Abstract
The aim of this study was to investigate and compare the microbial community structures of eubacteria and archaea in the pit mud of Chinese Luzhou-flavor liquor from the wall (Cw) and bottom (Cb) of cellar through nested PCR–denaturing gradient gel electrophoresis (DGGE). The Shannon–Wiener index (H) calculated from the DGGE profiles showed that the community diversities of eubacteria and archaea in samples from Cb were almost higher than that from Cw. In addition, cluster analysis of the DGGE profiles revealed that some differences were found in the microbial community structure in samples from different locations. The closely relative microorganisms of all eubacterial 16S rRNA gene sequences fell into four phyla (Firmicutes, Proteobacteria, Bacteroidetes and Actinobacteria), including 12 genera and 2 uncultured eubacteria. Moreover, 37.1 % eubacteria were affiliated with Clostridium. Particularly, genus Acinetobacter was absent in all samples from Cb but present in all samples from Cw. The closely relative microorganisms of all archaeal 16S rRNA gene sequences fell into four genera, which included Methanobrevibacter, Methanoculleus, Methanobacterium and Methanosaeta, while the dominant archaea in samples from Cw and Cb were similar. Results presented in this study provide further understanding of the spatial differences in microbial community structure in the pit mud, and is of great importance for the production and quality improvement of Luzhou-flavor liquor.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chinese liquor is one of the most distinctive products in China as well as one of the six famous distillates in the world. In general, it is typically divided into five categories: Luzhou-flavor style, light aroma style, soy sauce aroma style, sweet honey style, and miscellaneous style. Annual production of Luzhou-flavor style, manufactured from fermented grains in a soil cellar (called as pit), is the highest among the five styles. Pit is a rectangular underground pool constructed by pit mud, specific fermented clay, providing suitable habitat for the brewing microbiota. Microbial community structure plays a key role during the fermentation, which lies on micro-environment in pit and pit age (the use time of the cellar). Therefore, it is necessary to explore microbial community structure in the pit mud and understand the metabolic regulation during fermentation process.
Previous studies concerning microbial community structure have been performed based on culture-dependent methods. Wu et al. (2009) focused on microorganisms in cellars through traditional culture-dependent methods and more than 1,000 strains were obtained, among which 6 bacteria, 7 yeasts and 3 molds were selected for developing new sacchariferous starters according to their characteristics and abilities for liquor production. However, most of the microorganisms are uncultured or difficult to culture, and culture-dependent method is difficult to reveal the inner pattern comprehensively and objectively (Amann et al. 1995). Recently, molecular ecological methods, such as denaturing gradient gel electrophoresis (DGGE), temperature gradient gel electrophoresis (TGGE) (Muyzer and Smalla 1998; Muyzer 1999) and real-time PCR (Bowers et al. 2000) are widely used to analyze microbial community structures in complex environments. Among these methods, DGGE is a direct and reliable method to analyze the actual microbial community structure through band migration overcoming the disadvantages of culture-dependent methods (Wang et al. 2008). Moreover, DGGE has been used increasingly to analyze microbial community structure in various environments, such as wastewater treatment system (Yang et al. 2012), soils (Watanabe et al. 2006; Wang et al. 2009) and pit mud (Shi et al. 2011). For example, Shi et al. (2011) analyzed the microbial community features in different pit mud samples with the difference in pit age by PCR–DGGE, and the results showed that pit mud of different cellar ages would induce specific community evolvement of bacteria and methanogenic archaea. However, little is known about the eubacteria and archaea in pit mud as well as the difference in the different locations. Therefore, the aim of this study was to investigate the characteristics of eubacteria and archaea communities and the difference between Cw and Cb.
Materials and methods
Sampling
The samples were collected from Lu-zhou Laojiao Co., Ltd, located in Luzhou city, the southeast part of Sichuan province, which was one of the famous Luzhou-flavor style liquor manufactures in China. Samples were taken from two locations (the wall and bottom of cellar) of three individual pits that were 200 years old. Each sample plot was divided into four subplots and about 100g of pit mud was collected from each subplot. All samples were well mixed, then transferred to sterile polyethylene bags and stored at −20 °C until analyzed.
DNA extraction
Five grams of each sample was suspended in 25 mL of phosphate buffered saline (PBS, 0.1 mol/L, pH 8.0), and eddied for 5 min. The suspension liquid was centrifuged at 600g at 4 °C for 10 min and the precipitate was washed thrice by the same PBS buffer. The supernatant was pooled and centrifuged at 12,000g at 4 °C for 10 min, and the resulted pellet was washed three times by PBS, and then stored at −20 °C until DNA extraction.
The pellets were subjected to DNA extraction using the commercial Soil Genomic DNA Rapid Extraction Kit (Bioteke Corporation, Beijing, China) according to the manufacturer’s instruction. The crude DNA was detected quantitatively according to absorbance at 260 nm with micro-spectrophotometer K5500 (Beijing Kaiso Technology Development Co., Ltd. China), and the purity was assessed by electrophoresis on 0.6 % (w/v) agarose gels. The extracted DNA was subsequently used as a template for PCR to amplify 16S rRNA genes.
PCR amplification
All PCR primers were listed in Table 1. In order to increase sensitivity and facilitate DGGE analysis, a nested PCR technique was employed. For analysis of eubacterial diversity, primer pairs 27F and 1492R were used to amplify the nearly complete 16S rRNA encoding gene under conventional PCR conditions in the first step of PCR. Subsequently, this initial PCR product was diluted and used as a template for a nested PCR targeting the V3 region of the 16S rRNA gene with DGGE primers 357F with the GC clamp and 517R to create a DNA fragment suitable for DGGE analysis.
For analysis of archaeal diversity, PCR amplification of the 16S rRNA gene was performed using the universal primer PRA 46F and PREA 1100R in the first step, followed by nested PCR using the DGGE primers PARCH 340F with the GC clamp and PARCH 519R. The products were examined by electrophoresis on 1 % agarose gels before they were applied to DGGE.
DGGE analysis
The PCR products were analyzed by DGGE using the DCode Universal Mutation Detection System (Bio-Rad, Hercules, CA, USA). For determination of the eubacterial community, 8 % of the polyacrylamide gradient was superimposed onto a 30–60 % denaturant gradient, whereby 100 % was defined as 7 M urea and 40 % (v/v) formamide. Gels were carried out for 5 h at 150 V at 60 °C, and then gels were stained with SYBR Green I for 30 min (Molecular Probes, Eugene, OR, USA). For archaeal community, a 35–65 % denaturant gradient was applied to 8 % of the polyacrylamide gel. Gels were carried out for 5 h at 130 V at 60 °C, and then gels were stained with SYBR Green I for 30 min. The gels image was documented with a Gel Print TMXR system (Bio-Rad, USA) under UV illumination.
Excision of DGGE bands and sequencing
Representative bands observed in the DGGE profiles were excised from the gel using a sterile scalpel and eluted overnight at 4 °C in ultrapure water to allow DNA diffusion out of the polyacrylamide matrix. The solution was then used for further amplification. Excised bands were re-amplified using the GC-clamp primers described previously and re-run on DGGE gels to confirm their identity and improve purity prior to sequencing. After that, DGGE bands were re-amplified with no GC-clamp forward primer. The PCR products were purified with a universal PCR purification kit (Tiangen, Beijin, China) and sent to a commercial sequencing company for cloning and sequencing (Sangon, Shanghai, China). Then sequences obtained from this study, were compared to the 16S rRNA gene sequences available in GenBank (http://www.ncbi.nlm.nih.gov) to identify their closest phylogenetic relatives.
Statistical analysis
Both of cluster analyses and community diversity were performed using the Quantity One software, which was used to convert individual DGGE lane to densitometric profiles. The density of each band was calculated. The evenness index and Shannon–Wiener index were determined based on the number and relative intensity of the bands, and calculated using ZZSTAT V2010. Clustering of the sample profiles was done using the unweighted pair group method with arithmetic averages (UPGMA). Dendrogram was constructed with NTSYS-pc 2.10e software.
Results
Community diversity
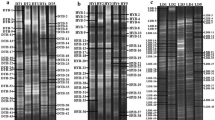
The DGGE finger-printing profiles of microbial community were shown in Fig. 1. Notable differences were observed in samples which were sampled from wall and bottom of cellar aged for 200-year. The eubacterial and archaeal community diversities were evaluated based on the analysis of the DGGE profiles (Table 2). The species richness (S) distinguished the six samples from each other effectively and showed that the number of bands in sample 1 (S: 28) was the highest and followed by sample 6. The evenness index (J) for the eubacterial community in Cw (0.977 < J < 0.985) appeared to have more homogeneous ecosystems than that in Cb (0.958 < J < 0.974). Remarkably, sample 1 exhibited the highest Shannon–Wiener index (H: 3.25) in the PCR–DGGE profiles, which indicated that the high number of different eubacterial species was presented in sample 1.
DGGE profiles of eubacteria (A) and archaea (B) from the V3 region of 16S rRNA obtained from 6 pit mud samples. Lanes corresponding to different samples are indicated by numbers at the top (Lanes 1, 2, 3 represent samples harvested from Cw-1, Cw-2, Cw-3, respectively; while lanes 4, 5, 6 represent samples collected from Cb-1, Cb-2, Cb-3, respectively). The bands indicated by the numbers were excised, re-amplified and subjected to sequencing
In terms of the species richness, archaeal DGGE profiles of samples from Cb (labelled by 4–6) exhibited a higher number of bands than that from Cw (labelled by 1–3). Obviously, sample 4 showed the highest number of bands (S: 23) and the highest Shannon–Wiener index (H: 3.022) in the PCR–DGGE profiles. In addition, samples from Cw-1 and Cw-3 showed higher evenness index than that from Cb-1 and Cb-3, respectively, while sample from Cw-2 showed slightly lower evenness index than that from Cb-2 (Table 2).
Cluster analysis of DGGE fingerprints
Cluster analysis was performed using DGGE data to estimate the order of relatedness among the pit mud samples (Fig. 2). Cluster analysis of the eubacterial DGGE profiles indicated that the sample 1 formed a group, and samples 2, 3 from Cw and samples 4, 5, 6 from Cb were clustered into different groups. In addition, it was interesting to find that the main microbial populations of samples 2 and 3 were similar to those samples from Cb (Fig. 2A).
Clustering analysis of DGGE profiles for eubacteria (A) and archaea (B). Similarity was calculated using Euclidean distance and clustering was done using UPGMA. Numbers 1–3 represent samples harvested from Cw-1, Cw-2, Cw-3, respectively; while 4–6 represent samples collected from Cb-1, Cb-2, Cb-3, respectively
Cluster analysis of the archaeal DGGE profiles showed that sample 4 formed a group, and samples 1, 2, 3 from Cw and samples 5, 6 from Cb were clustered into different groups. Moreover, all of the samples from Cw clustered within a group along with samples 5 and 6 (Fig. 2B).
Sequencing of DGGE bands
These representative bands of eubacterial PCR–DGGE were sequenced and the results were shown in Table 3. The similarity of all bands sequences was nearly >94 % comparing with those available in GenBank database. The closely relative microorganisms of all eubacterial 16S rRNA gene sequences fell into four phyla (Firmicutes, Proteobacteria, Bacteroidetes and Actinobacteria), including 12 genera, uncultured Clostridium sp. (bands 7 and 11) and uncultured Synergistetes bacterium (band 31) (Goodfellow et al. 2012). Among class Clostridia and order Clostridiales, these sequences belonged to four families, which included Clostridiaceae, Ruminococcaceae, Syntrophomonadaceae, and Peptococcaceae. Class Clostridia was dominant and accounted for 54.3 % of all the closely relative microorganisms, which fell into Clostridium (bands 1, 2, 6, 7, 10, 11, 21, 22, 24, 25, 27, 29 and 33), Ruminococcus (bands 9 and 26), Syntrophomonas (band 16), Sedimentibacter (band 18) and Desulfotomaculum (band 19), respectively. Three genera, including Lactobacillus (bands 4, 5 and 28), Virgibacillus (bands 12 and 13) and Bacillus (bands 14, 15 and 20) were affiliated with family Lactobacillaceae and family Bacillaceae in class Bacilli, respectively. In addition, Rhodococcus (bands 34 and 35) and Microbacterium (bands 30 and 32) were affiliated with class Actinobacteridae. Genus Acinetobacter (bands 8 and 23) and family Porphyromonadaceae (band 3) were affiliated with class γ-Proteobacteria and class Bacteroidia, respectively.
Differences of microbial community diversity among the samples in different cellars of the same pit age were observed. For No. 1 cellar, genus Rhodococcus was not detected in the sample from Cw, yet Acinetobacter, Sedimentibacter and Porphyromonadaceae were not detected in the sample from Cb. For No. 2 cellar, Synergistetes, Rhodococcus and Porphyromonadaceae bacterium were absent in the sample from Cw, additionally, Acinetobacter and P. bacterium were absent in the sample from Cb. For No. 3 cellar, P. bacterium was not observed in the sample from Cw, besides, Acinetobacter, Desulfotomaculum and P. bacterium were not observed in the sample from Cb.
For archaeal PCR–DGGE, selected bands were excised, sequenced and the results were listed in Table 4. Comparison of sequences from the excised bands and those available in the GenBank database revealed that all excised bands sequences were >98 % similar to 16S rRNA fragments already in the database. The closely relative microorganisms of all archaeal 16S rRNA gene sequences were affiliated with three families (Methanobacteriaceae, Menthanomicrobiaceae and Methanosaetaceae), and fell into four genera, including Methanobrevibacter, Methanoculleus, Methanobacterium, and Methanosaeta. Of these archaea, a great majority of the sequences identified belonged to genus Methanoculleus (accounted for 64.3 % of the band sequenced), following by genus Methanosaeta (bands 7, 8 and 9). The dominant archaeal species were almost the same in the samples from different cellars and different locations.
Discussion
In this study, PCR–DGGE was firstly employed to investigate the diversity of eubacterial and archaeal community structures in pit mud simultaneously, and the differences between Cw and Cb as well as the different cellars of the same pit age were examined. Cluster analysis for eubacteria and archaea indicated that the difference in eubacterial community structure of six samples was higher than archaeal community structure.
The results of sequencing showed that 37.1 % eubacteria were affiliated with genus Clostridium in all samples. Genus Clostridium, as one of important contributors to form representative aroma and flavor compounds in Luzhou-flavor type liquors, has been widely investigated. Genus Clostridium converted organic substances into organic acids, such as butyric and caproic acids, alcohols, CO2/H2 and minerals, forming ethyl butyrate and ethyl caproate when butyric and caproic acids reacted with alcohols by enzymatic and non-enzymatic catalysis (Zhang et al. 2005). As yet, no result on the diversity of Clostridium in the pit mud was reported. In fact, different strains of the same genus fulfilled different functions and synergistic metabolisms in co-culture. For example, Clostridium kluyveri was often used to convert alcohol into a mixture of acetic, butyric and caproic acids with methane bacteria (Bornstein and Barker 1948), whereas Clostridium ljungdahlii was able to co-metabolize CO and H2 to form acetate and ethanol (Worden et al. 1991). In addition, genus Ruminococcus was recognized as the major cellulolytic bacterial species (Koike and Kobayashi 2006). Some strains of genus Syntrophomonas detected such as S. zehnderi could degrade long-chain fatty acids in co-culture with Methanobacterium formicicum (Sousa et al. 2007). Genus Desulfotomaculum taxomomically was affiliated with the class clostridia and was grown in complex media with sulfate plus lactate or pyruvate that were incompletely oxidized into acetate (Klemps et al. 1985). Therefore, the Clostridiales is related to form major flavors compounds, such as ethyl caproate, ethyl acetate and ethyl lactate in Chinese Luzhou-flavor type liquor.
Genus Bacillus which was originated from Daqu, secreted various types of hydrolases such as amylases and protease (Ramos et al. 2011), and it was the dominant function bacteria in all types of Daqu (Xiu et al. 2012; Zheng et al. 2012). Bacillus species played an important role in liquefaction, saccharification (Owens et al. 1999), aroma producing (Zhu et al. 2010) as well as the production of heat- and acid-resistant cellulase. This enzyme degraded cellulose into glucose as carbon source and energy providing for other microorganisms in fermentation process (Gao et al. 2012). Likewise, genus Lactobacillus was also the functional microbes during liquor manufacture, which produced bacteriocin inhibiting the growth of pathogens and spoilage organisms (Zhang et al. 2005). Furthermore, lactic acid was also a precursor of ethyl lactate, which was one of the major flavor compounds.
Genus Microbacterium and genus Rhodococuus were both affiliated with Phylum Actinobacteria and were frequently found in soil, and the biodegradation potential of the genus Rhodoccus received increasing attention (Martínková et al. 2009), although the function was unknown in liquor brewing.
In our present research, the diversity was assessed to genus, and genus Methanoculleus was the dominant archaea in the pit mud aged for 200-year by the nested PCR–DGGE. The result agreed with conventional PCR–DGGE research. Meanwhile, it was consistent with one attained by fluorescence in situ hybridization, in which the order Methanomicrobiale was considered as the dominant community and was assessed roughly and quantitatively (data not shown). In fact, it is necessary to explore archaeal community diversity and interspecies interaction to reveal the regulation mechanisms of liquefaction, saccharification and fermentation under extremely environment, where the alcohol degree is more than 5 % (v/w) and titratable acidity is 2–3 %.
Moreover, there were some clear differences in the microbial community structure between Cw and Cb of the same pit age. Especially, some differences were found in the eubacterial community structure of different cellars, while archaeal community structure was almost the same. These may lie on the different micro-ecological environment, which is restricted by factors such as oxygen content, pH, process parameters etc.
In conclusion, the aim of this study was to investigate and compare the eubacterial and archaeal community diversity in Cw and Cb by nested PCR–DGGE. Results presented here showed the closely relative microorganisms of all eubacterial sequences included 12 genera and 2 uncultured eubacteria, and 37.1 % eubacteria were affiliated with Clostridium. The dominant archaea was Methanoculleus, accounting for 64.3 % of the bands sequenced. Moreover, there were some clear differences in the microbial community structure between Cw and Cb of the same pit age. These results may contribute to further understanding of the spatial differences in microbial community structure in the pit mud, and reveal the metabolic mechanisms involved in fermentation process.
References
Amann RI, Ludwig W, Schleifer KH (1995) Phylogenetic identification and in situ detection of individual microbial cells without cultivation. In: Microbiological reviews. American Society for Microbiology, New York, pp 143–169
Bornstein B, Barker H (1948) The nutrition of Clostridium kluyveri. J Bacteriol 55(2):223–230
Bowers HA, Tengs T, Glasgow HB, Burkholder JAM, Rublee PA, Oldach DW (2000) Development of real-time PCR assays for rapid detection of Pfiesteria piscicida and related dinoflagellates. Appl Environ Microbiol 66(11):4641–4648
Cunliffe M, Whiteley AS, Newbold L, Oliver A, Schäfer H, Murrell JC (2009) Comparison of bacterioneuston and bacterioplankton dynamics during a phytoplankton bloom in a fjord mesocosm. Appl Environ Microbiol 75(22):7173–7181
Gao J, Xiao DQ, Liu XP, Wang NQ, Zhang DW (2012) Identification and optimal degradation conditions for cellulase-degrading enzyme of a methanol-utilizing and cellulase-producing bacterium. Ind Microbiol 42(2):51–57 (in Chinese)
Goodfellow M, Kämpfer P, Busse HJ, Trujillo ME, Suzuki KI, Ludwig W, Whitman WB (2012) Bergey’s manual of systematic bacteriology. Springer, New York
Hu P, Zhou G, Xu X, Li C, Han Y (2009) Characterization of the predominant spoilage bacteria in sliced vacuum-packed cooked ham based on 16S rDNA-DGGE. Food Control 20(2):99–104
Kim TW, Lee JH, Park MH, Kim HY (2010) Analysis of bacterial and fungal communities in Japanese- and Chinese-fermented soybean pastes using nested PCR–DGGE. Curr Microbiol 60(5):315–320
Klemps R, Cypionka H, Widdel F, Pfennig N (1985) Growth with hydrogen, and further physiological characteristics of Desulfotomaculum species. Arch Microbiol 143(2):203–208
Koike S, Kobayashi Y (2006) Development and use of competitive PCR assays for the rumen cellulolytic bacteria: Fibrobacter succinogenes, Ruminococcus albus and Ruminococcus flavefaciens. FEMS Microbiol Lett 204(2):361–366
Martínková L, Uhnáková B, Pátek M, Nešvera J, Křen V (2009) Biodegradation potential of the genus Rhodococcus. Environ Int 35(1):162–177
Matsuyama T, Nakajima Y, Matsuya K, Ikenaga M, Asakawa S, Kimura M (2007) Bacterial community in plant residues in a Japanese paddy field estimated by RFLP and DGGE analyses. Soil Biol Biochem 39(2):463–472
Muyzer G (1999) DGGE/TGGE a method for identifying genes from natural ecosystems. Curr Opin Microbiol 2(3):317–322
Muyzer G, Smalla K (1998) Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Van Leeuwenhoek 73(1):127–141
Ovreås L, Forney L, Daae FL, Torsvik V (1997) Distribution of bacterioplankton in meromictic Lake Saelenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl Environ Microbiol 63(9):3367–3373
Owens JD, Allagheny N, Kipping G, Ames JM (1999) Formation of volatile compounds during Bacillus subtilis fermentation of soya beans. J Sci Food Agric 74(1):132–140
Ramos CL, de Almeida EG, Freire AL, Freitas Schwan R (2011) Diversity of bacteria and yeast in the naturally fermented cotton seed and rice beverage produced by Brazilian Amerindians. Food Microbiol 28(7):1380–1386
Shi S, Wang HY, Zhang WX, Deng Y, Lai DY, Fan A (2011) Analysis of microbial communities characteristics in different pit mud of Luzhou-flavor Liquor. Liquor Mak Sci Technol 5(203):38–41 (in Chinese)
Sousa DZ, Smidt H, Alves MM, Stams AJM (2007) Syntrophomonas zehnderi sp. nov., an anaerobe that degrades long-chain fatty acids in co-culture with Methanobacterium formicicum. Int J Syst Evol Microbiol 57(3):609–615
Wang HY, Zhang XJ, Zhao LP, Xu Y (2008) Analysis and comparison of the bacterial community in fermented grains during the fermentation for two different styles of Chinese liquor. J Ind Microbiol Biotechnol 35(6):603–609
Wang Y, Ke X, Wu L, Lu Y (2009) Community composition of ammonia-oxidizing bacteria and archaea in rice field soil as affected by nitrogen fertilization. Syst Appl Microbiol 32(1):27–36
Wang S, Nomura N, Nakajima T, Uchiyama H (2012) Case study of the relationship between fungi and bacteria associated with high-molecular-weight polycyclic aromatic hydrocarbon degradation. J Biosci Bioeng 113(5):624–630
Watanabe T, Asakawa S, Nakamura A, Nagaoka K, Kimura M (2006) DGGE method for analyzing 16S rDNA of methanogenic archaeal community in paddy field soil. FEMS Microbiol Lett 232(2):153–163
Wenhui Z, Zucong C, Lichu Y, He Z (2007) Effects of the long-term application of inorganic fertilizers on microbial community diversity in rice-planting red soil as studied by using PCR–DGGE. Acta Ecol Sin 27(10):4011–4018
Worden R, Grethlein A, Jain M, Datta R (1991) Production of butanol and ethanol from synthesis gas via fermentation. Fuel 70(5):615–619
Wu ZY, Zhang WX, Zhang QS, Hu C, Wang R, Liu ZH (2009) Developing new sacchariferous starters for liquor production based on functional strains isolated from the pits of several famous Luzhou-flavor liquor brewers. J I Brewing 115(2):111–115
Xiu L, Kunliang G, Hongxun Z (2012) Determination of microbial diversity in Daqu, a fermentation starter culture of Maotai liquor, using nested PCR–denaturing gradient gel electrophoresis. World J Microbiol Biotechnol 28:2375–2381
Yang Q, Wang J, Wang H, Chen X, Ren S, Li X, Xu Y, Zhang H, Li X (2012) Evolution of the microbial community in a full-scale printing and dyeing wastewater treatment system. Bioresource Technol 117:155–163
Zhang W, Qiao Z, Shigematsu T, Tang Y, Hu C, Morimura S, Kida K (2005) Analysis of the bacterial community in Zaopei during production of Chinese Luzhou-flavor liquor. J I Brewing 111(2):215–222
Zheng X-W, Yan Z, Han B-Z, Zwietering MH, Samson RA, Boekhout T, Nout M (2012) Complex microbiota of a Chinese “Fen” liquor fermentation starter, revealed by culture-dependent and culture-independent methods. Food Microbiol 31:293–300
Zhu BF, Xu Y, Fan WL (2010) High-yield fermentative preparation of tetramethylpyrazine by Bacillus sp. using an endogenous precursor approach. J Ind Microbiol Biotechnol 37(2):179–186
Acknowledgments
This work was financially supported by the National Science Foundation of China (31171742).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ding, XF., Wu, CD., Zhang, LQ. et al. Characterization of eubacterial and archaeal community diversity in the pit mud of Chinese Luzhou-flavor liquor by nested PCR–DGGE. World J Microbiol Biotechnol 30, 605–612 (2014). https://doi.org/10.1007/s11274-013-1472-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11274-013-1472-4