Abstract
Methylotrophic bacteria which are known to utilize C1 compounds including methane. Research during past few decades increased the interest in finding out novel genera of methane degrading bacteria to efficiently utilize methane to decrease global warming effect. Moreover, evaluation of certain known plant growth promoting strains for their methane degrading potential may open up a new direction for multiple utility of such cultures. In this study, efficient methylotrophic cultures were isolated from wetland paddy fields of Gujarat. From the overall morphological, biochemical and molecular characterization studies, the isolates were identified and designated as Bacillus aerius AAU M 8; Rhizobium sp. AAU M 10; B. subtilis AAU M 14; Paenibacillus illinoisensis AAU M 17 and B. megaterium AAU M 29. Gene specific PCR analysis of the isolates, P. illinoisensis, B. aerius, Rhizobium sp. and B. subtilis showed presence of pmoA gene encoding α subunit particulate methane monooxygenase cluster. B. megaterium, P. illinoisensis, Rhizobium sp. and Methylobacterium extrorquens showed presence of mmoX gene encoding α subunit of the hydroxylase component of the soluble methane monooxygenase cluster. P. illinoisensis and Rhizobium sp. showed presence mxaF gene encoding α subunit region of methanol dehydrogenase gene cluster showing that both isolates are efficient utilizers of methane. To the best of our knowledge, this is the first time report showing presence of methane degradation enzymes and genes within the known PGPB group of organisms from wet land paddy agro-ecosystem, which is considered as one of the leading methane producer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The atmospheric trace gas methane (CH4) is a prominent “greenhouse” gas. Methane accounts for 15–20 % of global warming (IPCC 2007). Up to 70–80 % of atmospheric CH4 is biogenic (Wahlen et al. 1989). Major sources of this input include natural wetlands, rice fields, enteric fermentation in animals, termites and landfills. The contribution from rice cultivation is estimated to range from 39 to 112 Tg CH4 y−1 (Khosa et al. 2011). About 80 % of the rice harvest is grown under the more productive flooded conditions (wetland paddy) and thereby major source of methane emission. The only known biological sink for atmospheric methane is its oxidation in aerobic soils by “methanotrophic (=methylotrophic) bacteria”, this may contribute up to 10–20 % to the total methane destruction (Reeburgh et al. 1993). Methanotrophs are classified as type I (γ-proteobacteria) and type II (α-proteobacteria), based on several characteristics including phylogeny, guanine and cytosine content of their DNA, intracellular membrane arrangement, carbon assimilation pathways and phospholipid fatty acids (PLFAs) composition (Hanson and Hanson 1996; Whittenbury et al. 1970). At present, there are 13 recognized genera of methanotrophs viz. Methylomonas, Methylosphaera, Methylomicrobium, Methylosarcina, Methylobacter, Methylocaldum, Methylococcus, Methylohalobius, Methylosoma Methylocystis, Methylocella, Methylocapsa and Methylosinus (Hanson and Hanson 1996; Bowman et al. 1997; Bodrossy et al. 1997; Dedysh et al. 2000, 2002; Wise et al. 2001; Heyer et al. 2005; Tsubota et al. 2005) consisting of both type I and type II methanotrophs.
All methanotrophs are having highly conserved key enzymes viz. particulate methane monooxygenase (pMMO), soluble methane monooxygenase (sMMO) and methanol dehydrogenase (MDH), so that detection of enzymatic activity may offer the possibility of detecting all known methanotrophs (Hanson and Hanson 1996). So application of methanotrophic bacteria having PGPR activity can present a great opportunity to reduce methane emission from agriculture sector and easily adoptable technology by farmers. Present research first time uncover five methane degrading bacterial strains within the known PGPB group of organisms from wet land paddy agro-ecosystem which are not previously reported.
Materials and methods
Isolation and screening of methane utilizing bacteria
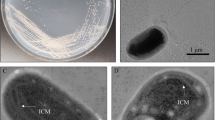
For isolation of methylotrophic bacteria from wet land paddy, total 15 soil samples were collected at flowering stage representing major traditionally rice growing areas of Middle and South Gujarat, India. Out of fifteen soil samples, 7 samples were collected from Main Rice Research Station, Nawagam, 4 samples from Agricultural Research Station for Irrigated Crops, Thasra, 2 each from farmer’s field at Tarapur and Vyara Taluka, respectively. For bacterial isolation method suggested by Hoppe et al. (2011) was followed with some modifications as follows. Instead of Ammonium Mineral Salt (AMS) medium, Nitrate Mineral Salt (NMS) broth (Sodium nitrate—2.0 g l−1, MgSO4·7H2O—0.2 g/l, KCl—0.04 g l−1, Calcium chloride—0.015 g l−1, Na2HPO4—0.21 g l−1, NaH2PO4—0.09 g l−1, FeSO4·7H2O—0.01 mg l−1, CuSO4·5H2O—5 μg l−1, H3BO4—10 μg l−1, MnSO4·5H2O —10 μg l−1, ZnSO4·7H2O—70 μg l−1, MoO3—10 g l−1) supplemented with 1 % methane gas was inoculated with soil samples and enriched for 15 days. After enrichment bacterial growth was purified on NMS agar supplemented with 1 % methanol. Additionally, all the suspected isolates were screened for growth in the evacuated tubes containing water as the basal media with 1 % methane in the head space of the tube and in second set water containing 1 % methanol as sole source of carbon was used. Growth of each isolates has been observed by colorimeter to screen their methylotrophic property.
In vitro conformation of methylotrophic activity
Suspected isolates (after preliminary screening) were tested for utilization of methane gas at the rate of 1 % and methanol at the rate of 1–5 % as sole source of carbon in evacuated tubes containing water (basal medium) + methane (head space of the tube) or methanol separately to confirm methylotrophic metabolism. Growth and survival of isolates were measured by recording colony counts from inoculated tubes at 10 days after inoculation on NMS agar and recorded as colony forming units (CFU) ml−1 after 10 days of inoculation.
Characterization and identification of potential methylotrophic isolates
Selected isolates after in vitro methylotrophic activity were characterized on the basis of morphological, cultural and biochemical characteristics (The prokaryotes 2006; Halt et al. 1994). Molecular characterization was carried out using PCR detection of genes representing methylotrophic metabolism and amplified ribosomal DNA restriction analysis (ARDRA) technique (Waturangi et al. 2011 and Horz et al. 2001). For identification and phylogenetic relationship of the potential isolates, 16S rDNA sequencing was carried out as described by Waturangi et al. 2011.
Genomic DNA extraction
Five methylotrophic isolates were grown in Luria broth for 24 h, and genomic DNA was extracted by C-TAB method. The integrity and concentration of purified DNA was determined by agarose gel electrophoresis. The total genomic DNA extracted was dissolved in sterile distilled water and stored at 4 °C.
Detection of bacterial methylotrophic metabolism genes
The presence of mmoX, pmoA and mxaF gene in the isolates, encoding sMMO, pMMO and methanol dehydrogenase which are key enzymes in bacterial methane metabolism pathway, was used for authentication of the isolates. The presence of genes in the isolates was detected by partial amplification of the genes using specific primers (Horz et al, 2001). Following primer pairs used for mmoX (534f—ccgctgtggaagggcatgaa & 1393r—cactcgtagcgctccggctc); pmoA (A189f—ggngactgggacttctgg & A682r—gaasgcngagaagaasgc)Footnote 1 and mxaF (1003f—gcggcaccaactggggctggt & 1561r—gggcagcatgaagggctccc), respectively. The 40 μl PCR reaction mixture contains DNA template 50 ng, 1X Taq buffer, 0.2 mM of each of dNTPs mixture, 1 μM of each primers, 1.5 mM·MgCl2, and 2 U of Taq DNA polymerase (Bangalore Genei, India). PCR amplification was performed in a thermal cycler (Eppendorf Master cycler, Germany) using the following conditions: initial denaturation at 95 °C for 1 min, 30 cycles consisting of 95 °C for 1 min (denaturation), annealing 62–52 °C touchdown—1 min for mmoX and pmoA, while 55 °C—1 min for mxaF gene, 72 °C for 1 min (primer extension), and final extension 72 °C for 5 min. PCR products were separated by electrophoresis on 1.5 % agarose gels stained with ethidium bromide and documented in AlphaImager TM1200 documentation and analysis system.
Amplified ribosomal DNA restriction analysis
16S rDNA amplification was performed in a thermal cycler (Eppendorf Master cycler, Germany) with a 25 μl reaction mixture containing 50 ng of genomic DNA, 0.2 mM of each dNTPs, 1 μM of each primer (Waturangi et al. 2011), 2.5 mM of MgCl2, and 1 U of Taq DNA polymerase (Bangalore Genei, India) and the buffer supplied with the enzyme. Approximately 1 μg of PCR-amplified 16S rDNA fragments were restricted with endonucleases TaqI, HinfI, and HaeIII (Fermentas, USA) separately at 37 °C for 3 h and resolved by electrophoresis in 3 % agarose gels. Banding patterns were visualized by ethidium bromide staining and documented in AlphaImager TM1200 documentation and analysis system. Strong and clear bands were scored for similarity and clustering analysis using the software, NTSYS-PC2 package (Numerical taxonomy analysis program package, Exeter software, USA). Similarity among the strains was calculated by Jaccard’s coefficient (Jaccard 1912), and dendrogram was constructed using UPGMA method (Nei and Li 1979).
DNA sequencing analysis
DNA fragment from five isolates were chosen to be sequenced based on the dominant isolates that isolated from soil samples. PCR product was purified using DNA Gel Extraction Kit (Banglore Genei, India). Purified products were sequenced using big dye terminator with ABI PRISMTM model 3130 Genetic Analyzer. The sequence data of bacterial isolates were compared to sequence from GenBank in National Center for Biotechnology Information (NCBI, www.ncbi.nlm.nih.gov) using BLASTN program for identification and phylogenetic analysis of the isolates.
In vitro enzyme activities for methane degradation
Qualitative and quantitative determination of enzyme activities for two key enzymes viz. methane monooxygenase and methanol dehydrogenase involved in methane oxidation were carried out according to standard literature.
Qualitative detection of soluble methane monooxygenase
For qualitative detection of sMMO enzyme, isolates were grown on NMS media with and without CuSO4 and incubated for 72 h at 30 °C. After incubation, few naphthalene crystals were sprinkled in the lid of the plate and the plates were stored in inverted position at 30 °C for 15 min in air followed by gentle spray of freshly prepared, ortho-dianisidine dye at the rate of 5 mg ml−1, for 2–3 s. The lids were replaced and the plates were stored for 15 min in the presence of the dye. If naphthol was produced by the colonies, a purple-red colour appeared upon contact with the dye. The colour once formed, remained stable for at least 24 h at room temperature (Graham et al. 1992).
Quantitative detection of soluble methane monooxygenase and methanol dehydrogenase
Preparation of cell free extract
Cell free extract was prepared by centrifugation of cell mass for 30 min at 10,000×g washed once by suspending in 0.05 M potassium phosphate buffer (pH 7.0), the washed cells were suspended in the same buffer, homogenized and disrupted by ultrasonic treatment of 50 Hz (10 s ml−1). The suspension was than centrifuged at 10,000×g for 30 min and the resulting supernatant was used as crude extract. Proteins were determined by Lowry’s method using bovine serum albumin as standard and methanol dehydrogenase activity was assayed by the method suggested by Anthony (1971).
Soluble methane monooxygenase activity
For quantitative determination of soluble methane monooxygenase a slightly modified version of the naphthalene oxidation assay of Koh et al. (1993) was followed. Each culture was transferred in 1 ml aliquots to 10 ml screw-cap tubes and 1 ml of pre-filtered saturated naphthalene solution was added to each tube. The samples were prepared in triplicate keeping sterile medium control as blank. The reaction mixtures were incubated at 200 rpm on incubator shaker at 25 °C for 1–3 h. After incubation, 100 μl of freshly prepared 4.21 mM tetrazotized-o-dianisidine solution was added to each tube and the intensity of coloured diazo-dye complex was immediately monitored by recording the A525 by spectrophotometry. The intensity of diazo-dye formation is proportional to the naphthol concentration (1-naphthol and 2-naphthol). The specific activity of sMMO was expressed as nanomoles of naphthol formed per milligram of cell protein per minute.
Methanol dehydrogenase activity
Qualitative detection of methanol dehydrogenase was carried out following method of Eggeling and Sahm (1980) with some modifications. The enzyme assays were performed in triplicate and data obtained were subjected to statistical analysis by using completely randomized design (CRD) (Panse and Sukhatme 1978).
Results
Isolation of methylotrophic bacteria from wetland paddy
In all total 74 isolates were obtained and during screening of isolates’ for their ability to survive by utilizing methane and methanol as sole carbon source isolate M 8 (from rhizospheric soil of organically grown rice cv. IR-64 at main rice research station, Nawagam, Gujarat, India), M 10 (from rhizospheric soil of rice grown at farmers’ field of Vyara, Surat, Gujarat, India), M 17 (from rhizospheric soil of rice grown at farmers’ field of Tarapur, Gujarat, India) and M 29 (from rhizospheric soil of organically grown rice cv. GR-11 at main rice research station, Nawagam, Gujarat, India) were found to multiply in 1 % methane + water as well as 1 % methanol + water. Whereas, isolate M 14 (from rhizospheric soil of rice grown at farmers’ field of Vyara, Surat, Gujarat, India) was found to multiply in by utilizing 1 % methane and failed to multiply on methanol substrate. So inspired from the results, these five potent isolates viz. M 8, M 10, M14, M 17 and M 29 were chosen for further study. As the reference standard Methylobacterium extrorquens strain (Cat. No. MTCC 298) obtained from Microbial Type Culture Collection Center, IMTECH, Chandigarh was used as these strain is reported to possess soluble methane monooxygenase and methanol dehydrogenase activities which are involved in bacterial methane metabolism.
Survival of isolates using methane gas and methanol as sole source of carbon
Results showed that five selected isolates were found capable of utilizing methane as sole source of carbon for their growth and survival. After 10 days of inoculation, there was fourfold increase in cell numbers confirming methanotrophic nature of isolates. (Figs. 1, 10). Results pertaining to survival of isolates on methanol in the concentration range of 1–5 % are presented in Table 4. Results showed that all the isolates were capable to survive and multiply on methanol concentration up to 5 % level and found to utilize them in their metabolism showing increase in cell numbers maximum after 10 days of inoculation. Isolate M 8, M 10, M 17 and M 29 were found to efficiently utilize methanol showing six fold increase in cell numbers from initial 102–108 at 10 DAI. Whereas, isolate M 14 and standard strain M. extrorquens MTCC 298 showed four fold increase (from 102 to 106) (Fig. 2).
Characterization of isolates
Colonial characteristics of all the isolates were noted in Table 1. Isolate M 8, M 14, M 17 and M 29 were Gram +ve medium sized rods occurring singly, whereas, isolate M 10 found to be Gram −ve short rod. All the isolates showed variable response during biochemical characterization as shown in Table 1 showing that they are different from each other.
Based on the biochemical characterization of selected methylotrophic isolates, two major groups were formed (Fig. 3). First cluster comprised three isolates viz. M 8, M 14 & standard strain MTCC 298 (M. extrorquens). While group II consisted of another three methylotrophic isolates viz. M 10, M 17 & M 29. Here the biochemical characters of the isolates distinguish them from each other and represent their different metabolism by utilizing variety of substrates.
Utilization of other C1 compounds
All the isolates showed good growth on 1 % formaldehyde which is the third product of methane metabolism after methane gas and methanol. Isolate M 17 and M 29 were found to utilize 1–5 % concentration of methyl acetate. Whereas, isolates M 8, M 10 and M 14 were not able to utilize methyl acetate. Isolate M 8 and M 29 were found to utilize 1 % concentration of trichloroethylene, whereas, isolate M 14 and M 17 tolerated 1–3 % level of trichloroethylene and isolate M 10 showed good growth up to 2 % trichloroethylene concentration (Table 2). These results support the presence of methane mono oxygenase enzyme in isolates which is required for oxidation of trichloroethyle like compounds.
Molecular characterization
An about 1,500 bp fragment of the 16S rRNA gene from all five selected isolates and standard strain was further amplified using the universal primer U27f and U1492r for restriction analysis. The results of the virtual restriction, chosen three tetra cutter restriction enzymes based on better outcomes obtained, namely TaqI, HinfI and HaeIII were employed for the characterization of methylotrophic isolates. The amplified products obtained from all isolates were subjected to restriction analysis yielding total of 60 fragments (Figs. 12, 13, 14). In order to assess the existence of species, specific restriction patterns were performed for each of the enzymes utilized and have showed different and distinguished pattern of native methylotrophic isolates. Pooled analysis of ARDRA for methylotrophic isolates is represented by dendrogram (Fig. 4). Coefficient of similarity for native methylotrophic isolates M 8 and M 14 showed some similarity (19 %) (Table 3).
Isolate M 17 showed 17 % similarity with M. extrorquens (MTCC 298) strain, isolate M 17 showed 13 % similarity with Isolate M 29, isolate M 10 showed 10 % similarity with isolate M 14. Isolate M 8 and M 14 showed 4 and 5 % similarity with M. extrorquens (MTCC 298) strain respectively. Isolate M 8 showed 5 % similarity with M 10, isolate M 14 showed 4 % similarity with isolate M 29.
From 16S rRNA partial gene sequence isolate M 8 was identified as Bacillus aerius with 100 % similarity and 100 % query coverage to B. aerius strain 24 K (Fig. 5). Isolate M 10 was identified as Rhizobium sp. showing 96 % identity with R. selenitireducens strain B1with 100 % query coverage which confirms the isolate M 10 belongs to Rhizobium genus but the species was not confirmed. (Figure 6). Isolate M 14 was also identified as B. subtilis with 99 % similarity and 100 % query coverage to B. subtilis strain DSM 10 (Fig. 7). The phylogenetic tree constructed showed one major clusters showing close similarity with B. subtilis. Isolate M 17 showed about 99 % similarity and 100 % query coverage with Paenibacillus illinoisensis strain JCM 9907 indicating M 17 may be the native species of P. illinoisensis (Fig. 8). Isolate M 29 showed about 99 % similarity and 100 % query coverage with B. megaterium QM B1551 (Fig. 9). The sequence analysis of partial 16S rRNA gene of isolate M 8, M 10, M 14, M 17 and M 29 have been deposited in NCBI, GeneBank under accession numbers KC787582, KC787583, KC855269, KC787584 and KC787585 respectively. The analysis named isolate M 8 as B. aerius AAU M 8, Isolate M 10 as Rhizobium sp. AAU M 10, isolate M14 as B. subtilis AAU M 14, isolate M 17 as P. illinoisensis AAU M 17 and M 29 as B. megaterium AAU M 29.
In vitro detection of methane monooxygenase and methanol dehydrogenase enzyme activities
Chosen isolates were further subjected to qualitative and quantitative detection of two key enzymes of methane degradation pathway viz. methane monooxygenase catalysing oxidation of methane to methanol and methanol dehydrogenase catalysing oxidation of methanol to formaldehyde (Murrell 1994).
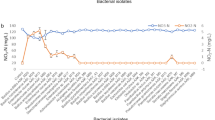
Methanotrophic colonies expressing sMMO turned deep purple when exposed successively to naphthalene and o-dianisidine. Isolate M 10, M 17, M 29 and M. extrorquens (MTCC 298) were found positive for sMMO activity in absence of copper ions in media. Moreover, when media was supplemented with CuSo4, sMMO activity was not detected (Fig. 11). The soluble methane monooxygenase activity of all the isolates ranged from 22.5 to 37.0 nmol min−1 mg of protein−1 (Table 4). Among all the isolates, P. illinoisensis showed higher soluble methane monooxygenase activity (37.0 nmol min−1 mg of protein−1) in cell free extract followed by B. megaterium (30.0 nmol min−1 mg of protein−1).
All the isolates were found positive for methanol dehydrogenase activity. Methanol dehydrogenase activity of all the isolates ranged from 35 to 75 nmol min−1 mg of protein−1 (Table 4). Among all the isolates, P. illinoisensis showed the highest methanol dehydrogenase activity (75 nmol min−1 mg of protein−1) in cell free extract followed by B. megaterium (63 nmol min−1 mg of protein−1).
Detection of methane metabolism genes
Among all the tested isolates B. aerius, Rhizobium sp., B. subtilis and P. illinoisensis showed ~518, 536, 518 and 527 bp respectively, indicating these isolates may possess pMMO enzyme responsible for capacity of methylotrophs to utilize methane (Fig. 15). Among the five tested isolates, Rhizobium sp., P. illinoisensis, B. megaterium and M. extrorquens strain gave single band of ~870 bp indicating these isolates may also have capacity to utilize methane (Fig. 16) using soluble methane monooxygenase enzyme. Overall, from the above results it was clear that isolate Rhizobium sp. and P. illinoisensis possess both sMMO and pMMO genes indicating they are better cultures for methane degradation. Similarly, Rhizobium sp. and P. illinoisensis showed single band of ~507 and ~579 bp size indicating presence of mxaF gene (Fig. 17), whereas other isolates and standard strain failed to show amplification of specific gene may be due to non-availability of selective primers to amplify methanol dehydrogenase genes from Bacillus sp.
Discussion
Isolation and screening of methylotrophic bacteria
As a source of isolation rhizospheric soils from wetland field at mid-season were selected as the chances of getting efficient methane degrading bacterial strains are maximum at this stage. Because of water logged conditions the methanogenic bacteria are producing methane at maximum rates during this time and due to abundance of favourable carbon source (methane) the methanotrophic bacteria are flourishing. Till date most of the efforts pertaining to isolate methylotrophic bacteria are concentrated on phyllospheric population but in present investigation the isolation was attempted from rhizospheric samples of wetland paddy fields by using enrichment culture technique. Hoppe et al. (2011) isolated a novel, pink-pigmented aerobic, facultatively methylotrophic bacterial strain from the phyllosphere of Funaria hygrometrica using Ammonium mineral salt (AMS) media supplemented with 0.5 % methanol as sole carbon source. Van-Aken et al. 2004 isolated Methylobacterium populi sp. nov. a novel aerobic, pink-pigmented, facultatively methylotrophic, methane-utilizing bacterium from poplar trees by enrichment of tissue culture explants in Nitrate mineral salt media supplemented with 1 % methanol as carbon source.
Characterization of isolates
All the isolates showed variable morphological and biochemical characteristics showing that they are different from each other (Fig. 3). The grouping based on the biochemical characterization supports the initial methylotrophic bacterial screening for utilization and growth in media containing methane and methanol as a sole source of carbon wherein the group II isolates M 10, M 17 & M 29 showed comparatively higher growth than group I isolates. Urakami and Komagata (1986) reported that some strains of methylotrophic bacteria can also utilize Larabinose, d-xylose, d-fucose, d-glucose, d-galactose, d-fructose, L-Aspartate, l-glutamate, adipate, Sebacate, d-tartarate, citrate, saccharte, mono-methylamine, trimethylamine, trimethylamine N-oxide, ethanolamine, butylamine, dimethylglycine and betaine, ammonia, nitrate and urea as source of nitrogen.
To further confirm the results of morphological and biochemical studies technique that is widely used for microbial diversity analysis i.e. ARDRA having potential to discriminate the bacteria at the species level was performed. Overall ARDRA results indicated that all the isolates have less than 50 % similarity and few have zero similarity showing all isolates are of different type and non-identical to each other, indicating good diversity in native methylotrophic isolates. These fact is supported by morphological and biochemical characterization studies as discussed previously. ARDRA was proved to be useful for relatedness of bacterial strains at different taxonomic levels, depending on selection of conserved or variable regions in the ribosomal genes for the analysis. It is widely used to check the clonal diversity of the isolates. This method involves amplification of the 16S rDNA region followed by digestion with one or more selected restriction enzymes (Heyndrickx et al. 1996). The sequencing of the 16S rRNA gene is another such technique used for phylogenetic placement, identification and diversity analysis of bacteria. So in present investigation ARDRA analysis and 16S rRNA analysis was carried out. Similarly, Raja et al. 2008 have carried out ARDRA profiling of amplified 16S rRNA genes digested with HaeIII enzyme, which discriminated 10 species of Methylobacterium strains isolated from phyllosphere, root nodules and internal tissues of selected plants into seven groups, but ARDRA could not easily discriminate more closely related Methylobacterium spp such as M. suomiense and M. aminovorans, M. populi and M. thyocyantum, M. fugisawaense and M. mesophilicum.
Detection of methane metabolism enzymes
All the isolates showed utilization of methane and methanol as sole carbon source. To confirm methanotrophic nature of isolates detection of enzymatic activity of highly conserved key enzymes viz. particulate pMMO, sMMO and MDH, may offer the possibility of detecting known methanotrophs (Hanson and Hanson 1996). The ability of methanotrophs to oxidize methane is due to the possession of the enzyme called methane monooxygenase (MMO). This enzyme oxidizes methane to methanol. The MMO enzyme is the subject of extensive biochemical and molecular research. There are two distinct forms of this enzyme, a membrane-bound pMMO and cytoplasmic sMMO (Hanson and Hanson, 1996). Among all the isolates, Here three isolates viz. Rhizobium sp., P. illinoisensis and B. megaterium along with standard methylotrophic strain M. extrorquens showed presence of soluble methane monooxygenase enzyme showing that the methane utilization showed by these isolates was due to presence of sMMO activity. Qualitative detection of sMMO activity was performed by naphthalene oxidation assay (Graham et al, 1992) on solid media as indicative from colonies turning to deep purple color due to development of a colored complex between 1-naphthol, formed when sMMO reacts with naphthalene and o-dianisidine (tetrazotized). Moreover, the isolates were found positive for sMMO activity in the absence of copper and in the presence of copper ions in media the isolates failed to show sMMO activity (Fig. 11). These results confirms the copper play vital role for expression of two forms of methane monooxygenase enzyme viz. sMMO present in the cytoplasm and membrane bound pMMO. In methanotrophs that have the genes for both pMMO and sMMO, their expression depends on the copper concentration in media. At high copper concentration, pMMO is expressed by incorporating Cu2+ into the active site of pMMO, while sMMO transcription in this condition is repressed by copper ions via an unknown mechanism and at low levels of copper ions in the growth medium sMMO is expressed (Kumaresan et al. 2009). Similar results were obtained by Koh et al. (1993) while working with the methylotrophic strain M. methanica 68-1 which showed higher rate of soluble methane monooxygenase activity in the absence of copper ions in media. Bowman et al. 1993 performed quantitative detection of sMMO activity by the naphthalene oxidation assay in NMS media for 25 methanotrophic bacteria obtained from groundwater samples contaminated with trichloroethylene and tetrachloroethylene and reported that most of the isolates have soluble methane monooxygenase activity in the range of <1–594 nmol h−1 mg−1 of protein.
Apart from methane and methanol, all the isolates showed utilization of formaldehyde which is the third important product in bacterial methane metabolism pathway. The reducing power required for the oxidation of methane to methanol and also for bacterial growth further derived by oxidation of methanol, via formaldehyde (HCHO) and formate (HCOOH) to finally carbon dioxide (CO2). Approximately 50 % of the formaldehyde produced is assimilated into cell carbon and the remainder is oxidized to CO2 and lost from the cells (Anthony 1982). All the isolates have also utilized trichloro ethylene which is used as an indicator molecule for detection of methane degrading enzymes within the microbial community as the first enzyme in methane metabolism pathway of bacteria i.e. methane monooxygenase is required for utilization of trichloroethylene. Aken et al. 2011 reported that M. populi sp. nov., a novel aerobic, pink-pigmented, facultatively methylotrophic, methane-utilizing bacterium isolated from poplar trees was able to utilize fructose, acetate, betadine, tartrate, ethanol methane and methylamine as carbon sources. Arfman et al. (1989) reported cells of Bacillus sp. C1 grown on methanol in batch culture oxidized methanol at a high rate (1,100–1,500 nmol min−1 mg of protein−1) only short chain (C1–C4) primary alcohols were oxidized, but compared to methanol (100 %) at lower relative rates (ethanol, 90 %; n-propanol, 57 %, n-butanol, 53 %). Secondary alcohols and formate were not oxidized. While the organism oxidized various alcohols but grew on only methanol. Rather than mere biochemical characterization studies, molecular techniques are most authenticated tools, providing significant information of species richness, diversity, and community analysis of microorganisms (Brusseau et al. 1994).
In all methylotrophic bacteria so far studied, methanol oxidation is catalyzed by MDH that catalyze the production of formaldehyde, the intermediate of both assimilative and dissimilative metabolism in methylotrophs. In methane-oxidizing bacteria (methanotrophs), MDH is the second enzyme in the methane oxidation pathway and it oxidizes the methanol produced from the oxidation of methane by methane monooxygenase (Murrell 1994). All the isolates showed methanol dehydrogenase enzyme activity which is the second key enzyme responsible for bacterial methane metabolism pathway which degrade second product of the pathway i.e. methanol. These results confirms the outcome of previous experiment wherein P. illinoisensis and B. megaterium were showing better survival at 1–5 % methanol concentration which may be due to higher rate of methanol dehydrogenase activity. Similarly, Arfman et al. (1989) reported that cell free extract of Bacillus sp. C1 were found to possess NAD dependent methanol dehydrogenase activities ranging from 1,000 to 1,200 nmol min−1 mg−1 of protein and also reported that carbon assimilation was by way of RuMP cycle of formaldehyde fixation. Duine et al. (1984) reported dye-linked methanol dehydrogenase activity from cell-free extracts of methanol-grown Nocardia sp. 239 in presence of NAD+. In the presence of NAD+ dehydrogenase activity was 33 nmol DCIP reduced min−1 mg−1 protein.
Detection of methane metabolism genes
Functional genes unique to the physiology and metabolism of organisms of interest have been explored in molecular studies. Functional genes for enzymes, such as methane monooxygenase (pMMO and sMMO; genes pmoA and mmoX, respectively) and methanol dehydrogenase (MDH; gene mxaF), involved in the methane oxidation pathway, have been used for detection of methanotrophs (McDonald et al. 2008). Gene probes that target functional genes have been developed for the pmoA gene (Sambrook et al. 1989) coding for the α-subunit of the pMMO and the mxaF gene (McDonald et al. 1996; McDonald and Murrell 1997) coding for the α-subunit of the MDH present in all methylotrophs (Hanson and Hanson 1996; McDonald et al. 1995). In present investigation all the isolates showed either presence of mmoX gene encoding the α subunit (mmoX) of the hydroxylase component of the sMMO cluster or pmoA gene encoding α-subunit of particulate methane monooxygenase cluster which are the two forms of methane mono oxygenase enzyme responsible for methane degradation. The soluble MMO is not universal to all methanotrophs and is found predominantly in the genera Methylosinus and Methylococcus (Stainthorpe et al. 1990). pmoA gene encoding one of the subunits of the particulate MMO, has also been examined recently (Holmes et al. 1995). This gene has been found in all methanotrophs studied and reported so far. The use of a second functional gene probe, mxaF, which is found in all methanotrophs including gram negative strains was recommended to confirm methylotrophic nature of isolates. The advantage of using the mxaF gene is that it can indicate larger group of organisms which assimilate C1 compounds for their role in environment. In present study, only two isolates showed presence of mxaF gene encoding α subunit of the methanol dehydrogenase, present in all methylotrophs. Though all the isolates showed methanol dehydrogenase activity, except Rhizobium sp. and P. illinoisensis, other isolates and standard strain failed to show amplification of specific gene may be due to non-availability of selective primers to amplify methanol dehydrogenase genes from Bacillus sp. Till date decoding of methane metabolism pathway in Paenibacillus species was restricted up to discovery of alcohol dehydrogenase gene (adh) from P. mucilaginosus 3016 (KEGG pathway entry no. pmq00680). Whereas, discovery of methane metabolism pathway in Bacillus species i.e. B. megaterium WSH-002 (KEGG pathway entry No. bmh00680 and B. subtilis subsp. subtilis 168 (KEGG pathway entry No. bsu00680) as well as in Rhizobium species i. e. R. etli CFN 42 (KEGG pathway entry No. ret 00680) was restricted up to the discovery of formaldehyde dehydrogenase gene which encodes enzyme responsible for oxidation of formaldehyde to CO2 for energy and assimilation for biosynthesis in methanotrophs (http://www.genome.jp/kegg/pathway/map/map01100.html). Horz et al. (2001) detected methanotrophic diversity on roots of submerged paddy using primer sets A189/A682 encoding pMMO, 534f/1393r encoding sMMO and f1003/r1561 encoding methanol dehydrogenase yielding PCR products of 550, 863 and 525 bp size respectively.
Conclusion
The results of present study clearly brought out that, all the five tested isolates from wetland paddy efficiently utilized methane and methanol as sole source of carbon and confirmed the presence of genes encoding enzymes responsible for methane degradation pathway of bacteria. Among all the isolates Rhizobium sp. and P. illinoisensis proved to be more efficient methane degraders followed by B. megaterium, B. aerius and B. subtilis. These novel indigenous methylotrophic PGPR based indicated wide scope and prospects as agriculturally beneficial bioinput to reduce methane emission from rice ecosystem with additional advantage of plant growth promotion for sustainable agriculture in a long run.
Notes
N: bases A, C, T, or G; S: bases G or C.
References
Aken B, Tehrani R, Schnoor JL (2011) Endophyte-assisted phytoremediation of explosives in poplar trees by Methylobacterium populi BJ001T. For sci 80:217–234. doi:10.1007/978-94-007-1599-8_14
Anthony C (1971) Prosthetic group of an alcohol dehydrogenase of a Pseudomonas. In: McCormick DB and Wright LD (eds) Methods in enzymology, vol XVIII. Elsevier, pp 808–809
Anthony C (1982) The biochemistry of methylotrophs. Academic Press, London
Arfman N, Watling EM, Clement W, Van Oosterwijk RJ, de Vries GE, Harder TW, Attwood MM, Dijkhuizen L (1989) Methanol metabolism in thermotolerant methylotrophic Bacillus strains involving a novel catabolic NAD-dependent methanol dehydrogenase as a key enzyme. Arch Microbiol 152:280–288
Bodrossy L, Holmes EM, Holmes AJ, Kovács KL, Murrell JC (1997) Analysis of 16S rRNA and methane monooxygenase gene sequences reveals a novel group of thermotolerant and thermophilic methanotrophs, Methylocaldum gen. nov. Arch Microbiol 168:493–503
Bowman JP, Jiménez L, Rosario I, Hazen TC, Sayler GS (1993) Characterization of the methanotrophic bacterial community present in a Trichloroethylene-contaminated subsurface groundwater site. Arch Microbiol 59(8):2380–2387
Bowman JP, McCammon SA, Skerratt JH (1997) Methylosphaera hansonii gen. nov., sp. nov., a psychrophilic, group I methanotroph from Antarctic marinesalinity, meromictic lakes. Microbiology 143:1451–1459
Brusseau GA, Bulygina ES, Hanson RS (1994) Phylogenetic analysis and development of probes for differentiating methylotrophic bacteria. Appl Environ Microbiol 60:626–636
Dedysh SN, Liesack W, Khmelenina VN, Suzina NE, Trotsenko YA, Semrau JD, Bares AM, Panikov NS, Tiedje JM (2000) Methylocella palustris gen. nov., sp. nov., a new methane-oxidizing acidophilic bacterium from peat bogs, representing a novel subtype of serine-pathway methanotrophs. Int J Syst Evol Microbiol 50:955–969
Dedysh SN, Khmelenina VN, Suzina NE, Trotsenko YA, Semrau JD, Liesack W, Tiedje JM (2002) Methylocapsa acidiphila gen. nov., sp. nov., a novel methane-oxidizing and dinitrogen-fixing acidophilic bacterium from Sphagnum bog. Int J Syst Evol Microbiol 52:251–261
Duine JA, Frank J, Berkhout MPJ (1984) NAD-dependent, PQQ-containing methanol dehydrogenase: a bacterial dehydrogenase in a multienzyme complex. FEBS Lett 168:217–221
Eggeling L, Sahm H (1980) Direct enzymatic assay for alcohol oxidase, alcohol dehydrogenase and formaldehyde dehydrogenase in colonies of Hansenula polymorpha. Appl Environ Microbiol 39(1):268–269
Graham DW, Korich DG, Leblanc RP, Sinclair NA, Arnold RG (1992) Applications of a colorimetric plate assay for soluble methane monooxygenase activity. Appl Environ Microbiol 58(7):2231–2236
Halt JG, Peter NR, Sneath HA, Staley JT, William ST (1994) Burgeys manual of detriminitave bacteriology, 9th edn. William and Wilkins, Baltimore, p 559
Hanson RS, Hanson TE (1996) Methanotrophic bacteria. Microbiol Rev 62:439–471
Heyer J, Berger U, Hardt M, Dunfield PF (2005) Methylohalobius crimeensis gen. nov., sp. nov., a moderately halophilic, methanotrophic bacterium isolated from hypersaline lakes of Crimea. Int J Syst Evol Microbiol 55:1817–1826
Heyndrickx M, Vauterin L, Vandamme P, Kersters K, De Vos P (1996) Applicability of combined amplified 16S rDNA restriction analysis (ARDRA) patterns in bacterial phylogeny and taxonomy. J Microbiol Meth 26:247–259
Holmes AJ, Costello A, Lidstrom ME, Murrell JC (1995) Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol Lett 132:203–208
Hoppe T, Peters K, Schmidt F (2011) Methylobacterium bullatum sp. nov., a methylotrophic bacterium isolated from Funaria hygrometrica. Syst Appl Microbiol 34:482–486. doi:10.1016/j.syapm.2010.12.005
Horz H, Yimga MT, Liesack W (2001) Detection of methanotroph diversity on roots of submerged rice plants by molecular retrieval of pmoA, mmoX, mxaF, and 16S rRNA and ribosomal DNA, including pmoA -based terminal restriction fragment length polymorphism profiling. Appl Environ Microbiol 67(9):4177–4185
Intergovernmental Panel on Climate Change (IPCC) (2007) Climate change, Synthesis Report. An assessment of the intergovernmental panel on climate change. Working group contributions to the fourth assessment report
Jaccard P (1912) The distribution of the flora in the alpine zone. New Phytol 11(2):37–50
Khosa MK, Sidhu BS, Benbi DK (2011) Methane emission from rice fields in relation to management of irrigation water. J Environ Biol 32:169–172
Koh S, Bowman JP, Sayler GS (1993) Soluble methane monooxygenase production and trichloroethylene degradation by a type I methanotroph, Methylomonas methanica 68-1. Appl Environ Microbiol 59(4):960–967
Kumaresan D, Abell GCJ, Bodrossy L, Stralis-Pavese N, Murrell JC (2009) Spatial and temporal diversity of methanotrophs in a landfill cover soil are related to soil abiotic factors. Environ Microbiol Rep 1:398–407
McDonald IR, Murrell JC (1997) The methanol dehydrogenase structural gene mxaF and its use as a functional gene probe for methanotrophs and methylotrophs. Appl Environ Microbiol 63:3218–3224
McDonald IR, Kenna EM, Murrell JC (1995) Detection of methanotrophic bacteria in environmental samples with the PCR. Appl Environ Microbiol 61:116–121
McDonald IR, Hall GH, Pickup RW, Murrell JC (1996) Methane oxidation potential and preliminary analysis of methanotrophs in blanket bog peat using molecular ecology techniques. FEMS Microbiol Ecol 21:197–211
McDonald IR, Bodrossy L, Chen Y, Murrell JC (2008) Molecular ecology techniques for the study of aerobic methanotrophs. Appl Environ Microbiol 74:1305–1315
Murrell JC (1994) Molecular genetics of methane oxidation. Biodegradation 5:145–159
Nei M, Li WH (1979) Mathematical models for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA 76:5269–5273
Panse VG and Sukhatme PV (1978) Statistical methods for agricultural workers, ICAR II Ed, New Delhi
Raja P, Balachandar D, Sundaram SP (2008) PCR finger printing for identification and discrimination of plant associated facultative methylobacteria. Ind J Biotechnol 7:508–514
Reeburgh WS, Whjalen SC, Alperin MJ (1993) The role of methylotrophy in the global methane budget. In: Murrell JC, Kelly DP (eds) Microbial Growth on C-1 Compounds Intercept, Ltd, Andover, pp 123–140
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory, Cold Spring Harbor
Stainthorpe AC, Salmond GP, Dalton CH, Murrell JC (1990) Screening of obligate methanotrophs for soluble methane monooxygenase genes. FEMS Microbiol Lett 72:345–348
Tsubota J, Eshinimaev BT, Khmelenina VN, Trotsenko YA (2005) Methylothermus thermalis gen. nov., sp. nov., a novel moderately thermophilic obligate methanotroph from a hot spring in Japan. Int J Syst Evol Microbiol 55:1877–1884
Urakami T, Komagata K (1986) Emendation of Methylobacillus Yordy a genus for methanol-utilizing bacteria. Int J Syst Bacteriol 36:502–511
Van-Aken B, Peres CM, Doty SL, Yoon JM, Schnoor JL (2004) Methylobacterium populi sp. nov., a novel aerobic, pink-pigmented, facultatively methylotrophic, methane-utilizing bacterium isolated from poplar trees (Populus deltoides x nigra DN34). Int J Syst Evol Microbiol 54:1191–1196. doi:10.1099/ijs.0.02796-0
Wahlen M, Tanaka N, Henry R, Deck B, Zeglen J, Vogel JS, Southon J, Shemesh A, Fairbanks R, Broecker W (1989) Carbon-14 in methane sources and in atmospheric methane: the contribution from fossil carbon. Science 245:286–290
Waturangi DE, Francisca I, Susanto CO (2011) Genetic diversity of methylotrophic bacteria from human mouth based on amplified ribosomal DNA restriction analysis (ARDRA). Hay J of Biosci 18(2):77–81. doi:10.4308/hjb.18.2.77
Whittenbury R, Phillips KC, Wilkinson JF (1970) Enrichment, isolation and some properties of methane-utilizing bacteria. J Gen Microbiol 61:205–218
Wise MG, McArthur JV, Shimkets LJ (2001) Methylosarcina fibrata gen. nov., sp. nov. and Methylosarcina quisquiliarum sp. nov., novel type I methanotrophs. Int J Syst Evol Microbiol 51:611–621
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jhala, Y.K., Vyas, R.V., Shelat, H.N. et al. Isolation and characterization of methane utilizing bacteria from wetland paddy ecosystem. World J Microbiol Biotechnol 30, 1845–1860 (2014). https://doi.org/10.1007/s11274-014-1606-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11274-014-1606-3