Abstract
The organophosphate pesticide chlorpyrifos (CP) has been used extensively since the 1960s for insect control. However, its toxic effects on mammals and persistence in environment necessitate its removal from contaminated sites, biodegradation studies of CP-degrading microbes are therefore of immense importance. Samples from a Pakistani agricultural soil with an extensive history of CP application were used to prepare enrichment cultures using CP as sole carbon source for bacterial community analysis and isolation of CP metabolizing bacteria. Bacterial community analysis (denaturing gradient gel electrophoresis) revealed that the dominant genera enriched under these conditions were Pseudomonas, Acinetobacter and Stenotrophomonas, along with lower numbers of Sphingomonas, Agrobacterium and Burkholderia. Furthermore, it revealed that members of Bacteroidetes, Firmicutes, α- and γ-Proteobacteria and Actinobacteria were present at initial steps of enrichment whereas β-Proteobacteria appeared in later steps and only Proteobacteria were selected by enrichment culturing. However, when CP-degrading strains were isolated from this enrichment culture, the most active organisms were strains of Acinetobacter calcoaceticus, Pseudomonas mendocina and Pseudomonas aeruginosa. These strains degraded 6–7.4 mg L−1 day−1 of CP when cultivated in mineral medium, while the consortium of all four strains degraded 9.2 mg L−1 day−1 of CP (100 mg L−1). Addition of glucose as an additional C source increased the degradation capacity by 8–14 %. After inoculation of contaminated soil with CP (200 mg kg−1) disappearance rates were 3.83–4.30 mg kg−1 day−1 for individual strains and 4.76 mg kg−1 day−1 for the consortium. These results indicate that these organisms are involved in the degradation of CP in soil and represent valuable candidates for in situ bioremediation of contaminated soils and waters.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
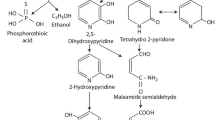
Organophosphate pesticides (OPs) are aliphatic, phenyl or heterocyclic esters of phosphoric acid that are chemically more reactive and less persistent than organochlorine pesticides and are in widespread use for chemical pest control (Singh and Walker 2006). Their mode of action depends on inhibition of acetylcholine esterase (AchE) which results in nervous and endocrine disruption in insects. However, they behave similarly in non-target organisms by blocking AchE, and cause convulsion, paralysis and death in insects and mammals (Cycoń et al. 2013). Chlorpyrifos (CP) (O,O-diethyl-3,5,6-trichloro-2-pyridyl phosphorothioate; CP) is a chlorinated OP which has been in use since 1960s, mainly for the control of foliar insects of cotton, paddy fields, pasture, and vegetable crops (Maya et al. 2011). CP is moderately persistent in the environment with a reported half-life of 2 weeks to over 1 year, depending on soil characteristics, organic content, climate and soil pH (Cycoń et al. 2013). CP degradation occurs through hydrolysis either by photolysis or microbial activity which converts it to diethylthiophosphoric acid (DETP) and 3, 5, 6-trichloro-2-pyridinol (TCP) (Cho et al. 2009). These metabolites can be broken down further, the proposed pathway suggests that DETP is converted to ethanol and phosphorothioic acid while TCP is metabolized by reductive dechlorination pathway (Singh and Walker 2006; Cao et al. 2013). TCP is more toxic and water soluble than CP that results in extensive contamination of soil and aquatic environments. Its persistence and antimicrobial activity tend to prevent proliferation of CP degrading microbes (Briceño et al. 2012). Therefore, biodegradation studies of CP and TCP are equally important.
Microbes in environment possess catabolic versatility to transform toxic compounds such as pesticides to innocuous state; this ability is termed as biodegradation. Biodegradation study for CP detoxification is needed to develop efficient and cheap biotechnological approaches for bioremediation of polluted environments. Several bacterial strains have been studied for CP-degradation, for example, Sphingomonas sp. (Li et al. 2007), Paracoccus sp. (Xu et al. 2008), Bacillus pumilus (Anwar et al. 2009) and Cupriavidus sp. (Lu et al. 2013) which have been isolated from contaminated soils, industrial wastewater as well as polluted sediments. Degradation of CP by microbial consortia derived from enrichment cultures has also been reported, including simultaneous degradation of chlorpyrifos and methyl parathion using a bacterial consortium (Pino and Peñuela 2011), and development of a CP-degrading consortium from four bacterial isolates namely Pseudomonas putida, Klebsiella sp., Pseudomonas stutzeri and Pseudomonas aeruginosa (Sasikala et al. 2012).
Enrichment culture techniques are normally used for isolation of pesticide degrading bacteria but they have been criticized for being biased in selection of rapidly growing bacteria whose role in in situ bioremediation is uncertain. Culture-independent molecular biological techniques have provided a solution to this problem with microbial ecology and diversity techniques used to study complex microbial populations in pesticide-contaminated environments. Enrichment culture methods have also been combined with molecular ecology techniques to characterize the interactions that occur between pesticide degrading bacteria during the enrichment process (Chanika et al. 2011; Ferreira et al. 2009) and study the ecological significance of pesticide degrading isolates (Zhou et al. 2014).
In this study we report the isolation of a number of CP-degrading bacteria from Pakistani soils using enrichment culture, and the potential of these isolates to remediate CP disappearance in liquid culture and in soils, both as individual isolates and as a consortium.
Materials and methods
Soil
The soils used in this experiment were collected from an agricultural field in Lahore (Punjab) having that had been cultivated with a rotation of rice and vegetables, and had a history of CP application for more than 10 years. Samples were collected at 10–20 cm depth from five different areas of same field, mixed together, transferred to sterile plastic bags and stored at 4 °C. Before use, the soil was dried and sieved (2 mm mesh).
Chemicals and media
Technical grade pesticides [CP (98 %), Profenofos (88 %), Triazophos (85 %), Cypermethrin (92 %) and Bifenthrin (97 %), Carbofuran (70 %)] were obtained from Ali Akbar group, Pakistan. Analytical grade CP, TCP and other pesticides were purchased from Sigma-Aldrich. HPLC grade organic solvents were procured from Merck. 10 % (w/v) stock solutions of pesticides were made in methanol or acetone and to enhance solution’s solubility in water it was mixed with 0.5 % (w/v) Triton X. The same ratio of methanol/acetone and Triton X was added to the Controls.
Microbial isolations were carried out in minimal salt medium (MSM, pH 6.8–7.0) containing (gL−1) K2HPO4, 1.5; KH2PO4, 0.5; NaCl, 0.5; (NH4)2 SO4, 0.5; MgSO4·7H2O, 0.2 and 100× trace element solution, 10 mL. 100× trace element solution was composed of (mg L−1) Na2EDTA·2H20, 500; FeCl2·4H2O, 143; ZnCl2, 4.7; MnCl2·4H2O, 3.0; H3BO3, 30; CoCl2·6H2O, 20; CuCl2·2H2O, 1.0; NiCl2·6H2O, 2.0; Na2MoO4·2H2O, 3.0; and CaCl2·2H2O, 100. For medium solidification 1.5 % of agar was used.
Enrichment, isolation and screening of CP-degrading strains
10 g of soil was added to 50 mL MSM containing 50 mg L−1 CP and cultured in 250 mL Erlenmeyer flasks on rotary shaker at 150 rpm incubated at 30 °C. After a week, 5 mL culture was transferred to fresh MSM with same CP concentration. Afterwards two successive transfers were carried out in fresh MSM with 100 mg L−1 CP as the only source of carbon by subculturing 5 mL inocula every time and incubating for one week each. After final transfer, tenfold dilutions of cultures were prepared and 100 µL of each dilution was spread on minimal agar plates containing CP (50 mg L−1). Isolated colonies were purified on CP supplemented nutrient agar. Isolates were screened on the basis of growth on minimal agar with CP as sole source of Carbon. Growth of strains was also monitored in MSM containing 20–100 mg L−1 CP in shake flasks; after 10 days incubation residues were quantified by HPLC and growth was determined as OD600. Four strains, designated LCp4, LCp5, LCp8 and LCp9 (Table 1) were selected for further CP degradation studies.
Taxonomic identification of the bacterial strain
The 16S rRNA gene was amplified using universal primers 27f (AGAGTT TGATCCTGGCTCAG) and 1492R (TACGGTTACCTTGTTACGACTT) in a reaction mixture (50 µL) comprising 20 pmol each primer, 25 µL PCR Master Mix (Thermoscientific) and 20 ng of template DNA. The PCR reaction was performed with following cyclic profile: initial denaturation at 94 °C for 1 min, annealing at 50 °C for 1 min, extension at 72 °C for 2 min and final extension at 72 °C for 10 min. The amplified PCR products were purified with thermoscientific GeneJET gel Extraction Kit and sequenced from Macrogen Korea, using ABI 3730 automated DNA sequencer.
Biodegradation studies
Inoculum preparation
Seed culture of each isolate was grown in nutrient broth containing 50 mg L−1 CP, harvested by centrifugation at 4,600×g for 5 min, washed with MilliQ H2O to set an OD600 of 1.0. Colony forming units (cfu mL−1) of these suspensions were quantified by dilution plate counting. For degradation studies cell concentration of 1.6 × 107cfu mL−1 or g−1 for pure cultures and 4 × 106 cfu mL−1 or g−1 of each strain for consortium was sued to keep equal cell number.
Biodegradation of CP and TCP in liquid medium
Shake flask studies were carried out to determine the CP degrading capacity of the isolated strains. Erlenmeyer flasks (250 mL) containing MSM (100 mL) amended with 100 mg L−1 of CP were inoculated with bacterial cell suspension in triplicates. TCP degradation was estimated by inoculating MSM supplemented with TCP (25 mg L−1). Where required, glucose was added as a supplementary carbon source (1 % w/v). The flasks were incubated at 30 °C with shaking at 150 rpm and an uninoculated flask was used as control. For growth studies 1 mL culture was withdrawn for 10 days at interval of 2 days and growth was evaluated as OD600. After 0, 2, 4, 6, 8 and 10 days, culture from control and treated flasks was centrifuged and supernatant was used for residual pesticide estimation. After extraction of supernatant with equal volume of ethyl acetate the pooled organic phase was dried over anhydrous sodium sulfate and evaporated on a rotary evaporator at 40 °C under vacuum. The residue was dissolved in methanol and final volume was made to 2 mL for HPLC analysis. The average recoveries of CP and TCP in liquid media were always found to be between 93 and 98 %.
Substrate range
The ability of consortium to degrade different pesticides was determined by inoculating MSM supplemented with additional pesticides such as Profenophos, Triazophos (organophosphates), Cypermethrin, Bifenthrin (pyrethroids) or Carbofuran (carbamate) at concentration of 100 mg L−1. The flasks were incubated at 30 °C with shaking at 150 rpm and uninoculated flasks were used as control. The extraction and estimation method was same as described above.
Biodegradation of Chlorpyrifos in soil
Degradation analysis of CP in soils was carried out in sterilized soil inoculated with individual strains and consortium (SS+inoculum) and non-inoculated non-sterilized (nSS) and sterilized soils (SS). Samples of 100 g soil were placed in sterile containers and spiked with CP up to 200 mg kg−1 concentration by adding an acetone solution of CP to a small portion (10 g) of soil and then mixing it with the rest of the soil after solvent evaporation. Soils were inoculated and incubated at 30 °C. Water content was maintained at 40 % of water holding capacity.
Soil samples were removed at different time intervals (0, 7, 14, 21, 28, 35 and 42 days) and analyzed for residual CP and TCP. Subsamples of 5 g soils were extracted twice with 15 mL of acidified acetone [acetone + water + conc. H3PO4 (98:1:1) by shaking on an end-over-end shaker for 4 h and then centrifuging at 4,000 rpm for 30 min. The combined extracts were evaporated using a rotary evaporator at 45 °C. The residue was redissolved in 1 mL of methanol–water (90:10 v/v) and filtered through a 0.45 µm PVDF filter for HPLC analysis. Average recovery of CP from soil ranged from 94 to 98.5 %.
High performance liquid chromatography (HPLC) analysis
Residual pesticides were analyzed on a SYKAM HPLC equipped with a HPLC Pump S 1122, programmable variable-wavelength UV detector S3210, Autosampler S 5200, delivery system S1122 and Phenomenex C18 reversed-phase column (150 mm). Detector output was processed by clarity chromatography data system. Pesticide concentration analysis was conducted using gradient elution conditions with initial solvent conditions of 100 % solvent A [methanol: H2O: Acetic Acid] (20:80:0.5) to 100 % solvent B [methanol: H20: Acetic Acid] (80:20:0.5) in 30 min at a flow rate of 1 mL min−1. Sample injection volume was 20 µL and detection was performed at 230 nm. The retention times of CP and TCP were 6.09 and 2.74 min, respectively. Whereas, Triazophos, Profenophos, Carbofuran, Cypermetrin and Bifenthrin were detected at 5.89, 7.52, 2.1, 5.95 and 9.60 min respectively. The calibration curves of pesticides were made from the serial dilutions of the samples dissolved in 100 % methanol. The linear range and the equation of linear regression were obtained sequentially at 0, 50, 100, 150, 250 and 300 µg mL−1. Mean areas generated from the standard solution were plotted against concentration to establish the calibration equation. The concentration of pesticide residues was determined on the basis of the peak areas in the chromatograms. The limit of detection was found to be 0.08–0.2 µg L−1 or kg−1 and limit of quantification ranged from 0.5 to 1.6 µg L−1 or kg−1.
Denaturing gradient gel electrophoresis (DGGE) analysis
DNA extraction from soils and enrichment cultures
DNA was extracted from soil and bacterial culture using Fastprep DNA extraction from soil adapted from (Yeates 1998). 300 mg of soil (or pellet from 1 mL of enrichment culture) was processed by bead beating in lysis buffer (0.1 M NaHPO4–Na EDTA, pH 8.0), and precipitated with 7.5 M potassium acetate, supernatant was mixed with equal volume of binding Matrix (MP Biomedicals), collected by centrifugation and resuspended in 100–200 µL TE buffer. Purified DNA was diluted tenfold in MilliQ H2O prior to use.
Denaturing gradient gel electrophoresis (DGGE) and band sequencing
Partial eubacterial 16S rRNA gene fragments (V3) were amplified using primers GC-341f and 518r (Kawasaki et al. 2011) using Xp Cycler (Bioer) thermal cycler. 25 µL reaction mixture comprised 10 pmol of each primer, 12.5 µL PCR Master Mix (Thermoscientific) and 10 ng of template DNA. A touchdown protocol was applied with an initial denaturation at 95 °C (5 min), followed by 10 cycles of 94 °C (30 s), 65 °C (30 s, −1 °C per cycle) and 72 °C (30 s); and then 20 cycles for 95 °C (30 s), 50 °C (30 s) and 68 °C (30 s), followed by a final extension at 68 °C (5 min).
DGGE was carried out using a D-code 16 cm × 16 cm electrophoresis chamber (Bio-Rad) (Cunliffe and Kertesz 2006), casting the gels with a linear denaturant gradient of 30–60 % [40 % (v/v) formamide, 7 M urea]. Gels were loaded with DGGE PCR products (~200 ng per sample), run at 63 V and 60 °C for 16 h, stained with SYBR Gold (0.2 µL/20 mL TAE) (Invitrogen) and analyzed with Dolphin-1D software (Wealtec). Selected bands were excised from the gel and left soaked overnight in MilliQ H2O at 4 °C. After reamplification using identical primers but lacking the GC clamp, the PCR product was purified and sequenced.
Sequence analysis and accession numbers
DNA sequence analysis was done by Macrogen Korea, using an ABI 3730 automated DNA sequencer. DNA sequence homologies were carried out with BLASTN and Ribosomal database project (rdp.cme.msu.edu/). Multiple alignments and distance matrix analyses were conducted using the MEGA 5.0 software package (Tamura et al. 2011). A phylogenetic tree was constructed using neighbor-joining analysis with 1,000 replicates of bootstrap analysis. GenBank accession numbers were assigned for 16S rRNA gene sequences of the isolates (KC119333–KC119335 and KF263488) and of the DGGE bands (KF559226–KF559252).
Degradation rate analysis
The CP degradation rate constant (k) was determined using the kinetic model Ct = C0 × e_kt, where C0 is the initial concentration of CP at time zero, Ct is the concentration of CP at time t, t is the degradation period in days and k is the rate constant (d−1). The half-life (T1/2) of CP was determined using the algorithm T1/2 = ln2/k. Correlation coefficient (R2) and Regression Equation were calculated from linear equation between ln(Ct/C0) of chemical data and time. The results from three replicates of each treatment were also evaluated using analysis of variance and statistical analysis. The significance (p < 0.05) of differences was assessed by post hoc comparison of means using the least significant differences (LSD) test using the IBM SPSS statistics 20.
Results
Enrichment culturing using chlorpyrifos as carbon source
Survival and proliferation of the bacterial community during enrichment in liquid culture with CP as sole carbon source was characterized by profiling of partial 16S rRNA genes. Analysis of DGGE profiles indicated substantial difference between enrichment cultures and soils, while fewer differences were seen between control and treated cultures (Fig. 1). Higher numbers of bands were present in soil as compared to cultures grown with and without CP. More bands were observed in the first week profile than in the fingerprints from later weeks suggesting a selection for chlorpyrifos-resistant organisms. Three different groups of bands were observed in DGGE gel. First, some of the bands were present in fingerprints of all enrichment steps; these represented dominant bacterial species in CP treated soil that can utilize CP as C source hence proliferating in CP-supplemented media. Sequence analysis showed that these were affiliated with Pseudomonas sp. (B2, B6), Novosphingobum sp. (B13), Burkholderia sp. (B14) and Sphingomonas sp. (B15). A second group of bands were not present at initial steps but appeared in the fingerprint of final enrichment steps. The corresponding organisms appeared to be favored by enrichment process and sequencing showed their similarity to Nitrosomonas sp. (B3), Burkholderia sp. (B9, B20), Pseudomonas sp. (B4, B5), Stenotrophomonas sp. (B16, B18), Agrobacterium sp. (B19, B26), and Paracoccus sp. (B27). The third group of bands corresponded to organisms that were present in the soil or initial enrichment step, but became very weak or disappeared by the final enrichment steps. Sequences of these bands showed similarity to Flavobacterium sp. (B1), Bacillus sp. (B7 and B10), uncultured γ-Proteobacteria (B11), Pseudoxanthomonas sp. (B17), Lysobacter sp. (B21), uncultured bacterium (B22, B23) and Rhodococcus sp. (B25). Bands appearing in the fingerprints of later steps are result of continual selection pressure resulting in proliferation of potential CP degrading bacteria. The disappearance of bands present only in fingerprints of initial steps can be due to the loss of bacteria that need to use soil particles as a solid substratum to attach to, or their displacement by bacteria with better CP-degrading abilities.
16S rDNA denaturing gradient gel electrophoresis (DGGE) analysis of bacterial community in soils (lane 1–2) and in control (C) and treated (T) enrichment cultures. Lanes 3–6, 7–10, 11–14 and 15–18 represent first, second, third and fourth enrichment cycles respectively. Cultures were enriched in replicates (T1, T2, and T3). B1–B27 are bands eluted from the gel and sequenced
Phylogenetic relationships of these sequences to other closely related species is shown in Fig. 2. Most of the sequences belonged to members of the Proteobacteria and Firmicutes with one band each of Bacteroidetes and Actinobacteria. The Phylogenetic tree showed a shift in microbial community composition with members of Bacteroidetes, Firmicutes, α- and γ-Proteobacteria and Actinobacteria present at initial steps whereas β-Proteobacteria appeared in later steps and only Proteobacteria (highlighted band numbers) were dominant among the members selected by enrichment culturing.
Phylogenetic analysis of partial 16S rRNA gene (V3 region) sequence from excised DGGE bands of enriched CP degrading bacterial community to other related organisms. The tree was constructed with MEGA 5.0 software package using neighbor-joining analysis and 1,000 replicates for bootstrap analysis. Bar 0.02 represents sequence divergence
Isolation, screening and identification of CP degrading strains
From the enrichment culture, a range of morphologically different bacterial strains were isolated that were able to grow on mineral medium containing up to 100 mg L−1 CP. The eight fastest growing isolates were selected for further study (Table 1). Biochemical characterization and 16S rRNA gene analysis showed that bacterial strains belonged to the Stenotrophomonas, Paracoccus, Pseudomonas, Acinetobacter, Sphingomonas, and Burkholderia genera. Strains of Acinetobacter calcoaceticus LCp4, Stenotrophomonas maltophilia LCp5, Pseudomonas mendocina LCp8 and P. aeruginosa LCp9 showed the highest CP degradation potential and were selected for consortium development and biodegradation studies.
Biodegradation of CP and TCP in mineral salt medium
Significant differences (p < 0.05) were observed in growth kinetics of individual bacterial isolates. For isolates LCp4, LCp8 and LCp9 and a mixed consortium of all four strains, the most effective growth was observed over 2–6 days without an initial lag phase, and maximum growth was attained after 8 days of incubation (Fig. 3A1). Isolate LCp5 multiplied comparatively slowly where a steady increase in growth was observed for 6 days. Biodegradation (percentage depletion of CP) was estimated by HPLC (Fig. 3A2), and confirmed that all isolates were able to degrade CP but with different degradation rates. Individual isolates degraded 60–75 % of CP while the consortium’s efficiency was higher. After 10 d of incubation 74.6, 60.5, 69.8 and 67.5 % of initial concentration of CP was utilized by A. calcoaceticus LCp4, S. maltophilia LCp5, P. mendocina LCp8 and P. aeruginosa LCp9 respectively, while the consortium degraded 92 % of initial concentration of CP after 10 days (Fig. 3A2). In control flasks CP degradation by abiotic losses was insignificant, with only 3–4 % CP depletion observed at the end of 10 days. With glucose as an additional C source bacterial growth was considerably stimulated, giving approximately two times faster growth rate at the start of incubation period (0–4 days). Maximum OD600 was obtained at 8 days of incubation while most effective growth was observed during the first 4 days (Fig. 3B1). CP degradation improved in the presence of glucose reaching 90, 73.5, 81.4 and 78 % for LCp4, LCp5, LCp8 and LCp9 respectively in 10 days. Meanwhile, the consortium (LCp4, LCp5, LCp8 and LCp9) succeeded in removing 87 % of applied CP in 6 days, degrading it completely in 10 days (Fig. 3B2). HPLC analysis also revealed that A. calcoaceticus LCp4, P. mendocina LCp8, P. aeruginosa LCp9 and consortium were able to mineralize CP completely, with only transient accumulation of the CP metabolic product TCP. However, in culture extracts of S. maltophilia LCp5 TCP was accumulated and did not disappear. Similar results were observed for degradation of TCP in MSM where LC8 and LCp9 degraded 83.2 and 78.4 % of applied TCP while LCp4 and consortium utilized it completely. In LCp4 inoculated and control media insignificant TCP degradation was observed (Fig. 4).
Growth of bacterial isolates and degradation kinetics during growth with chlorpyrifos. Strains were grown in mineral salt medium (MSM) supplemented with chlorpyrifos (CP) (100 mg L−1) as sole source of carbon. Cultures were grown without (A1, A2) or with (B1, B2) glucose (1 %) as an additional source of carbon. Symbols: (white circle) non inoculated control; medium inoculated with (diamond) Stenotrophomonas maltophilia LCp5; (triangle) Pseudomonas mendocina LCp8; (black circle) Pseudomonas aeruginosa LCp9 (square); Acinetobacter calcoaceticus LCp4; and (black triangle) Consortium of LCp4, LCp5, LCp8 and LCp9. Symbols are the means of three replicates and error bars represent the standard deviation of the mean
Degradation of trichloropyridinol (TCP) in mineral salt medium. strains were grown in mineral salt medium (MSM) supplemented with TCP (25 mg L−1) as sole source of carbon. Symbols: (white circle) non inoculated control; medium inoculated with (diamond) S. maltophilia LCp5; (black circle) P. aeruginosa LCp9; (triangle) P. mendocina LCp8; (square) A. calcoaceticus LCp4 and (black triangle) Consortium of LCp4, LCp5, LCp8 and LCp9. Symbols are the means of three replicates and error bars represent the standard deviation of the mean
Substrate range
In order to evaluate the capacity of the consortium to utilize structurally different pesticides, the cultures were supplemented with various organophosphates (Triazophos and Profenophos), Pyrethroids (Cypermethrin and Bifenthrin) and a carbamate (Carbofuran) as sole source of C. The consortium was able to degrade 86 and 83 % of Triazophos and Profenophos within 10 days, which was expected as these compounds have a similar P–O–C bond to as CP, and could be attacked by organophosphate hydrolase (OPH). Carbofuran was also degraded efficiently by the consortium, with 90 % disappearance during incubation period. However, the consortium’s efficiency to degrade pyrethroids was lower than for OPs and carbamates, with only 55 and 50 % of Cypermethrin and Bifenthrin degraded after 10 days (Fig. 5).
Degradation of different pesticides by consortium in mineral salt medium. Symbols: (diamond) Carbofuran; (square) Triazophos; (triangle) Profenophos; (white circle) Cypermethrin and (black circle) Bifenthrin. The solid line represents treated samples while broken line is for control samples. Initial concentrations were 100 mg L−1. Symbols are the means of three replicates and error bars represent the standard deviation
Biodegradation of Chlorpyrifos in soil
Studies of the degradation dynamics of CP in various microbiologically active soils revealed significant differences (p < 0.05) in CP degradation potential between bacterial isolates (Fig. 6; Table 2). CP depletion was highest in sterilized soil inoculated with A. calcoaceticus LCp4, with rate constant of 0.0478 d−1, following first order kinetics and the average rate of CP degradation of 4.40 mg kg−1 d−1. Soils inoculated with P. mendocina LCp8 and P. aeruginosa LCp9 showed similar CP degradation dynamics with rate constants of 0.0400 and 0.0361 d−1 respectively. LCp8 and LCp9 degraded 85.7 and 89 % of applied CP within 42 days with average rates of 4.08 and 4.24 mg kg−1 d−1. S. maltophilia LCp5 was able to degrade 77.5 % of added CP with rate constant and average rate of 0.0303 d−1 and 3.69 mg kg−1 d−1. T 1/2 values for CP degradation in these soil treatments were 16.5, 22.87, 19.2 and 17.32 d for strains LCp4, LCp5, LCp8 and LCp9 respectively. With the consortium the average CP disappearance rate was 4.76 mg kg−1 d−1 within 42 d with a rate constant of 0.0702 d−1, following first order kinetics, with T 1/2 of 9.87 d (Table 2). For non-inoculated, non-sterilized soil, CP depletion was characterized by a lag phase of 7-days during which only 3 % of applied CP was degraded. CP disappearance then increased considerably, with a rate constant of 0.0111 d−1, resulting in utilization of 42.2 % of initial amount at the end of incubation time. No lag phase was observed in sterilized soil samples inoculated with individual bacterial strains and their consortium. In sterilized soil CP disappearance was insignificant, and 80 % of applied CP still persisted at the end of incubation period. The average rate of CP degradation in sterilized soil was 0.84 mg kg−1 d−1 with rate constant of 0.0053 d−1 and T 1/2 was 130.7 d.
CP degradation dynamics of chlorpyrifos in different soil treatments. Symbols: (white circle) non-inoculated sterilized soil; (black diamond) non-sterilized soil; (white diamond) SS+ S. maltophilia LCp5; (white triangle) SS+ P. mendocina LCp8; (black circle) SS+ P. aeruginosa LCp9; (square) SS+ A. calcoaceticus LCp4; (black triangle) SS+ Consortium of LCp4 + LCp5 + LCp8 + LCp9. Chlorpyrifos initial concentration was 200 mg kg−1. Symbols are the means of three replicates and error bars represent the standard deviation
Discussion
CP has been in widespread use for almost 50 years and because of its toxicity and recalcitrance its removal from environment is of immense importance. This study was designed to isolate potential CP-degraders from contaminated soil and to monitor the survival of bacterial isolates during enrichment process. CP-degrading bacterial strains were isolated from agricultural soils which had been treated with CP and other pesticides over an extended period, indicating the ability of the organisms to adapt to the environmental conditions. The isolates that showed maximum CP degradation were identified as A. calcoaceticus, S. maltophilia, P. mendocina and P. aeruginosa. Bacteria belonging to these genera have been documented to play important roles in xenobiotic and pesticide metabolism. Lakshmi et al. (2008) studied degradation of both CP and TCP by P. aeruginosa in soil and aqueous media. Pseudomonas strains have also been studied for degradation of Cadusafos (Abo-Amer 2012), Cypermethrin (Zhang et al. 2011), Fenamiphos and CP (Chanika et al. 2011). Acinetobacter strains have been described as responsible for degradation of methyl parathion (Liu et al. 2007). Similarly, Stenotrophomonas has been studied for its role in degradation of fenvelerate (Chen et al. 2011a) and CP (Dubey and Fulekar 2012).
These strains degraded 60–75 % CP in mineral medium within 10 days, the degradation rate (6.0–7.5 mg L−1 d−1) of these strains was comparable to maximum degradation rate Vmax (7.4–12.1 5 mg L−1 d−1) of CP-degrading strains studied earlier by Maya et al. (2011). The consortium of these strains was more efficient and removed 92 % of CP in same period. Lakshmi et al. (2009) also observed enhanced CP degradation by consortium consisted of P. aeruginosa, Bacillus cereus, Klebsiella sp. and Serratia marscecens. The increased degradation of CP by consortium can be due to its ability to remove CP without accumulation of toxic intermediate TCP. In the consortium the inability of S. maltophilia LCp5 to degrade TCP was complemented by other TCP degrading strains to remove the toxic product that positively influenced the degradation process. Similar enhanced pesticide degradation using microbial consortia has been reported before for degradation of atrazine and diuron (Smith et al. 2005; Sorensen et al. 2008). Enhanced degradative abilities of consortia can be attributed to requirement for growth factors and nutrients provided by secondary strain, and cross-feeding of metabolites from the degradation pathway. For example, in a study coculture of B. cereus ZH-3 and Streptomyces aureus HP-S-01 significantly enhanced cypermethrin degradation; HP-S-01 was able to mineralize the toxic metabolite 3-phenoxybenzaldehyde (3-PBA) that ZH-3 was unable to utilize, thus stabilizing the consortium by cross feeding of metabolites (Chen et al. 2012).
Addition of glucose as a supplementary C source significantly enhanced CP degradation by the consortium, which utilized 87 % of applied CP in 6 days under these conditions compared to 71.8 % in mineral medium without additional C source in the same period (Fig. 3B2). Similar enhanced degradation of pesticide has been reported in the presence of glucose in earlier studies (Cycon et al. 2009; Pino and Peñuela 2011). However, contrasting results were reported by Singh et al. (2004) where addition of C source suppressed CP degradation by Enterobacter sp. for first three days and CP degradation started after depletion of C source. Glucose as an immediate energy source enhanced cell growth which in turn increased CP degradation. It can also be assumed that consortium not only metabolized CP but also co-metabolized it in the presence of other C source. Chen et al. (2012) reported the similar results for degradation of Cypermethrin, where addition of glucose enhanced its degradation.
A different feature of consortium revealed during the study was its ability to degrade carbofuran along with various OPs and pyrethroids other than CP. Since the general chemical structure of OPs is alike, the consortium’s capability of degrading other OPs was anticipated. Cycoń et al. (2013) also reported degradation of various OPs by diazinon degrading bacteria. The soil used for isolation of CP-degrading bacteria had also received occasional applications of carbamates and pyrethroids and this could explain the parallel adaptation of strains to pesticides of different structures. The consortium might contain organisms with specific enzyme systems adapted for degradation of structurally different pesticides. As reported earlier, bacterial phosphotriesterases are capable of hydrolyzing P–O, P–S, P–CN and C–O bonds in esters and lactones (Wang et al. 2012). Chanika et al. (2011) have reported isolation of a P. putida strain that was able to hydrolyze both carbamates and organophosphates. The degradation of pyrethroids by consortium to a lesser extent as compared to OPs and carbamates can be related to generation of toxic compound 3-phenoxy benzoic acid (3-PBA). 3-PBA is persistent towards microbial degradation and has antimicrobial activities that might have limited consortium’s capability to degrade pyrethroids (Chen et al. 2011b). There are no earlier reports of consortium able to degrade different classes of compounds, though an attempt has been made to develop a genetically modified organism with co-expression of organophosphate hydrolase (opd) and carboxylesterase genes, for degradation of organophosphates, carbamates and synthetic pyrethroids (Lan et al. 2006).
Results of CP-degradation in soil exhibited that consortium utilized CP more quickly in sterilized soil as compared to soils inoculated with a single bacterial strain and non-inoculated non-sterilized soil. The consortium degraded 100 % of added CP within 42 days as compared to 76–90 % CP degradation by individual strains. Singh et al. (2006) also observed enhanced fenamiphos degradation in soil using bacterial consortium. The lag phase observed in the case of non-sterilized soil corresponds to the time required for propagation of small community of CP degrading bacteria to attain sufficient level for efficient degradation of pesticide (Cycon et al. 2009). Slower degradation of CP in soils as compared to liquid media might be due to influence of various soil parameters such as soil moisture, organic content and soil structure. CP being hydrophobic in nature tends to adsorb at soil surfaces and solid phase of organic matter thus limiting its availability to microbes and slowing down the process of degradation (Liang et al. 2011). Soil moisture also influence pesticide degradation by regulating microbial mobility and proliferation and solute diffusion (Singh et al. 2006). As higher CP concentration (200 mg kg−1) was applied to soil as compared to liquid media (100 mg kg−1), it might have hindered the initial catabolic activities of microbes in soil. As studied by Fang et al. (2009) that high CP concentration provoked inhibitory effects on microbes, though duration of repressing effects on microbial functional diversity was temporary.
Combination of enrichment culture and DGGE was used to analyze the survival of bacteria in CP rich environment. Similarity of band sequences to 16S rDNA sequence of CP degrading strains confirmed that Pseudomonas sp. Sphingomonas sp. and Acinetobacter sp. were among dominant bacteria in soil while others such as Stenotrophomonas sp. and Paracooccus sp. were selected by enrichment procedure. In earlier studies ecological significance of pesticide degrading strains isolated from enrichment cultures had been demonstrated (Chanika et al. 2011; Breugelmans et al. 2007).
In conclusion, the CP-degrading bacterial isolates obtained in this study showed enhanced CP-degradation potential when developed in a consortium. The consortium utilized CP as sole source of carbon and also co-metabolized it in the presence of glucose, and was able to bioremediated soil with CP concentration as high as 200 mg kg−1. Moreover, the consortium harbored the ability to degrade structurally different pesticides. DGGE analysis confirmed that these isolates were dominant members of the enrichment. These results suggest that the microbial consortium could be a promising candidate for remediation of CP-contaminated environment.
References
Abo-Amer AE (2012) Characterization of a strain of Pseudomonas putida isolated from agricultural soil that degrades cadusafos (an organophosphorus pesticide). World J Microbiol Biotechnol 28:805–814
Anwar S, Liaquat F, Khan QM, Khalid ZM, Iqbal S (2009) Biodegradation of chlorpyrifos and its hydrolysis product 3,5,6-trichloro-2-pyridinol by Bacillus pumilus strain C2A1. J Hazard Mater 168:400–405
Breugelmans P, D’Huys PJ, De Mot R, Springael D (2007) Characterization of novel linuron-mineralizing bacterial consortia enriched from long-term linuron-treated agricultural soils. FEMS Microbiol Ecol 62:374–385
Briceño G, Fuentes MS, Palma G, Jorquera MA, Amoroso MJ, Diez MC (2012) Chlorpyrifos biodegradation and 3,5,6-trichloro-2-pyridinol production by actinobacteria isolated from soil. Int Biodeterior Biodegrad 73:1–7
Cao L, Xu J, Wu G, Li M, Jiang J, He J, Li S, Hong Q (2013) Identification of two combined genes responsible for dechlorination of 3, 5, 6-trichloro-2-pyridinol (TCP) in Cupriavidus pauculus P2. J Hazard Mater 260:700–706
Chanika E, Georgiadou D, Soueref E, Karas P, Karanasios E, Tsiropoulos NG, Tzortzakakis EA, Karpouzas DG (2011) Isolation of soil bacteria able to hydrolyze both organophosphate and carbamate pesticides. Bioresour Technol 102:3184–3192
Chen S, Hu M, Liu J, Zhong G, Yang L, Rizwan-ul-Haq M, Han H (2011a) Biodegradation of beta-cypermethrin and 3-phenoxybenzoic acid by a novel Ochrobactrum lupini DG-S-01. J Hazard Mater 187:433–440
Chen S, Yang L, Hu M, Liu J (2011b) Biodegradation of fenvalerate and 3-phenoxybenzoic acid by a novel Stenotrophomonas sp. strain ZS-S-01 and its use in bioremediation of contaminated soils. Appl Microbiol Biotechnol 90:755–767
Chen S, Luo J, Hu M, Lai K, Geng P, Huang H (2012) Enhancement of cypermethrin degradation by a coculture of Bacillus cereus ZH-3 and Streptomyces aureus HP-S-01. Bioresour Technol 110:97–104
Cho KM, Math RK, Islam SMA, Lim WJ, Hong SY, Kim JM, Yun MG, Cho JJ, Yun HD (2009) Biodegradation of chlorpyrifos by lactic acid bacteria during kimchi fermentation. J Agirc Food Chem 57:1882–1889
Cunliffe M, Kertesz MA (2006) Effect of Sphingobium yanoikuyae B1 inoculation on bacterial community dynamics and polycyclic aromatic hydrocarbon degradation in aged and freshly PAH-contaminated soils. Environ Pollut 144:228–237
Cycon M, Wojcik M, Piotrowska-Seget Z (2009) Biodegradation of the organophosphorus insecticide diazinon by Serratia sp. and Pseudomonas sp. and their use in bioremediation of contaminated soil. Chemosphere 76:494–501
Cycoń M, Żmijowska A, Wójcik M, Piotrowska-Seget Z (2013) Biodegradation and bioremediation potential of diazinon-degrading Serratia marcescens to remove other organophosphorus pesticides from soils. J Environ Manage 117:7–16
Dubey KK, Fulekar MH (2012) Chlorpyrifos bioremediation in Pennisetum rhizosphere by a novel potential degrader Stenotrophomonas maltophilia MHF ENV20. World J Microbiol Biotechnol 28:1715–1725
Fang H, Yu Y, Chu X, Wang X, Yang X, Yu J (2009) Degradation of chlorpyrifos in laboratory soil and its impact on soil microbial functional diversity. J Environ Sci 21:380–386
Ferreira EPdB, Dusi AN, Costa JR, Xavier GR, Rumjanek NG (2009) Assessing insecticide and fungicide effects on the culturable soil bacterial community by analyses of variance of their DGGE fingerprinting data. Eur J Soil Biol 45:466–472
Kawasaki A, Watson ER, Kertesz MA (2011) Indirect effects of polycyclic aromatic hydrocarbon contamination on microbial communities in legume and grass rhizospheres. Plant Soil 358:169–182
Lakshmi CV, Kumar M, Khanna S (2008) Biotransformation of chlorpyrifos and bioremediation of contaminated soil. Int Biodeterior Biodegrad 62:204–209
Lakshmi CV, Kumar M, Khanna S (2009) Biodegradation of chlorpyrifos in soil by enriched cultures. Curr Microbiol 58:35–38
Lan WS, Gu JD, Zhang JL, Shen BC, Jiang H, Mulchandani A, Chen W, Qiao CL (2006) Coexpression of two detoxifying pesticide-degrading enzymes in a genetically engineered bacterium. Int Biodeterior Biodegrad 58:70–76
Li X, He J, Li S (2007) Isolation of a chlorpyrifos-degrading bacterium, Sphingomonas sp. strain Dsp-2, and cloning of the mpd gene. Res Microbiol 158:143–149
Liang B, Yang C, Gong M, Zhao Y, Zhang J, Zhu C, Jiang J, Li S (2011) Adsorption and degradation of triazophos, chlorpyrifos and their main hydrolytic metabolites in paddy soil from Chaohu Lake, China. J Environ Manage 92:2229–2234
Liu F, Hong M, Liu D, Li Y, Shou P, Yan H, Shi G (2007) Biodegradation of methyl parathion by Acinetobacter radioresistens USTB-04. J Environ Sci 19:1257–1260
Lu P, Li Q, Liu H, Feng Z, Yan X, Hong Q, Li S (2013) Biodegradation of chlorpyrifos and 3, 5, 6-trichloro-2-pyridinol by Cupriavidus sp. DT-1. Bioresour Technol 127:337–342
Maya K, Singh R, Upadhyay S, Dubey SK (2011) Kinetic analysis reveals bacterial efficacy for biodegradation of chlorpyrifos and its hydrolyzing metabolite TCP. Process Biochem 46:2130–2136
Pino N, Peñuela G (2011) Simultaneous degradation of the pesticides methyl parathion and chlorpyrifos by an isolated bacterial consortium from a contaminated site. Int Biodeterior Biodegrad 65:827–831
Sasikala C, Jiwal S, Rout P, Ramya M (2012) Biodegradation of chlorpyrifos by bacterial consortium isolated from agriculture soil. World J Microbiol Biotechnol 28:1301–1308
Singh BK, Walker A (2006) Microbial degradation of organophosphorus compounds. FEMS Microbiol Rev 30:428–471
Singh BK, Walker A, Morgan JA, Wright DJ (2004) Biodegradation of chlorpyrifos by Enterobacter strain B-14 and its use in bioremediation of contaminated soils. Appl Environ Microbiol 70:4855–4863
Singh B, Walker A, Wright D (2006) Bioremedial potential of fenamiphos and chlorpyrifos degrading isolates: influence of different environmental conditions. Soil Biol Biochem 38:2682–2693
Smith D, Alvey S, Crowley DE (2005) Cooperative catabolic pathways within an atrazine-degrading enrichment culture isolated from soil. FEMS Microbiol Ecol 53:265–273
Sorensen SR, Albers CN, Aamand J (2008) Rapid mineralization of the phenylurea herbicide diuron by Variovorax sp. strain SRS16 in pure culture and within a two-member consortium. Appl Environ Microbiol 74:2332–2340
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739
Wang S, Zhang C, Yan Y (2012) Biodegradation of methyl parathion and p-nitrophenol by a newly isolated Agrobacterium sp. strain Yw12. Biodegradation 23:107–116
Xu G, Zheng W, Li Y, Wang S, Zhang J, Yan Y (2008) Biodegradation of chlorpyrifos and 3,5,6-trichloro-2-pyridinol by a newly isolated Paracoccus sp. strain TRP. Int Biodeterior Biodegrad 62:51–56
Yeates Gillings (1998) Rapid purification of DNA from soil for molecular biodiversity analysis. Lett Appl Microbiol 27:49–53
Zhang C, Wang S, Yan Y (2011) Isomerization and biodegradation of beta-cypermethrin by Pseudomonas aeruginosa CH7 with biosurfactant production. Bioresour Technol 102:7139–7146
Zhou G, Wang Y, Ma Y, Zhai S, Zhou L, Dai Y, Yuan S (2014) The metabolism of neonicotinoid insecticide thiamethoxam by soil enrichment cultures, and the bacterial diversity and plant growth-promoting properties of the cultured isolates. J Environ Sci Health B 49:381–390
Acknowledgments
Education Commission (HEC), Islamabad, Pakistan id being acknowledged for providing financial support through Indigenous scholarship and International research support initiative program (IRSIP).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Akbar, S., Sultan, S. & Kertesz, M. Bacterial community analysis in chlorpyrifos enrichment cultures via DGGE and use of bacterial consortium for CP biodegradation. World J Microbiol Biotechnol 30, 2755–2766 (2014). https://doi.org/10.1007/s11274-014-1699-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11274-014-1699-8