BIOLOG assay to explore the microbial community structures and functional diversity and community structures associated with the rhizosphere of Lanzhou lily plants grown in field conditions, and we elucidated the differences in the community structure according to the health status of plants. There have been few previous studies of the interactions between Lanzhou lily and the rhizospheric microbial community in natural ecosystems, and earlier studies employed culture-dependent methods (Bai et al. 2013; Li and Li 1996). To be best of our knowledge, the present study is the first culture-independent characterization of the microbial diversity associated with healthy and wilted RS samples from Lanzhou lily plants grown under field conditions. In this study, we identified the bacterial and fungal communities using the same DNA extracts. We only studied a small number of samples, so caution is advised when drawing conclusions based on such a limited dataset. Nevertheless, the results obtained demonstrated that the microbial composition and functional diversity associated with the RS from healthy and wilted Lanzhou lily plants grown under fields was distinct. In addition, two wild Bacilli strains with strong antagonistic effects towards Lanzhou lily wilt pathogens were screened as potential biocontrol agents.
Our results support the hypothesis that the microbial communities and functional diversity varied according to the health status of Lanzhou lily plants. In particular, it was interesting that the difference in bacterial diversity between the healthy and wilted RS samples was more distinct than that in of the fungal diversity (Fig. 5). It has been reported that pathogenic fungus Verticillium dahliae caused cotton wilt affected the bacterial composition of cotton rhizospheres (Zhang et al. 2011b). Thus, we assume that the wilt disease might affect the structural change of bacterial communities. Moreover, biolog results clearly indicated that the AWCD values of microbial functional diversity and the degree of carbon substrate utilization in healthy soils were greater than that of the wilted soil (Figs. 6, 7). The high variation in the microbial community structures (Fig. 5a) may contribute to the different patterns of carbon substrates utilization between healthy and wilted RS (Fig. 8). However, by the EcoPlate we used in this study, AWCD values do not represent the activity of all bacteria or fungi (Preston-Mafham et al. 2002), but these results provide direct evidence for the functional activities of microbes colonizing rhizosphere soils. According to the differences in the OTUs and alpha diversity indices, the healthy Lanzhou lily plants exhibited lower rhizosphere-associated bacterial diversity than the diseased plants, whereas the opposite trend was observed for fungi based on the OTUs (Table 1). This finding is in contrast to previous studies of Picea mariana based on Sanger sequencing, which showed that healthy seedlings grown in a nursery had greater bacterial diversity than diseased seedlings, while the diseased seedlings had higher fungal diversity than the healthy seedlings (Filion et al. 2004). Moreover, it has been (Li et al. 2014a) reported that healthy greenhouse tomato RS samples harbored higher bacterial diversity compared with diseased RS samples according to 454 pyrosequencing. Differences in the plant growth conditions, plant disease-causing agents, microbial identification methods, and plant genotypes may explain these contrasting results.
The dominant phyla among the bacteria and fungi sequenced from the Lanzhou lily rhizosphere have also been reported as important rhizosphere-inhabiting organisms in other culture-independent studies. For example, within the bacterial community, the phylum Proteobacteria was the predominant rhizosphere colonizer and similar results have been obtained using rhizosphere samples from Populus deltoides, maize, lettuce, and cucumber grown under field conditions (Gottel et al. 2011; Peiffer et al. 2013; Schreiter et al. 2014; Tian and Gao 2014), mainly because Proteobacteria are generally fast-growing r-strategists with the ability to utilize a broad range of root-derived carbon substrates (Philippot et al. 2013). In the healthy RS, we detected a reduction in the relative abundance of Proteobacteria, which contradicts previous observations (Li et al. 2014a), but the underlying mechanism is difficult to explain. Moreover, Acidobacteria, Actinobacteria, Bacteroidetes, and Firmicutes are common phyla in the rhizosphere of various plants (Inceoglu et al. 2011; Knief et al. 2012; Lundberg et al. 2012; Uroz et al. 2010; Weinert et al. 2011). Chloroflexi, Planctomycetes, Gemmatimonadetes, Verrucomicrobia, and Crenarchaeota were also found in the rhizosphere of the Zigongdongdou soybean cultivar (Liang et al. 2014). However, bacteria do not monopolize the nutrient-rich rhizosphere because saprotrophic fungi have been identified in this niche, including representatives of all the major terrestrial phyla, i.e., Ascomycota, Basidiomycota, and Zygomycota (Gomes et al. 2003; Renker et al. 2004; Vujanovic et al. 2007). The saprotrophic fungi found in the rhizosphere may be involved in the degradation of simple root exudates as well as the more complex compounds present in sloughed root cells (Buee et al. 2009).
Among the most important differences observed between the bacterial and fungal communities associated with the rhizosphere of healthy and wilted Lanzhou lily plants, the OTUs classified as Candidatus nitrososphaera, OD1, OP11, OP3, Leptospiraceae, Asteroleplasma, TM6, TM7, Gymnascella aurantiaca, and Leucoagaricus leucothites were associated only with the healthy RS, whereas the OTUs related to Armatimonadetes, Chlamydiales, Fusobacteria, Gemmatimonadetes, NKB19, Pezizales, and Mrakia were detected only in the wilted RS. Candidatus nitrososphaera is a genus of ammonia-oxidizing archaea in the phylum Thaumarchaeota (Crenarchaeota), which comprises two currently recognized species called N. viennensis (Tourna et al. 2011) and Candidatus N. gargensis, where the sequenced genome of Candidatus N. gargensis contains a gene set for ammonia oxidation and CO2 fixation (Spang et al. 2012). The good growth of Lanzhou lily in fields may be related to the presence of more Crenarchaeota in healthy RS due to nitrification, which would increase the probability of plants taking up more nitrogen. Partial genomic analysis of a single cell (ZG1) belonging to candidate division OP11 provided evidence for antibiotic resistance, antibiotic production (bacteriocin), and extracellular bactericidal peptidases (Youssef et al. 2011). In healthy RS, the presence of this group may be related to pathogen inhibition. It has been reported that a catechin produced by Leucoagaricus leucothites (Vittad.) Wasser exhibited strong antimicrobial activity against some foodborne and spoilage bacteria (Aslim and Ozturk 2011). This fungus may also contribute to the inhibition of unknown bacterial pathogens. Members of candidate division TM6 and TM7 are present in aquatic habitats related to human activities (Hugenholtz et al. 2001; Marcy et al. 2007; McLean et al. 2013), but little is known about their interactions within agroecosystems. Among the OTUs related to wilted RS, two of our Armatimonadetes isolates, i.e., Armatimonas rosea YO-36T (Tamaki et al. 2011) and Fimbriimonas. ginsengisoli Gsoil 348T (Im et al. 2012), have also been isolated from similar mesophilic soil environments where they had close associations with plant root systems, but no previous studies have reported the relationships between Armatimonadetes clones and pathogenicity in plants or animals (Lee et al. 2014). Unfortunately, little information is available about the functions of the other OTUs associated with the RS samples.
At the genus level (Fig. 4), among the bacteria identified in the present study, Flavisolibacter, Opitutus, Adhaeribacter, Steroidobacter, Bacillus, and Lactococcus were more abundant in healthy RS, whereas Skermanella, Planctomyces, Thermomonas, and Phyllobacterium were more frequent in wilted RS. Among these genera, Bacillus spp. is the most common plant growth promoting rhizobacteria (PGPR) biocontrol agent sources (Santoyo et al. 2012). Bacillus and Lactococcus are members of Firmicutes, which have been shown to be more abundant in suppressive soil, disease-suppressive soils, i.e. are exceptional ecosystems in which crop plants suffer less from specific soil-borne pathogens than expected owing to the activities of other soil microorganisms (Mendes et al. 2011), and some Bacillus spp. are effective in controlling Fusarium wilt by colonizing the roots of plants (Cao et al. 2011; Zhang et al. 2011a). By screening of potential bacterial biocontrol agents against Lanzhou lily wilt pathogens, our results showed that both the antagonistic strains are Bacilli and with biofilm-forming abilities (Fig. 9), and it has been reported that biocontrol of tomato wilt disease by wild B.subtilis isolates depends on conserved genes mediating biofilm formation (Chen et al. 2012). So we consider that the relatively higher abundance of Bacillus in Firmicutes in healthy RS may contribute to disease suppression, thereby promoting Lanzhou lily growth. Studies have shown that Phyllobacterium strains occur in leaf nodules and in the rhizosphere (Lambert et al. 1990; Mantelin et al. 2006), and thus they may be considered plant growth-promoting bacteria (Larcher et al. 2003). Among the fungi identified in our samples, Thielaviopsis and Chaetomium were relatively more abundant in healthy RS, whereas Geomyces and Rhizoctonia were more frequent in wilted RS. Thielaviopsis includes several important agricultural pathogens and the most widespread is T. basicola, which is the causal agent of black root rot diseases in economically important crop species (Coumans et al. 2009; Kumar et al. 2013). Chaetomium is a genus that encompasses species with important roles in the decomposition of plants and other cellulose-rich materials, and they are readily isolated from dung, plant debris, and soil. Many strains of the type species C. globosum have been reported to have potent effects in disease control via the production of antifungal compounds (Cuomo et al. 2015; Park et al. 2005). Species in the genus Geomyces are common in the soils of temperate and high-latitude ecosystems, and they can even thrive in cold, low-nutrient polar environments (Hayes 2012). Asterric acid derivatives with antibacterial or antifungal activities were detected in an unidentified Geomyces isolate from a soil sample (Li et al. 2008). In the genus Rhizoctonia, R. solani is an important soil-borne fungal phytopathogen with a global distribution, which is responsible for damping off, root rot, and many other pathogenic conditions (Hane et al. 2014). In addition, Verticillium (sample YD), Penicillium, and Ilyonectria (Neonectria) (sample CD) are common pathogens that cause plant vascular wilt (Tarkka et al. 2008), bulb rot (Dugan et al. 2013), and root rot (Vitale et al. 2012) diseases, respectively. Principal coordinates analysis based on the OTUs showed that wilt disease had a great impact on the microbial community, where there were contrasting differences in the healthy and wilted RS in terms of their bacterial and fungal communities, i.e., CD and YD has similar bacterial communities whereas their fungal communities were different. This agreed with the heatmap results, which showed that CD and YD had different potential pathogens, whereas the healthy samples, CH and YH, exhibited the opposite trend to CD and YD in terms of their two communities, although the reason for these differences is unknown.
We identified that Fusarium spp. were the dominant pathogens in fields by culture-dependent methods, but unexpectedly we found that Fusarium was only dominant in the healthy sample YH by high-throughput sequencing, whereas Rhizoctonia was present in both the wilted samples, while YD had a higher relative abundance of Verticillium and CD had higher relative abundances of Penicillium and Ilyonectria (Neonectria), all of which are related to root diseases, although they have not been reported as pathogens in Lanzhou lily plants. Thus, we cannot confirm the main wilt-causing pathogen(s) involved and it is possible that several might have contributed to the wilt disease caused by root rot. We found that the potential pathogen Thielaviopsis was present in both of the healthy samples, as well as Fusarium in YH. Thus, it appears reasonable to suggest that the microbial representatives identified in this study with generally recognized beneficial impact on plants, such as Bacillus, may have significant roles in biological control to combat soilborne pathogens. However, little information is available regarding the eco-functions in the soil or rhizosphere of many OTUs or genera, so we should not ignore their presence in this niche.
Conclusion
A huge range of highly diverse microorganisms are associated with plant roots, where they play crucial roles in plant growth and health. Our results suggest that the major differences in the healthy and wilted Lanzhou lily RS samples in terms of the microbial composition and functional diversity were linked to the health status of plants. These results indicate that the growth of healthy plants may be facilitated by relative higher abundance of Bacillus contribute to disease suppression in continuous cropping systems while maintaining a balanced soil composition. Therefore, further research is required to determine the biocontrol efficacy of Bacillus spp. on Lanzhou lily wilt pathogens and to identify the optimal biological control strategies for use in Lanzhou lily cultivation fields.
References
Aslim B, Ozturk S (2011) Phenolic composition and antimicrobial and antioxidant activities of Leucoagaricus leucothites (Vittad.) Wasser. J Med Food 14:1419–1424. doi:10.1089/jmf.2010.0259
Bai B, Yang H, He S, Yu A (2013) Studies on morphology and biological characteristics of Botrytis elliptica (Berk) Cooke causing foliage blight of Lilium davidii Duch. var. unicolor (Hoog) cotton China vegetables (in Chinese). 16:78–84
Berendsen RL, Pieterse CM, Bakker PA (2012) The rhizosphere microbiome and plant health. Trends Plant Sci 17:478–486
Berg G (2009) Plant–microbe interactions promoting plant growth and health: perspectives for controlled use of microorganisms in agriculture. Appl Microbiol Biotechnol 84:11–18
Berg G, Smalla K (2009) Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol Ecol 68:1–13
Berg G, Krechel A, Ditz M, Sikora RA, Ulrich A, Hallmann J (2005) Endophytic and ectophytic potato-associated bacterial communities differ in structure and antagonistic function against plant pathogenic fungi. FEMS Microbiol Ecol 51:215–229. doi:10.1016/j.femsec.2004.08.006
Buee M, De Boer W, Martin F, van Overbeek L, Jurkevitch E (2009) The rhizosphere zoo: an overview of plant-associated communities of microorganisms, including phages, bacteria, archaea, and fungi, and of some of their structuring factors. Plant Soil 321:189–212
Campbell CD, Grayston SJ, Hirst DJ (1997) Use of rhizosphere carbon sources in sole carbon source tests to discriminate soil microbial communities. J Microbiol Methods 30:33–41
Cao Y, Zhang Z, Ling N, Yuan Y, Zheng X, Shen B, Shen Q (2011) Bacillus subtilis SQR 9 can control Fusarium wilt in cucumber by colonizing plant roots. Biol Fertil Soils 47:495–506
Caporaso JG et al (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336
Caporaso JG et al (2011) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci 108:4516–4522
Chao A, Lee S-M (1992) Estimating the number of classes via sample coverage. J Am Stat Assoc 87:210–217
Chen Y, Yan F, Liu H, Chai Y, Guo J (2012) Biofilm formation of Bacillus subtilis on tomato roots enhances biocontrol efficacy against tomato bacterial wilt disease caused by Ralstonia solanacearum. Phytopathology 102:21–22
Coumans JV, Poljak A, Raftery MJ, Backhouse D, Pereg-Gerk L (2009) Analysis of cotton (Gossypium hirsutum) root proteomes during a compatible interaction with the black root rot fungus Thielaviopsis basicola. Proteomics 9:335–349
Cuomo CA, Untereiner WA, Ma L-J, Grabherr M, Birren BW (2015) Draft genome sequence of the cellulolytic fungus Chaetomium globosum. Genome Announc 3:e00021-00015
Degnan PH, Ochman H (2012) Illumina-based analysis of microbial community diversity. ISME J 6:183–194. doi:10.1038/ismej.2011.74
Dugan FM, Lupien SL, Vahling-Armstrong CM, Chastagner GA, Schroeder BK (2013) Host range of Penicillium spp. (blue mold) rotting bulb crops. Phytopathology 103:37
Edgar RC (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998
Filion M, Hamelin RC, Bernier L, St-Arnaud M (2004) Molecular profiling of rhizosphere microbial communities associated with healthy and diseased black spruce (Picea mariana) seedlings grown in a nursery. Appl Environ Microb 70:3541–3551
Fitter A, Gilligan C, Hollingworth K, Kleczkowski A, Twyman R, Pitchford J (2005) Biodiversity and ecosystem function in soil. Funct Ecol 19:369–377
Garbeva P, Van Veen J, Van Elsas J (2004) Microbial diversity in soil: selection of microbial populations by plant and soil type and implications for disease suppressiveness. Annu Rev Phytopathol 42:243–270
Glenn TC (2011) Field guide to next-generation DNA sequencers. Mol Ecol Resour 11:759–769
Gomes NCM, Fagbola O, Costa R, Rumjanek NG, Buchner A, Mendona-Hagler L, Smalla K (2003) Dynamics of fungal communities in bulk and maize rhizosphere soil in the tropics. Appl Environ Microb 69:3758–3766
Gomez E, Ferreras L, Toresani S (2006) Soil bacterial functional diversity as influenced by organic amendment application. Bioresour Technol 97:1484–1489. doi:10.1016/j.biortech.2005.06.021
Gottel NR et al (2011) Distinct microbial communities within the endosphere and rhizosphere of Populus deltoides roots across contrasting soil types. Appl Environ Microb 77:5934–5944
Hackett CA, Griffiths BS (1997) Statistical analysis of the time-course of Biolog substrate utilization. J Microbiol Methods 30:63–69
Hane JK, Anderson JP, Williams AH, Sperschneider J, Singh KB (2014) Genome sequencing and comparative genomics of the broad host-range pathogen Rhizoctonia solani AG8. PLoS Genet. doi:10.1371/journal.pgen.1004281
Hayes MA (2012) The geomyces fungi: ecology and distribution. Bioscience 62:819–823. doi:10.1525/bio.2012.62.9.7
Hiltner L (1904) Über neuere Erfahrungen und Probleme auf dem Gebiete der Bodenbakteriologie unter besonderer Berücksichtigung der Gründüngung und Brache. Arbeiten der Deutschen Landwirtschaftlichen Gesellschaft 98:59–78
Hugenholtz P, Tyson GW, Webb RI, Wagner AM, Blackall LL (2001) Investigation of candidate division TM7, a recently recognized major lineage of the domain bacteria with no known pure-culture representatives. Appl Environ Microb 67:411–419. doi:10.1128/Aem.67.1.411-419.2001
Im W-T, Hu Z-Y, Kim K-H, Rhee S-K, Meng H, Lee S-T, Quan Z-X (2012) Description of Fimbriimonas ginsengisoli gen. nov., sp. nov. within the Fimbriimonadia class nov., of the phylum Armatimonadetes. Antonie Van Leeuwenhoek 102:307–317
Inceoglu O, Abu Al-Soud W, Salles JF, Semenov AV, van Elsas JD (2011) Comparative analysis of bacterial communities in a potato field as determined by pyrosequencing. PLoS ONE. doi:10.1371/journal.pone.0023321
Keel C, Défago G (1997) Interactions between beneficial soil bacteria and root pathogens: mechanisms and ecological impact. In: Gange AC, Brown VK (eds) Multitrophic interactions in terrestrial systems. Blackwell Science, Oxford, pp 27–47
Kent AD, Triplett EW (2002) Microbial communities and their interactions in soil and rhizosphere ecosystems. Annu Rev Microbiol 56:211–236
Knief C et al (2012) Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice. ISME J 6:1378–1390. doi:10.1038/ismej.2011.192
Kuczynski J, Stombaugh J, Walters WA, González A, Caporaso JG, Knight R (2012) Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr Protoc Microbiol. doi:10.1002/9780471729259.mc01e05s27
Kumar V, Joshi SG, Bell AA, Rathore KS (2013) Enhanced resistance against Thielaviopsis basicola in transgenic cotton plants expressing Arabidopsis NPR1 gene. Transgenic Res 22:359–368. doi:10.1007/s11248-012-9652-9
Lambert B, Joos H, Dierickx S, Vantomme R, Swings J, Kersters K, Van Montagu M (1990) Identification and plant interaction of a Phyllobacterium sp., a predominant Rhizobacterium of young sugar beet plants. Appl Environ Microbiol 56:1093–1102
Larcher M, Muller B, Mantelin S, Rapior S, Cleyet-Marel JC (2003) Early modifications of Brassica napus root system architecture induced by a plant growth-promoting Phyllobacterium strain. New Phytol 160:119–125
Lee KC, Dunfield PF, Stott MB (2014) The phylum Armatimonadetes. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (eds) The prokaryotes: other major lineages of bacteria and the archaea. Springer, Berlin, pp 447–458. doi:10.1007/978-3-642-38954-2_388
Li C, Li J (1996) Identification of the causal agent of Lily wilt. Acta Phytopathol Sin 02:97
Li Y, Sun BD, Liu SC, Jiang LH, Liu XZ, Zhang H, Che YS (2008) Bioactive asterric acid derivatives from the Antarctic ascomycete fungus Geomyces sp. J Nat Prod 71:1643–1646. doi:10.1021/Np8003003
Li JG, Ren GD, Jia ZJ, Dong YH (2014a) Composition and activity of rhizosphere microbial communities associated with healthy and diseased greenhouse tomatoes. Plant Soil 380:337–347
Li XY et al (2014b) Transcriptome analysis of carbohydrate metabolism during bulblet formation and development in Lilium davidii var. unicolor. BMC Plant Biol 14:1–12. doi:10.1186/s12870-014-0358-4
Liang JG et al (2014) Comparison of the rhizosphere bacterial communities of Zigongdongdou soybean and a high-methionine transgenic line of this cultivar. PLoS ONE. doi:10.1371/journal.pone.0103343
Lundberg DS et al (2012) Defining the core Arabidopsis thaliana root microbiome. Nature 488:86. doi:10.1038/Nature11237
Magoč T, Salzberg SL (2011) FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963
Mantelin S et al (2006) Emended description of the genus Phyllobacterium and description of four novel species associated with plant roots: Phyllobacterium bourgognense sp. nov., Phyllobacterium ifriqiyense sp. nov., Phyllobacterium leguminum sp. nov. and Phyllobacterium brassicacearum sp. nov. Int J Syst Evol Microbiol 56:827–839. doi:10.1099/ijs.0.63911-0
Marcy Y et al (2007) Dissecting biological “dark matter” with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc Natl Acad Sci USA 104:11889–11894. doi:10.1073/pnas.0704662104
McLean JS et al (2013) Candidate phylum TM6 genome recovered from a hospital sink biofilm provides genomic insights into this uncultivated phylum. Proc Natl Acad Sci USA 110:E2390–E2399. doi:10.1073/pnas.1219809110
Mendes R et al (2011) Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332:1097–1100. doi:10.1126/science.1203980
Metzker ML (2010) Sequencing technologies—the next generation. Nat Rev Genet 11:31–46
Park J-H, Choi GJ, Jang KS, Lim HK, Kim HT, Cho KY, Kim J-C (2005) Antifungal activity against plant pathogenic fungi of chaetoviridins isolated from Chaetomium globosum. FEMS Microbiol Lett 252:309–313
Peiffer JA et al (2013) Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc Natl Acad Sci 110:6548–6553
Pessi IS, Elias SD, Simoes FL, Simoes JC, Macedo AJ (2012) Functional diversity of microbial communities in soils in the vicinity of Wanda Glacier, Antarctic Peninsula. Microb Environ 27:200–203
Philippot L, Raaijmakers JM, Lemanceau P, van der Putten WH (2013) Going back to the roots: the microbial ecology of the rhizosphere. Nat Rev Microbiol 11:789–799
Prashar P, Kapoor N, Sachdeva S (2014) Rhizosphere: its structure, bacterial diversity and significance. Rev Environ Sci Bio Technol 13:63–77
Preston-Mafham J, Boddy L, Randerson PF (2002) Analysis of microbial community functional diversity using sole-carbon-source utilisation profiles—a critique. FEMS Microbiol Ecol 42:1–14
Raaijmakers JM, Paulitz TC, Steinberg C, Alabouvette C, Moenne-Loccoz Y (2009) The rhizosphere: a playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil 321:341–361
Renker C, Blanke V, Borstler B, Heinrichs J, Buscot F (2004) Diversity of Cryptococcus and Dioszegia yeasts (Basidiomycota) inhabiting arbuscular mycorrhizal roots or spores. FEMS Yeast Res 4:597–603. doi:10.1016/j.femsyr.2004.01.001
Santoyo G, Orozco-Mosqueda MD, Govindappa M (2012) Mechanisms of biocontrol and plant growth-promoting activity in soil bacterial species of Bacillus and Pseudomonas: a review. Biocontrol Sci Techn 22:855–872. doi:10.1080/09583157.2012.694413
Schloss PD et al (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microb 75:7537–7541
Schreiter S et al (2014) Effect of the soil type on the microbiome in the rhizosphere of field-grown lettuce. Front Microbiol 5:144. doi:10.3389/fmicb.2014.00144
Shang QH, Zhao X, Li YY, Xie ZK, Wang RY (2014) First report of Fusarium tricinctum causing stem and root rot on Lanzhou lily (Lilium davidii var. unicolor) in China. Plant Dis 98:999–1000. doi:10.1094/Pdis-11-13-1146-Pdn
Shi SG, Ma FW, Li YH, Feng FJ, Shang ZZ (2012) Overexpression of l-galactono-1, 4-lactone dehydrogenase (GLDH) in Lanzhou lily (Lilium davidii var. unicolor) via particle bombardment-mediated transformation. In Vitro Cell Dev Biol-Plant 48:1–6. doi:10.1007/s11627-011-9383-2
Spang A et al (2012) The genome of the ammonia-oxidizing Candidatus Nitrososphaera gargensis: insights into metabolic versatility and environmental adaptations. Environ Microbiol 14:3122–3145
Tamaki H et al (2011) Armatimonas rosea gen. nov., sp. nov., of a novel bacterial phylum, Armatimonadetes phyl. nov., formally called the candidate phylum OP10. Int J Syst Evol Microbiol 61:1442–1447
Tarkka M, Schrey S, Hampp R (2008) Plant associated soil micro-organisms. In: Nautiyal CS, Dion P (eds) Molecular mechanisms of plant and microbe coexistence. Springer, Berlin, pp 3–51. doi:10.1007/978-3-540-75575-3_1
Tian Y, Gao L (2014) Bacterial diversity in the rhizosphere of cucumbers grown in soils covering a wide range of cucumber cropping histories and environmental conditions. Microb Ecol 68:794–806
Tourna M et al (2011) Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc Natl Acad Sci 108:8420–8425
Uroz S, Buee M, Murat C, Frey-Klett P, Martin F (2010) Pyrosequencing reveals a contrasted bacterial diversity between oak rhizosphere and surrounding soil. Environ Microbiol Rep 2:281–288. doi:10.1111/j.1758-2229.2009.00117.x
Van Loon L, Bakker P, Pieterse C (1998) Systemic resistance induced by rhizosphere bacteria. Annu Rev Phytopathol 36:453–483
Vitale A, Aiello D, Guarnaccia V, Perrone G, Stea G, Polizzi G (2012) First report of root rot caused by Ilyonectria (=Neonectria) macrodidyma on Avocado (Persea americana) in Italy. J Phytopathol 160:156–159. doi:10.1111/j.1439-0434.2011.01869.x
Vujanovic V, Hamelin RC, Bernier L, Vujanovic G, St-Arnaud M (2007) Fungal diversity, dominance, and community structure in the rhizosphere of clonal Picea mariana plants throughout nursery production chronosequences. Microb Ecol 54:672–684. doi:10.1007/s00248-007-9226-1
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microb 73:5261–5267
Wang R, Wang G, Zhao Q, Zhang Y, An L, Wang Y (2010) Research Expression, purification and characterization of the Lily symptomless virus coat protein from Lanzhou Isolate. Virol J 7:1–7. doi:10.1186/1743-422x-7-34
Weinert N et al (2011) PhyloChip hybridization uncovered an enormous bacterial diversity in the rhizosphere of different potato cultivars: many common and few cultivar-dependent taxa. FEMS Microbiol Ecol 75:497–506. doi:10.1111/j.1574-6941.2010.01025.x
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press, San Diego, pp 315–322
Youssef NH, Blainey PC, Quake SR, Elshahed MS (2011) Partial genome assembly for a candidate division OP11 single cell from an anoxic spring (Zodletone Spring, Oklahoma). Appl Environ Microbiol 77:7804–7814. doi:10.1128/AEM.06059-11
Zhang N et al (2011a) A new bioorganic fertilizer can effectively control banana wilt by strong colonization with Bacillus subtilis N11. Plant Soil 344:87–97
Zhang Y, Du BH, Jin ZG, Li ZH, Song HN, Ding YQ (2011b) Analysis of bacterial communities in rhizosphere soil of healthy and diseased cotton (Gossypium sp.) at different plant growth stages. Plant Soil 339:447–455. doi:10.1007/s11104-010-0600-2
Acknowledgments
This research was supported by the Natural Science Foundation of China (No. 31370447) and the Hundred Talents Program of the Chinese Academy of Sciences “Molecular mechanism of biological control of plant diseases.”
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
11274_2016_2051_MOESM2_ESM.tif
Fig. S1 UniFrac UPGMA cluster (at phylum level) of fungal communities associated with healthy and wilted RS from Lanzhou lily plants. The figure was constructed on the basis of Illumina sequencing data (TIFF 216 kb)
Rights and permissions
About this article
Cite this article
Shang, Q., Yang, G., Wang, Y. et al. Illumina-based analysis of the rhizosphere microbial communities associated with healthy and wilted Lanzhou lily (Lilium davidii var. unicolor) plants grown in the field. World J Microbiol Biotechnol 32, 95 (2016). https://doi.org/10.1007/s11274-016-2051-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-016-2051-2