Abstract
Methanotrophs of the genus Methylocystis are frequently found in rice paddies. Although more than ten facultative methanotrophs have been reported since 2005, none of these strains was isolated from paddy soil. Here, a facultative methane-oxidizing bacterium, Methylocystis iwaonis SD4, was isolated and characterized from rhizosphere samples of rice plants in Nanjing, China. This strain grew well on methane or methanol but was able to grow slowly using acetate or ethanol. Moreover, strain SD4 showed sustained growth at low concentrations of methane (100 and 500 ppmv). M. iwaonis SD4 could utilize diverse nitrogen sources, including nitrate, urea, ammonium as well as dinitrogen. Strain SD4 possessed genes encoding both the particulate methane monooxygenase and the soluble methane monooxygenase. Simple and rapid genetic manipulation methods were established for this strain, enabling vector transformation and unmarked genetic manipulation. Fast growth rate and efficient genetic tools make M. iwaonis SD4 an ideal model to study facultative methanotrophs, and the ability to grow on low concentration of methane implies its potential in methane removal.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Methane is the second most prevalent greenhouse gas after carbon dioxide and has a significant impact on global climate change (Knief 2019). Currently, the average concentration of methane in the atmosphere is above 1.8 ppmv and is increasing year by year (Stavert et al. 2022). Methanotrophs are the bacteria that can grow on methane as their sole source of carbon and energy (Whittenbury et al. 1970). These bacteria act as a natural biofilter of methane in diverse methanogenic environments and play a crucial role in global methane cycle. Methanotrophic bacteria are found in three phyla: Proteobacteria, Verrucomicrobia, and candidate phylum NC10 (Guerrero-Cruz et al. 2021). Traditionally, it was believed that aerobic methanotrophs could only use C1 compounds like methane and methanol for growth. However, in 2005, Dedysh et al. demonstrated that Methylocella species are facultatively methanotrophic. They found that three Methylocella strains (Methylocella palustris K, M. silvestris BL2, and M. tundrae T4) can use some multi-carbon compounds such as acetate, pyruvate, succinate, malate, and ethanol for growth in addition to methane and other C1 compounds. This study provided definitive evidence for the existence of facultative methanotrophs (Dedysh et al. 2000, 2004, 2005; Dunfield et al. 2003). Notably, Methylocella strains are different from almost all other methanotrophic bacteria in possessing only soluble methane monooxygenase (sMMO) rather than particulate methane monooxygenase (pMMO) (Farhan Ul Haque et al. 2020).
Shortly after the discovery of facultative methanotrophy in Methylocella, approximately ten facultative methanotrophs have been isolated. For example, Dunfield et al. found that Methylocapsa aurea isolated from forest soil could use acetate as the sole growth substrate (Dunfield et al. 2010). Dedysh and Belova et al. reported that some members of the Methylocystis genus, including M. bryophila H2 (Dedysh et al. 2007), M. heyeri H2s (Belova et al. 2011), and M. bryophila S284 (Belova et al. 2013) were able to facultatively grow on acetate; Im et al. characterized a novel facultative Methylocystis sp. strain SB2 capable of growth on methane as well as acetate or ethanol (Im et al. 2011). Compared to the diverse substrates of facultative Methylocella, facultative Methylocapsa aurea and Methylocystis can only grow on one or two kind of multi-carbon compounds (acetate and ethanol). Moreover, the preferable carbon source for facultative Methylocella is acetate (Dedysh et al. 2005), while facultative Methylocapsa aurea and Methylocystis grow better on methane (Dedysh et al. 2019; Farhan Ul Haque et al. 2020; Semrau et al. 2011). In addition to the facultative methanotrophs from Proteobacteria, methanotrophs from Verrucomicrobia have been found to grow using hydrogen and carbon dioxide via Knallgas reaction in the absence of methane (Carere et al. 2017; Mohammadi et al. 2017).
Paddy fields are a major contributor to methane emissions, accounting for approximately 10% to 15% of global methane emissions. Most of the methane generated in rice fields is consumed by methane-oxidizing bacteria, with only around 15% to 30% being released to the atmosphere (Curry 2007). Methanotrophic bacteria play a crucial role in reducing methane emissions from paddy fields (Lee et al. 2014). Many previous studies show that the methanotrophs from Methylocystis genus are generally abundant in paddy soil (Bao et al. 2014; Cao et al. 2022; Hara et al. 2022; Ishii et al. 2011). These methanotrophs have the ability to colonize the surface and interior of rice roots, conducting methane oxidation and potentially promoting rice growth (Bao et al. 2014). Notably, in 2015, Leng et al. reported that some uncultivated Methylocystis strains in paddy soil could utilize acetate as a secondary carbon source (Leng et al. 2015). Considering the facultative methanotrophy in several Methylocystis species as well as the common existence of acetate in paddy soil, it is reasonable that acetate-utilizing Methylocystis strains are widely distributed in paddy soil. However, no pure culture of facultative methanotroph has been isolated from paddy soil so far. Additionally, how facultative Methylocystis strains use acetate or ethanol for growth is still far from clear, and current knowledges are mainly obtained from genomic and transcriptomic analyses (Vorobev et al. 2014).
In this study, a facultative methanotroph Methylocystis iwaonis SD4 was isolated and characterized from the rhizosphere of rice plants. This strain exhibited high growth rate on methane and could also grow slowly by utilizing acetate or ethanol as alternative substrate. Furthermore, an efficient genetic manipulation method was developed for M. iwaonis SD4, facilitating plasmid transformation as well as unmarked gene knock-out and knock-in. These results make M. iwaonis SD4 as a desired material to investigate how facultative Methylocystis utilize acetate and how they thrive in paddy soil.
Materials and methods
Isolation and cultivation of methanotrophs
The rice samples were collected from the Baima experimental field of Nanjing Agricultural University in Nanjing City, Jiangsu Province, China (31°36′N, 119°10′E). This field has been consistently used for rice cultivation. According to previous method (Zhang et al. 2019), rice root samples (Nipponbare) with soil attached were washed to remove excess soil from the surface, followed by three washes in a phosphate buffered solution (PBS). The last wash solution was collected as the rice rhizosphere soil sample. The rhizosphere soil samples were inoculated in 100 ml glass serum bottles containing 20 ml of nitrate mineral salts (NMS) medium (Dedysh et al. 2014) at a 5% inoculum ratio. The bottles were sealed with butyl rubber stoppers and aluminum seals (Bussmann et al. 2006). Shaking cultures were conducted at 30 ℃ in the presence of 20% (v/v) methane. After 5 days of incubation, 2% of the enriched broth was transferred to 20 ml of fresh NMS medium for further enrichment cultivation (Zhu et al. 2020). After three transfers, the enriched cultures were diluted and plated onto NMS plates. These plates were subsequently incubated in a jar at 30 ℃ with 20% v/v methane for 7 days. Individual colonies were selected and repeatedly sub-cultured in order to obtain pure culture isolates. Strain SD4 was deposited in Jiangsu Province Agricultural Culture Collection Center of China (JSACC) under accession No. of JSACC32501.
Morphological and biochemical characterization
The strains were cultured on NMS plates for the observation of colony morphology characteristics. Cells grown on NMS plates for 2 days were collected for observing cell morphology, size, and flagella using transmission electron microscopy. (HT7700, Hitachi, Japan). Similarly, cells cultured in NMS liquid medium were subjected to centrifugation, fixation, thin-sectioning, staining, and observation using transmission electron microscopy (H-7650, Hitachi, Japan) to study cell ultrastructure (Belova et al. 2011).
The effects of temperature and initial pH on the growth of strain SD4 were tested in liquid NMS medium. The sensitivity of strain SD4 to chloramphenicol, bleomycin, streptomycin, spectinomycin, kanamycin, gentamicin, erythromycin, ampicillin, and tetracycline was also assessed in liquid NMS medium. The activity of sMMO was detected using the naphthalene oxidation assay with the cells grown under copper-limited condition (Graham et al. 1992).
Phylogenetic and genomic analysis
Genomic DNA was extracted from fresh cultures using a Rapid Bacterial Genomic DNA Isolation Kit (Sangon Biotech, Shanghai, China). The gene of 16S rRNA was PCR amplified with primer pair 27f/1492r (Weisburg et al. 1991). Phylogenetic trees were reconstructed using the neighbor-joining (NJ) method. The MEGA software was used for tree reconstruction with 1000 bootstraps (Tamura et al. 2013). The genome of strain SD4 was sequenced using a combination of PacBio RS and Illumina NovaSeq 6000 sequencing platforms at Shanghai BIOZERON Co., Ltd (Shanghai, China) The genome was assembled using ABySS (http://www.bcgsc.ca/platform/bioinfo/software/abyss) with multiple-Kmer parameters. The GC content of strain SD4 was calculated based on the whole genome sequence. Average nucleotide identity (ANI) was calculated between the genomes using ANI calculator (https://www.ezbiocloud.net/tools/ani) (Lee et al. 2016). Digital DNA-DNA hybridization (dDDH) was calculated using the Genome-to-Genome Distance Calculator (http://ggdc.dsmz.de/ggdc.php) (Goris et al. 2007).
Growth of strain SD4 on different carbon and nitrogen sources
The range of potential growth substrates was investigated using liquid medium NMS by adding 0.1% (w/v for solid compounds and v/v for liquid compounds) filter-sterilized carbon sources. These included formate, methylamine, ethanol, sodium acetate, pyruvate, succinate, citrate, malate, urea, glucose, sucrose, and glycerol. Methanol was assessed as the sole carbon source at concentrations ranging from 0.05 to 2% (v/v) in liquid NMS medium. Additionally, the growth of strain SD4 in liquid NMS medium under low methane concentrations of 100 ppmv, 500 ppmv and 1.9 ppmv (atmospheric methane) was tested according to the method established by He et al. with the headspace in each bottle refreshed daily (He et al. 2023).
The nitrogen sources for the growth of strain SD4 were examined in nitrogen-free liquid NMS medium supplemented with 0.1% (w/v) filter-sterilized compounds, including nitrate, nitrite, urea, ammonium, glycine, alanine, serine, glutamine, lysine, arginine, asparagine, aspartate and yeast extract. The strain was also incubated in nitrogen-free liquid medium at varying oxygen levels of 2%, 4%, 10%, and the normal level of 21% to evaluate its nitrogen fixing ability (Dam et al. 2013).
Electroporation of strain SD4
Electroporation of strain SD4 was carried out according to a previous protocol with some modification (Yan et al. 2016). Briefly, a full loop of cell biomass was evenly spread on an NMS1 plate and grown at 30 °C in a sealed jar containing 20% methane for 2 days; the cells were collected using inoculating loop and washed twice with deionized water (50 mL each time); the cells were finally concentrated in 100 μl deionized water; 400–600 ng DNA fragment (less than 5 μl) was mixed with competent cells (50 μl), transferred to a 1-mm-gap cuvette (Bio-Rad) and placed on ice for 5 min; electroporation was performed using a Gene Pulser II system (Bio-Rad) set at 1500 V, 25 μF and 150 Ω; immediately after electroporation, 1 mL of NMS liquid medium (30 °C) was added to the cells; the resuspended cells were transferred to 10 mL of NMS medium in a 100 mL glass bottle, which was then added with 20% (V/V) methane and incubated at 30 °C with shaking at 180 rpm for 12 h; 1 mL of the cells culture was centrifuged at 5000 rpm for 3 min to collect cells, which were then spread on NMS plates containing kanamycin (20 mg/l) and cultured in the presence of methane at 30 °C for about 7 days.
Gene knockout and knockin
To delete mmoX through marker exchange method, the left flanking fragment of mmoX (LF), kanamycin resistance marker (Kanr) and the right flanking fragment of mmoX (RF)were assembled in the order of LF, Kanr and RF via overlap extension PCR (Shevchuk et al. 2004). The fused fragment LF-Kanr-RF was directly electroporated into strain SD4 and selected on the NMS plate supplemented with kanamycin. The target transformant was verified by PCR amplification and sMMO activity assay (Graham et al. 1992). The mutant strain was named SD4-XK. The insertion of the fragment Ptac-dTomato at mmoX site was performed similarly, generating mutant SD4-XKT. The fragments of Ptac-dTomato and Kanr were amplified from plasmid pAWP89 (Puri et al. 2015). LF and RF were amplified from the genome of strain SD4. These four fragments were fused by PCR to generate fragment LF-Kanr-Ptac-dTomato-RF, which was electroporated into strain SD4 and selected by kanamycin. The target transformant was verified by PCR amplification and fluorescence microscopy. The primers used in this work are listed in Table S1.
pheS.AG based unmarked mutagenesis in strain SD4
Two mutations (T258A and A311G) were introduced into the pheS gene of strain SD4 via overlap extension PCR, generating pheSAG; then, the native promoter and ribosome binding site (RBS) of pheSAG were replaced with promoter Ptac and the RBS of mmoX, resulting in the fragment Ptac-RBSmmox-pheSAG; three fragments of Ptac-RBSmmox-pheSAG, the RBS of lacZ and the coding region of Kanr were assembled via PCR to give a operon PK in which the transcription of pheSAG and kanamycin resistance gene was controlled by Ptac (Liu et al. 2020). To perform unmarked deletion of mmoY, the LF of mmoY, a direct repeat fragment (DR), PK, and the upstream fragment of mmoY (RF) were separately amplified and then assembled in the order of LF, DR, PK and RF using overlap extension PCR. The mmoY deletion amplicon was electroporated into SD4 strain and selected by kanamycin. Verification was performed through PCR amplification. A positive transformant was streaked onto NMS agar plates and incubated at 30 °C for two days; then the cells were collected and spread onto NMS plates containing 0.5 mM p-chloro-phenylalanine (p-Cl-Phe) (Sigma-Aldrich, product No. C6506). Verification of the mmoY deletion was done through PCR amplification and sMMO activity assay. The mutant strain was named SD4-∆Y.
Results
Isolation of strain SD4
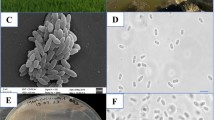
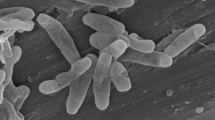
Individual colonies of methanotrophic bacteria obtained from rice rhizosphere soil were checked for purity and tested for the ability to grow on acetate. A strain named SD4 exhibited obvious growth on liquid NMS medium supplemented with acetate. On agar plates, this strain produced small (diameter 0.5–2 mm), circular, raised, smooth, opaque, and complete-edged creamy-colored colonies (Fig. 1A). The cells were Gram-negative and short rods with a polar flagellum (Fig. 1B). When grown on methane, the cells contained a well-developed system of type II intracytoplasmic membranes (ICM), which were aligned parallel to the cytoplasmic membrane (Fig. 1C). When grown on acetate, these ICM were maintained, albeit in a reduced form (Fig. 1D). In addition, inclusion bodies were observed within the cells.
Morphological characteristics of strain SD4. (A) Colony morphology of strain SD4 cells grown on NMS plate with methane. (B) Transmission electron micrograph of negatively stained cells of strain SD4 grown on NMS agar plate with methane. Bar, 1 µm. (C and D) Electron micrographs of ultrathin sections of strain SD4 cells grown in liquid media with methane and acetate as carbon sources. Bar, 0.2 µm
Strain SD4 exhibited an optimal growth temperature of 30 °C and an optimal pH of 7.0 when grown on methane (Fig. S1). The cells of strain SD4 grown in copper-limited condition showed sMMO activity. Additionally, strain SD4 was sensitive to kanamycin (15 mg/l), ampicillin (15 mg/l), and gentamicin (30 mg/l), but resistant to chloramphenicol (5 mg/l), bleomycin (20 mg/l), streptomycin (50 mg/l), spectinomycin (50 mg/l), erythromycin (10 mg/l), and tetracycline (10 mg/l).
Phylogenetic and genotypic characteristics
Phylogenetic analysis of the 16S rRNA genes sequences showed that strain SD4 belongs to the family Methylocystaceae in the class Alphaproteobacteria (Fig. 2). The closest taxonomically described methanotroph to strain SD4 was Methylocystis iwaonis SS37A-ReT (99.86% 16S rRNA gene sequence identity) (Kaise et al. 2023). The other close strains were Methylocystis parvus OBBPT (98.86%), Methylosinus sporium ATCC 35069T (98.37%), Methylosinus pucelana MTS (98.01%), Methylosinus trichosporium OB3bT (97.66%), Methylocystis bryophila H2sT (97.41%) and Methylocystis bryophila S284 (97.36%).
The genome of strain SD4 (GenBank accession No. CP143314) comprises a single circular chromosome and four plasmids, with a chromosome length of 3,705,154 bp. The sMMO operon is present in one copy, while the pMMO operon has three copies (VN910_RS02755 to VN910_RS02765, VN910_RS05405 to VN910_RS05415, VN910_RS07985 to VN910_RS07995). One of these three copies (VN910_RS05405 to VN910_RS05415) of the pMMO operon encodes pMMO2, which is responsible for high-affinity methane oxidation in Methylocystis species (Baani and Liesack 2008). Comparative assessment of the average nucleotide identity (ANI) and digital DNA-DNA hybridization (dDDH) values were employed to confirm the taxonomic position of strain SD4 (Table S1). The ANI and dDDH values between strains SD4 and SS37A-ReT are 96.55% and 71.10%, respectively. By contrast, the ANI and dDDH values between strain SD4 and other similar strains ranged from 74.81 to 81.07% and 21.8 to 25.10%, respectively, which were significantly lower than the proposed species thresholds (ANI, 95–96% and dDDH, 70%). Additionally, the DNA G + C content of strain SD4 (63.3 mol%) is similar to that of strain SS37A-ReT (63.2 mol%). Therefore, strain SD4 was identified as a member of Methylocystis iwaonis.
Growth of strain SD4 on different carbon sources
M. iwaonis SD4 was able to grow on methane, methanol, acetate or ethanol (Fig. 3A), while no growth was detected in the presence of formate, methylamine, pyruvate, succinate, citrate, malate, urea, glucose, sucrose or glycerol. Highest growth rate was observed using methane as carbon source. When cultured in 50 mL liquid NMS in 250 mL bottle containing 20% (v/v) methane at 30 °C and 180 rpm, strain SD4 achieved a maximum OD600 of 0.72 within 32 h, with a specific growth rate of 0.1155 h−1 (equivalent to a doubling time of 6 h) (Fig. 3A). Methanol was the second preferable carbon source for this strain. However, growth was sustained only when the methanol concentration was below 2% (v/v). The optimal concentrations of methanol for strain SD4 growth were 0.1–0.2% (Fig. 3B). Significantly slower growth was found with acetate or ethanol as carbon source (Fig. 3A). Specifically, the maximum OD600 of strain SD4 in acetate-supplemented medium reached 0.24 after 9–10 days, and the maximum OD600 was 0.14 after 5–6 days in ethanol-supplemented medium.
Growth of strain SD4 on different carbon sources. (A) Growth curve of SD4 on methane, methanol, acetate, ethanol as the sole energy and carbon source. (B) Growth curve of SD4 with different concentrations of methanol. (C) The time of resume exponential growth of strain SD4 translocation from acetate to methane. In the treatment of control, the cells were kept for the same period of time in NMS medium without any carbon source. (D) The growth of strain SD4 at low concentrations of methane. All data points are means ± SD of three separate experiments
We further examined whether acetate utilization allows strain SD4 to maintain the methane oxidation machinery. As shown in Fig. 3C, the cells harvested from acetate-supplemented medium could resume their exponential growth after a lag-phase of about 65 h after transfer to a fresh methane-supplemented medium. In the control treatment, the cells were kept for the same period of time in NMS medium without any carbon source and then these cells were collected and transferred to methane medium; exponential growth of these cells was observed after approximately 80 h. In comparison, if the cells were precultured on methane, they rapidly entered exponential growth after transfer to methane medium.
Since rice paddy effluents contains elevated methane in the 500 ppmv range (He et al. 2023), we then tested the growth of strain SD4 at low concentration of methane. As shown in Fig. 3D, strain SD4 demonstrated the ability to grow on methane in the range of concentrations between 100 and 500 ppmv, with maximum OD600 of 0.08 and 0.13, respectively. In contrast, no trace growth was detected under atmospheric methane condition.
Growth of strain SD4 on different nitrogen sources
Growth of strain SD4 on different nitrogen sources was investigated in liquid medium with 20% (v/v) methane. Strain SD4 was able to thrive on diverse nitrogen sources, including nitrate, urea, ammonium (NH4Cl), yeast extract, glycine, or alanine. No growth was observed in the addition of nitrite, serine, glutamine, lysine, arginine, asparagine or aspartate. Furthermore, strain SD4 exhibited the ability to fix N2 (Fig. S2). However, this ability was affected by O2 concentration. The maximum OD600 of strain SD4 could reach 0.24 under 2% O2 (v/v) condition, while no obvious growth was observed under atmospheric oxygen concentration (21%), suggesting the nitrogenase of strain SD4 was sensitive to oxygen.
Development of genetic tools for strain SD4
As described in Material and Method, a simple and rapid electroporation protocol was established for strain SD4 according to a previous method developed for type I methanotrophs. The broad-host-range vector pAWP89 extracted from E. coli was electroporated into strain SD4 at a frequence of about 9 × 106 CFU/μg DNA. Based on this high transformation efficiency, we attempted to perform gene knockout through marker exchange. As shown in Fig. 4A, the deletion construction that contained flanking region of mmoX (about 800 bp each side) and antibiotic resistant marker were fused through PCR and then electroporated into strain SD4. Approximately 2 × 104 transformants were obtained per μg deletion construction. Ten transformants were randomly selected and verified through PCR and sMMO activity assay, which showed that all tested transformants were positive (Fig. 4B). Similarly, a foreign gene dTomato for red fluorescence protein was inserted at mmoX site (Fig. 4C).
Gene knockout and knockin in strain SD4. (A) Flowchart of gene knockout through marker exchange. (B) The deletion of mmoX was validated via PCR amplification and sMMO activity assay. (C) The insertion of red fluorescence protein gene at mmoX site was confirmed by PCR amplification and fluorescence microscopy
Considering that an ideal genetic manipulation should not leave antibiotic resistant marker in the host, we sought to create a counter-selectable marker to rescue antibiotic resistance marker. Since pheS encoding the α-subunit of phenylalanyl-tRNA synthetase has been successfully developed as a counter-selectable marker in diverse bacteria (Ishikawa et al. 2018; Miyazaki 2015), the pheS of strain SD4 was modified to generate pheSAG through introducing two-point mutations (T252A and A306G). Theoretically, PheSAG could recognize p-Cl-Phe as a substrate and incorporated p-Cl-Phe into proteins, making the proteins inactive; Therefore, expression of pheSAG in a cell will lead to cell death in the presence of p-Cl-Phe. An unmarked gene knockout method was used according to previous reports (Fig. 5A, B), in which integration of foreign fragment into chromosome was selected by antibiotic resistant marker (Kanr) and elimination of Kanr and pheSAG was selected by p-Cl-Phe. As expected, the cells harboring pheSAG were very sensitive to p-Cl-Phe (0.5 mM) (Fig. 5C). Over 90% colonies grown on p-Cl-Phe plate were positive based on PCR amplification and sMMO activity assay (Fig. 5D).
Counter-selective marker pheSAG based unmarked gene deletion in strain SD4. (A) The construction of PK cassette. The pheSAG was fused with kanamycin resistance gene to construct a PK cassette. (B) PK cassette based markerless deletion strategy. To delete a target fragment (dotted line), a direct repeat of the downstream of deletion region (DR, ∼450-bp) and PK cassette were inserted upstream of the deletion region by electroporation and selected by kanamycin. Recombination deletion of PK cassette and the target fragment was selected by 0.5 mM p-Cl-Phe. (C) The sensitivity of strains SD4 and SD4-PZ toward p-Cl-Phe. Cell cultures were serially diluted 1:10. Each serial dilution was spotted onto agar plates containing p-Cl-Phe at different concentrations. (D) Confirmation of mmoY deletion by PCR amplification and sMMO activity assay
Discussion
In paddy soil, anaerobic decomposition of organic matter usually generates significant amount of acetate, with concentration as high as several millimolar, especially when rice straw and root is anaerobically degraded (Conrad 2007; Rui et al. 2009). Previous study suggested the existence of uncultivated facultative Methylocystis in paddy soil (Leng et al. 2015), which is confirmed by our work. Since methane availability is variable in paddy soil, acetate utilization can be a survival strategy of facultative Methylocystis, an advantage over obligate methanotrophs. This may partially explain why Methylocystis can thrive in paddy soil. In addition, we predict that paddy soil also hosts the facultative methanotrophs from other genera, such as Methylocella and Methylocapsa.
Among the facultative methanotrophs documented so far, five are Methylocella, seven come from Methylocystis and two come from Methylocapsa (Table 1). These facultative methanotrophs are classified into two types by Dedysh et al. based on the range of carbon source: the champion of facultative methanotrophs (Methylocella) and the limited facultative methanotrophs (Methylocapsa and Methylocystis) (Dedysh et al. 2019). Facultative Methylocella have more diverse substrates than that of facultative Methylocapsa and Methylocystis. Notably, Methylocella only harbor sMMO and use acetate rather methane as preferable carbon source (Dedysh et al. 2005), while all of the facultative Methylocapsa and Methylocystis contain pMMO and exhibit a clear preference for growth on methane. Among eight limited facultative methanotrophs seven strains are Methylocystis, implying this genus has abundant facultative strains. However, the results of Belova et al. demonstrated that not all Methylocystis are facultative (Belova et al. 2011).
Compared to other facultative Methylocystis, M. iwaonis SD4 contained both pMMO and sMMO, could utilize both acetate and ethanol, and showed N2-fixing ability and exhibited fast growth rate; these features make strain SD4 a good candidate to study facultative Methylocystis. Some crucial knowledge gaps still exist in facultative Methylocystis. For example, the genetic and biochemical background of facultative growth remains unclear; their survival strategy and behavior in paddy soil should be investigated in the natural environment; it is also interesting to explore whether they can interact with rice. Genetic manipulation tools are not available for facultative Methylocystis,which is a bottleneck in filling these knowledge gaps. The genetic methods established in this work will accelerate the study of facultative Methylocystis species.
Conclusion
We isolated and characterized a facultative methanotrophic bacterium M. iwaonis SD4 from rice rhizosphere. This strain is capable of utilizing acetate and ethanol as sole carbon and energy sources for growth. Rapid and simple genetic tools were developed for this strain, making M. iwaonis SD4 a promising model for studying facultative Methylocystis species.
References
Baani M, Liesack W (2008) Two isozymes of particulate methane monooxygenase with different methane oxidation kinetics are found in Methylocystis sp. strain SC2. Proc Natl Acad Sci 105:10203–10208. https://doi.org/10.1073/pnas.0702643105
Bao Z, Okubo T, Kubota K, Kasahara Y, Tsurumaru H, Anda M, Ikeda S, Minamisawa K (2014) Metaproteomic identification of diazotrophic methanotrophs and their localization in root tissues of field-grown rice plants. Appl Environ 80:5043–5052. https://doi.org/10.1128/AEM.00969-14
Belova SE, Baani M, Suzina NE, Bodelier PL, Liesack W, Dedysh SN (2011) Acetate utilization as a survival strategy of peat-inhabiting Methylocystis spp. Environ Microbiol 3:36–46. https://doi.org/10.1111/j.1758-2229.2010.00180.x
Belova SE, Kulichevskaya IS, Bodelier PL, Dedysh SN (2013) Methylocystis bryophila sp. nov., a facultatively methanotrophic bacterium from acidic Sphagnum peat, and emended description of the genus Methylocystis (ex Whittenbury, et al 2013 1970) Bowman et al. 1993. Int J Syst Evol Microbiol 63:1096–1104. https://doi.org/10.1099/ijs.0.043505-0
Bussmann I, Rahalkar M, Schink B (2006) Cultivation of methanotrophic bacteria in opposing gradients of methane and oxygen. FEMS Microbiol Ecol 56:331–344. https://doi.org/10.1111/j.1574-6941.2006.00076.x
Cao W, Cai Y, Bao Z, Wang S, Yan X, Jia Z (2022) Methanotrophy alleviates nitrogen constraint of carbon turnover by rice root-associated microbiomes. Front Microbiol 13:885087. https://doi.org/10.3389/fmicb.2022.885087
Carere CR, Hards K, Houghton KM, Power JF et al (2017) Mixotrophy drives niche expansion of verrucomicrobial methanotrophs. ISME J 11:2599–2610. https://doi.org/10.1038/ismej.2017.112
Conrad R (2007) Microbial ecology of methanogens and methanotrophs. Adv Agron 96:1–63. https://doi.org/10.1016/S0065-2113(07)96005-8
Curry CL (2007) Modeling the soil consumption of atmospheric methane at the global scale. Global Biogeochem Cycles 404:35–342. https://doi.org/10.1029/2006GB002818
Dam B, Dam S, Blom J, Liesack W (2013) Genome analysis coupled with physiological studies reveals a diverse nitrogen metabolism in Methylocystis sp. strain SC2. PLoS One 8:e74767. https://doi.org/10.1371/journal.pone.0074767
Danilova OV, Oshkin IY, Belova SE, Miroshnikov KK, Ivanova AA, Dedysh SN (2023) One step closer to enigmatic USCα methanotrophs: isolation of a Methylocapsa-like bacterium from a subarctic soil. Microorganisms. 11:2800. https://doi.org/10.3390/microorganisms11112800
Dedysh SN, Dunfield PF (2014) Cultivation of methanotrophs. In: McGenity T, Timmis K, Nogales B (eds) Hydrocarbon and lipid microbiology protocols. Springer, Berlin, pp 231–247
Dedysh SN, Dunfield PF (2019) Facultative methane oxidizers. In: McGenity T (ed) Genomics and ecophysiology of hydrocarbon-degrading microbes. Springer, Cham, pp 279–297
Dedysh SN, Liesack W, Khmelenina VN, Suzina NE, Trotsenko YA, Semrau JD, Bares AM, Panikov NS, Tiedje JM (2000) Methylocella palustris gen. nov., sp. nov., a new methane-oxidizing acidophilic bacterium from peat bogs, representing a novel subtype of serine-pathway methanotrophs. Int J Syst Evol Microbiol 50:955–969. https://doi.org/10.1099/00207713-50-3-955
Dedysh SN, Berestovskaya YY, Vasylieva LV, Belova SE, Khmelenina VN, Suzina NE, Trotsenko YA, Liesack W, Zavarzin GA (2004) Methylocella tundrae sp. nov., a novel methanotrophic bacterium from acidic tundra peatlands. Int J Syst Evol Microbiol 54:151–156. https://doi.org/10.1099/ijs.0.02805-0
Dedysh SN, Knief C, Dunfield PF (2005) Methylocella species are facultatively methanotrophic. J Bacteriol 187:4665–4670. https://doi.org/10.1128/JB.187.13.4665-4670.2005
Dedysh SN, Belova SE, Bodelier PL, Smirnova KV, Khmelenina VN, Chidthaisong A, Trotsenko YA, Liesack W, Dunfield PF (2007) Methylocystis heyeri sp. nov., a novel type II methanotrophic bacterium possessing ‘signature’ fatty acids of type I methanotrophs. Int J Syst Evol Microbiol 57:472–479. https://doi.org/10.1099/ijs.0.64623-0
Dunfield PF, Khmelenina VN, Suzina NE, Trotsenko YA, Dedysh SN (2003) Methylocella silvestris sp. nov., a novel methanotroph isolated from an acidic forest cambisol. Int J Syst Evol Microbiol 53:1231–1239. https://doi.org/10.1099/ijs.0.02481-0
Dunfield PF, Belova SE, Vorob’ev AV, Cornish SL, Dedysh SN (2010) Methylocapsa aurea sp. nov., a facultative methanotroph possessing a particulate methane monooxygenase, and emended description of the genus Methylocapsa. Int J Syst Evol Microbiol 60:2659–2664. https://doi.org/10.1099/ijs.0.020149-0
Farhan Ul Haque M, Crombie AT, Murrell JC (2019) Novel facultative Methylocella strains are active methane consumers at terrestrial natural gas seeps. Microbiome 7:1–17. https://doi.org/10.1186/s40168-019-0741-3
Farhan Ul Haque M, Xu H-J, Murrell JC, Crombie A (2020) Facultative methanotrophs–diversity, genetics, molecular ecology and biotechnological potential: A mini-review. Microbiology 166:894–908. https://doi.org/10.1099/mic.0.000977
Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM (2007) DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57:81–91. https://doi.org/10.1099/ijs.0.64483-0
Graham D, Korich D, LeBlanc R, Sinclair N, Arnold R (1992) Applications of a colorimetric plate assay for soluble methane monooxygenase activity. Appl Environ Microbiol 58:2231–2236. https://doi.org/10.1128/aem.58.7.2231-2236.1992
Guerrero-Cruz S, Vaksmaa A, Horn MA, Niemann H, Pijuan M, Ho A (2021) Methanotrophs: discoveries, environmental relevance, and a perspective on current and future applications. Front Microbiol 12:678057. https://doi.org/10.3389/fmicb.2021.678057
Hara S, Wada N, Hsiao SS-Y, Zhang M et al (2022) In vivo evidence of single 13C and 15N isotope–labeled methanotrophic nitrogen-fixing bacterial cells in rice roots. Mbio 13:e01255–e01322. https://doi.org/10.1128/mbio.01255-22
He L, Groom JD, Wilson EH, Fernandez J, Konopka MC, Beck DA, Lidstrom ME (2023) A methanotrophic bacterium to enable methane removal for climate mitigation. Proc Natl Acad Sci 120:e2310046120. https://doi.org/10.1073/pnas.2310046120
Im J, Lee SW, Yoon S, DiSpirito AA, Semrau JD (2011) Characterization of a novel facultative Methylocystis species capable of growth on methane, acetate and ethanol. Environ Microbiol 3:174–181. https://doi.org/10.1111/j.1758-2229.2010.00204.x
Ishii S, Ikeda S, Minamisawa K, Senoo K (2011) Nitrogen cycling in rice paddy environments: past achievements and future challenges. Microbes Environ 26:282–292. https://doi.org/10.1264/jsme2.me11293
Ishikawa M, Yokoe S, Kato S, Hori K (2018) Efficient counterselection for Methylococcus capsulatus (Bath) by using a mutated pheS gene. Appl Environ Microbiol 84:e01875–e01918. https://doi.org/10.1128/AEM.01875-18
Jung G-Y, Rhee S-K, Han Y-S, Kim S-J (2020) Genomic and physiological properties of a facultative methane-oxidizing bacterial strain of Methylocystis sp. from a wetland. Microorganisms 8:1719. https://doi.org/10.3390/microorganisms8111719
Kaise H, Sawadogo JB, Alam MS, Ueno C, Dianou D, Shinjo R, Asakawa S (2023) Methylocystis iwaonis sp. nov., a type II methane-oxidizing bacterium from surface soil of a rice paddy field in Japan, and emended description of the genus Methylocystis (ex Whittenbury et al. 1970) Bowman et al. 1993. Int J Syst Evol Microbiol 73:005925. https://doi.org/10.1099/ijsem.0.005925
Knief C (2019) Diversity of methane-cycling microorganisms in soils and their relation to oxygen. Curr Issues Mol Biol 33:23–56. https://doi.org/10.21775/cimb.033.023
Lee HJ, Kim SY, Kim PJ, Madsen EL, Jeon CO (2014) Methane emission and dynamics of methanotrophic and methanogenic communities in a flooded rice field ecosystem. FEMS Microbiol Ecol 88:195–212. https://doi.org/10.1111/1574-6941.12282
Lee I, Ouk Kim Y, Park S-C, Chun J (2016) OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol 66:1100–1103. https://doi.org/10.1099/ijsem.0.000760
Leng L, Chang J, Geng K, Lu Y, Ma K (2015) Uncultivated Methylocystis species in paddy soil include facultative methanotrophs that utilize acetate. Microb Ecol 70:88–96. https://doi.org/10.1007/s00248-014-0540-0
Liu Y, He X, Zhu P, Cheng M, Hong Q, Yan X (2020) pheS AG Based Rapid and Efficient Markerless Mutagenesis in Methylotuvimicrobium. Front Microbiol 11:441. https://doi.org/10.3389/fmicb.2020.00441
Miyazaki K (2015) Molecular engineering of a PheS counterselection marker for improved operating efficiency in Escherichia coli. Biotechniques 58:86–88. https://doi.org/10.2144/000114257
Mohammadi S, Pol A, van Alen TA, Jetten MS, Op den Camp HJ (2017) Methylacidiphilum fumariolicum SolV, a thermoacidophilic ‘Knallgas’ methanotroph with both an oxygen-sensitive and-insensitive hydrogenase. ISME J 11:945–958. https://doi.org/10.1038/ismej.2016.171
Puri AW, Owen S, Chu F, Chavkin T, Beck DA, Kalyuzhnaya MG, Lidstrom ME (2015) Genetic tools for the industrially promising methanotroph Methylomicrobium buryatense. Appl Environ Microbiol 81:1775–1781. https://doi.org/10.1128/AEM.03795-14
Rui J, Peng J, Lu Y (2009) Succession of bacterial populations during plant residue decomposition in rice field soil. Appl Environ Microbiol 75:4879–4886. https://doi.org/10.1128/AEM.00702-09
Semrau JD, DiSpirito AA, Vuilleumier S (2011) Facultative methanotrophy: false leads, true results, and suggestions for future research. FEMS Microbiol Lett 323:1–12. https://doi.org/10.1111/j.1574-6968.2011.02315.x
Shevchuk NA, Bryksin AV, Nusinovich YA, Cabello FC, Sutherland M, Ladisch S (2004) Construction of long DNA molecules using long PCR-based fusion of several fragments simultaneously. Nucleic Acids Res 32:e19–e19. https://doi.org/10.1093/nar/gnh014
Stavert AR, Saunois M, Canadell JG, Poulter B et al (2022) Regional trends and drivers of the global methane budget. Glob Chang Biol 28:182–200. https://doi.org/10.1111/gcb.15901
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. https://doi.org/10.1093/molbev/mst197
Vorobev A, Jagadevan S, Jain S, Anantharaman K, Dick GJ, Vuilleumier S, Semrau JD (2014) Genomic and transcriptomic analyses of the facultative methanotroph Methylocystis sp. strain SB2 grown on methane or ethanol. Appl Environ Microbiol 80:3044–3052. https://doi.org/10.1128/AEM.00218-14
Wang J, Geng K, Farhan Ul Haque M, Crombie A, Street LE, Wookey PA, Ma K, Murrell JC, Pratscher J (2018) Draft genome sequence of Methylocella silvestris TVC, a facultative methanotroph isolated from permafrost. Genome Announc 6:10. https://doi.org/10.1128/genomeA.00040-18
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703. https://doi.org/10.1128/jb.173.2.697-703.1991
Whittenbury R, Davies SL, Davey J (1970) Exospores and cysts formed by methane-utilizing bacteria. Microbiology 61:219–226. https://doi.org/10.1099/00221287-61-2-219
Yan X, Chu F, Puri AW, Fu Y, Lidstrom ME (2016) Electroporation-based genetic manipulation in type I methanotrophs. Appl Environ Microbiol 82:2062–2069. https://doi.org/10.1128/AEM.03724-15
Zhang J, Liu Y-X, Zhang N, Hu B et al (2019) NRT1. 1B is associated with root microbiota composition and nitrogen use in field-grown rice. Nat Biotechnol 37:676–684. https://doi.org/10.1038/s41587-019-0104-4
Zhao T, Zhang L, Zhang Y, Xing Z, Peng X (2013) Characterization of Methylocystis strain JTA1 isolated from aged refuse and its tolerance to chloroform. J Environ Sci 25:770–775. https://doi.org/10.1016/s1001-0742(12)60046-1
Zhu P, Cheng M, Pei D, Liu Y, Yan X (2020) Methylomonas rhizoryzae sp. nov., a type I methanotroph isolated from the rhizosphere soil of rice. Antonie Van Leeuwenhoek 113:2167–2176. https://doi.org/10.1007/s10482-020-01487-2
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 32270127). The authors are grateful to Chun Qin for his help in the operation of electron microscopy.
Funding
This work was supported by the National Natural Science Foundation of China (No. 32270127, 32170126) and the Fundamental Research Funds for the Central Universities (KYCYXT2022003).
Author information
Authors and Affiliations
Contributions
YW: Strain isolation and characterization, Writing. YW: Genetic tools development, Writing. KZ: Electroporation method, Writing. HZ: Genome analysis, Writing. MC: Writing – review & editing. BW: Writing – review & editing. XY: Design of this work, Writing – review & editing.
Corresponding author
Ethics declarations
Competing interest
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, Y., Wang, Y., Zhou, K. et al. Isolation of a facultative methanotroph Methylocystis iwaonis SD4 from rice rhizosphere and establishment of rapid genetic tools for it. Biotechnol Lett 46, 713–724 (2024). https://doi.org/10.1007/s10529-024-03495-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-024-03495-y