Abstract
Trichoderma asperellum parasitizes a large variety of phytopathogenic fungi. The mycoparasitic activity of T. asperellum depends on the secretion of complex mixtures of hydrolytic enzymes able to degrade the host cell wall and proteases which are a group of enzymes capable of degrading proteins from host. In this study, a full-length cDNA clone of aspartic protease gene, TaAsp, from T. asperellum was obtained and sequenced. The 1,185 bp long cDNA sequence was predicted to encode a 395 amino acid polypeptide with molecular mass of 42.3 kDa. The cDNA of TaAsp was inserted into the pPIC9K vector and transformed into yeast Pichia pastoris GS115 for heterologous expression. A clearly visible band with molecular mass about 42 kDa in the SDS-PAGE gel indicated that the transformant harboring the gene TaAsp had been successfully translated in P. pastoris and produced a recombinant protein. Enzyme characterization test showed that the optimum fermentation time for P. pastoris GS115 transformant was 72 h. Enzyme activity of the recombinant aspartic proteinase remained relatively stable at 25–60 °C and pH 3.0–9.0, which indicated its good prospect of application in biocontrol. The optimal pH value and temperature of the enzyme activity were pH 4.0 and 40 °C, and under this condition, with casein as the substrate, the recombinant protease activity was 18.5 U mL−1. In order to evaluate antagonistic activity of the recombinant protease against pathogenic fungi, five pathogenic fungi, Fusarium oxysporum, Alternaria alternata, Cytospora chrysosperma, Sclerotinia sclerotiorum and Rhizoctonia solani, were applied to the test of in vitro inhibition of their mycelial growth by culture supernatant of P. pastoris GS115 transformant.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Biological control of soil-borne plant pathogens by antagonistic microorganisms is a potential nonchemical means of plant disease control. Trichoderma fungi are well known for their antagonism against several soil–phytopathogens fungi. Their mycoparasitic activity is facilitated by antifungal products or secondary metabolites, including peptide and nonpeptide toxins, and a battery of lytic enzymes, mainly chitinases, glucanases and proteases, released in the presence of a suitable host (Harman et al. 2004; Schuster and Schmoll 2010). During mycoparasitism process, fungal proteases may play an important role in cell wall lysis, because fungal cell walls also contain protein besides chitin and/or β-glucan fibrils which are embedded in a protein matrix (Wessels 1986). In comparison with chitinases and glucanases, little was known about the proteases secreted by Trichoderma strains, despite the fact that it is also involved in the biocontrol process. However, the study of the components of the proteolytic system of Trichoderma spp. and their contribution to biocontrol has been receiving increasing attention in recent years. Fungal proteases may be significantly involved in antagonistic activity, not only by breaking down the host cell wall (Kapteyn et al. 1996), but also by acting as proteolytic inactivators of pathogen enzymes involved in the plant infection process (Elad and Kapat 1999; Suarez et al. 2004). Understanding the biocontrol mechanisms of action and regulation of serine proteases can stimulate the development of approaches for detecting and increasing the biocontrol activity of related fungi. Recently, many studies about proteinases involving in biocontrol activities were reported (Olmedo-Monfil et al. 2002; Pozo et al. 2004; Delgado-Jarana et al. 2002; Viterbo et al. 2004; Suarez et al. 2005; Szekeres et al. 2004; Delgado-Jarana et al. 2000; Grinyer et al. 2005; Yan and Qian 2009; Liu and Yang 2007).
In this paper, the cloning and sequence analyses of the aspartic protease gene of T. asperellum and its heterologous expression in a laboratory strain of Pichia pastoris are described. The temperature stability and pH dependence for protease activity were determined. The antifungal activity of this protease against five phytopathogenic fungi was assessed in vitro. It is a novel attempt that T. asperellum aspartic protease was functionally expressed in a heterologous host, and this effort can be further extended to studies of fungal development and biocontrol applications. This study will also contribute to search for more environmentally and toxicologically safe efficacious fungicides.
Materials and methods
Strain, vectors and culture media
Trichoderma asperellum strain T4 was stored at the Microbial Genetic Engineering Laboratory, Harbin Institute of Technology (Harbin, China). Escherichia coli Top 10 was used for cloning and nucleotide sequencing. P. pastoris host strain GS115 and the pPIC9K Vector (Invitrogen, Carlsbad, USA) were used as expression system. The PCR primers were synthesized by Sangon Biotech (Shang Hai, China) Co., Ltd. Five strains of plant-pathogenic fungi, Fusarium oxysporum, Alternaria alternata, Cytospora chrysosperma, Sclerotinia sclerotiorum and Rhizoctonia solani, were stored at the Microbial Genetic Engineering Laboratory, Harbin Institute of Technology (Harbin, China).
Trichoderma asperellum and five strains of plant-pathogenic fungi were cultured on potato dextrose agar, and P. pastoris was cultured in yeast extract peptone dextrose medium. E. coli Top 10 were routinely cultured in a Luria–Bertani medium. Culture media, such as regeneration dextrose medium, buffered glycerol-complex medium and buffered methanol-complex medium (BMMY) were prepared as described in Pichia Expression Kit Manual.
Minimal dextrose medium (MD) consists of 1.34 % yeast nitrogen Base, 4 × 10−5 % biotin, 2 % dextrose, 1.5 % agar, which is used in the screening of transformants GS115-TaAsp.
The conventional chemicals were purchased from Sangon Biotech Co., Ltd., Shang Hai, China.
Cloning of aspartic protease TaAsp gene
Liu et al. (2010) constructed cDNA library from T. asperellum mycelia and obtained 3,114 ESTs. Among these ESTs, the EST EU 816200 represented aspartic protease gene TaAsp. The full length cDNA sequence of TaAsp was obtained directly from the library clone containing the EST EU 816200.
Primers for amplification of Aspartic protease TaAsp gene were designed as follows, Asp5: 5′-ATGAAGAGCGCATTGATTGCCGC-3′; Asp3: 5′-TTATTTGGCCTTGG CAAGACCGAC-3′). Genomic DNA was extracted from T. asperellum mycelium according to the molecular cloning procedures described by Jones and Hancock (1987). Polymerase chain reaction (PCR) was carried out in Gen AmPCR System 2400 (Perkin Ilmer, Waltham, USA) for amplification of the TaAsp gene; the PCR product was inserted into the pMD18-T vector (TaKaRa Biotechnology Company, Dalian, China) for sequencing. To obtain the cDNA sequence of TaAsp, total RNA was extracted from T. asperellum mycelium using Trizol reagent (Invitrogen, Carlsbad, USA) and digested with Dnase I (Promega, Madison, USA). The PCR product was inserted into a pMD18-T vector for sequencing.
Bioinformatics analysis of TaAsp gene
The open reading frame (ORF) of the TaAsp gene was searched using the ORF program (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). Catalytic domain of the TAASP protein was identified by InterProScan (http://www.ebi.ac.uk/Tools/InterProScan/) and the theoretical molecular mass and isoelectric point of the protein were calculated using the ProtParam tool (http://us.expasy.org/tools/protparam.html). Signal peptide was predicted using SignalP 3.0 Server (http://www.cbs.dtu.dk/services/SignalP/).Alignments of the deduced amino acid sequence of TaAsp with the related protein sequences were performed by the NCBI nucleotide blast programs (http://www.ncbi.nlm.nih.gov/) to elucidate its phylogenic relationship.
Construction of TaAsp expression system
In order to constructing the TaAsp expression system in P. pastoris GS115, shuttle plasmid pPIC9K was used. Primers with EcoRI (TaAsp5) and Not I (TaAsp3) sites (TaAsp5: 5′-ATCGGA ATTCGGCGTCCACAAGATGAAGCTGC-3′, TaAsp3: 5′-CGATGCGGCCGCTTTGGCCTTGGC AAGACCGACAG-3′) were designed for the cloning of TaAsp cDNA sequence. The full length of TaAsp cDNA obtained from the 1.4 kb PCR product by digestion with EcoRI and NotI (TaKaRa Biotech. Co., Dalian, China) was ligated into the expression pPIC9K vector. The plasmid was then transformed into E. coli Top 10 cells. With the empty plasmid pPIC9K as control, recombinant expression plasmid pPIC9K-TaAsp was verified by restriction enzyme digestion. The plasmid DNA was isolated from the bacterial cells using the SiMax™ Plasmid DNA Miniprep Kit (SBS Genetech, Beijing, China) and linearized using Sac I restriction enzyme (TaKaRa Biotech. Co., Dalian, China). The competent cells of P. pastoris GS115 were prepared just before the transformation as instructed by the supplier (Invitrogen, Carlsbad, USA). The linearized plasmid DNA (5–15 μg) was transformed into 80 μL yeast competent cells using Electroporator 2510 (Eppendorf, GER) with a pre-set program PIC (1,150 V, 6.10 ms). For regenerating the transformants, 1 mL of pre-cooled sorbitol was added into the mixture. Afterwards, 200 μL portions of the regeneration mixture were plated onto the Minimal-Dextrose medium plates at 30 °C until individual colonies appeared (Maeda 1989; Waterham et al. 1997; Masuda et al. 2004).
Production of the recombinant pPIC9K-TaAsp and SDS-PAGE analysis of recombinant aspartic proteinase TAASP
One colony of P. pastoris GS115 pPIC9K-TaAsp was inoculated into 2 mL of buffered minimal glycerol medium containing 100 μg of ampicillin mL−1 and grown overnight at 30 °C in HZQ-C air bath rotary shaker (HDL Co. Ltd., Beijing, China). The overnight culture was transferred to 25 mL of the same medium and grown again to an A600 value of 2.0–6.0 (UV1000 UV/Visible Spectrophotometer, Techcomp Co., Shang Hai, China). By centrifuging at room temperature, the collected cells were resuspended by BMMY to an A600 value of 1.0, placed in a rotary shaker under the condition of 250 rpm, 28–30 °C. Protein production was induced by addition of 100 % methanol to a final concentration of 0.5 mM every 24 h. Growth of P. pastoris GS115 pPIC9K was also conducted as control groups. The sample of induced transformant was salted by 80 % (NH4)2SO4 overnight; precipitate was dissolved with 1 mL buffer after 10 min centrifuge (12,000 rpm). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed in a 10 % polyacrylamide gel (Laemmli 1970). Proteins were fixed in the gels by soaking in a solution containing 40 % (v/v) methanol and 10 % (v/v) acetic acid for approximately 1 h and subsequently visualized by Coomassie blue staining (Coomassie Brilliant Blue G-250, Thermo Fisher Sci. Inc., Rockford, IL). Detection of proteinase activity was carried out with minor modifications (Pastor et al. 2001). Instead of containing SDS-PAGE, indicator agarose gels containing substrate were used.
Northern blot analysis
For expression study of TaAsp in transformants, the transformants were cultured in medium containing 1 % methanol for 12, 36, 60, and 72 h, respectively. Total RNA was extracted using a yeast RNA mini kit (Watson Biotechnologies Co., China). The total RNA (20 mg) was separated on a 1.2 % agarose gel containing 1.5 % formaldehyde and blotted onto a nylon membrane. Digoxigenin High Prime DNA Labeling and Detection Starter Kit II (Roche Molecular Biochemicals, Germany) were used for the preparation of the probe and detection of the transcripts of the TaAsp gene. The TaAsp gene digested by HindIII and EcoRI from plasmid (pPIC9K-TaAsp) was labeled with digoxigenin as a probe. Probes for hybridization were prepared by the random primer extension method. Northern blot analysis was conducted according to the manufacturer’s instruction.
Properties of recombination aspartic proteinase TAASP
To measure aspartic proteinase activity, the yeast transformants were induced by 1 % (v/v) methanol at 30 °C for 1–7 days, at 1 day intervals. To determine enzyme properties, the supernatant of culture was collected by centrifuging and used to measure the protease activity. The yeast cells were centrifuged at 3,100g, 4 °C for 10 min. The culture supernatant (1.0 mL) and 1 % casein solution (1.0 mL) in different buffer (pH 3.0–9.0) were preincubated at 40 °C for 5 min, respectively, and then mixed. The mixture was incubated at 40 °C for 10 min and 2 mL of 0.4 mol L−1 trichloroacetic acid solution was added to the mixture immediately to stop the reaction. The reaction mixture was centrifuged at 9,500g and 4 °C for 10 min. The culture supernatant (1.0 mL) was mixed with 5 mL of 0.4 mol L−1 sodium carbonate and 1 mL folin-phenol reagent. The mixture was incubated at different temperatures (25–60 °C) for 20 min. The tyrosine content in the culture supernatant was determined colorimetrically at 650 nm using folin-phenol reagent (Lowry et al. 1951). One unit of protease is defined as the amount of enzyme that catalyzes the release of 1 mg of l-tyrosine per min under the above assay conditions.
To determine the optimum temperature for enzyme activity, the reaction mixtures (pH 4.0) were incubated at 25–60 °C for 20 min at 5 °C intervals. The optimum pH of the expressed TaAsp was measured at 40 °C between pH 3 and 9 at an interval of 1 pH unit. The pH of the reaction mixture was adjusted using a series of buffers: 0.01 M citric acid–0.02 M Na2HPO4 (pH 3–5), 0.02 M Na2HPO4–NaH2PO4 (pH 6–8) and 0.02 M glycine–NaOH (pH 9–10). To investigate the effect of ions on enzyme activity, TAASP was measured in reaction buffer that was supplemented with 5 mM of a metal ion in the form of a chloride or sulfate salt. Several different buffer solutions were prepared, which were mixed with one metal salt (KCl, BaCl2, CaC12, NaCl, LiCl, MgSO4, MnSO4, FeSO4, CuSO4 or ZnSO4) respectively. The relative activity of TAASP was calculated as the activity in the ion-enhanced buffer divided by that under normal conditions (as the control).
All these experiments were performed in triplicate, and average values were calculated based on results from three independent experiments.
All of the above experiments were completed, at a minimum, in triplicate, and average values were calculated based on results from three independent experiments.
Antifungal assays
The experiment was carried out in 90 mm × 15 mm petriplates containing 15 mL PDA. The transformants were inoculated in BMMY with 1 % methanol. The supernatant of fermentation was collected by centrifugation of 2,500 rpm, 5 min, and standed at 4 °C after ultrafiltration through 0.45 μm filter membrane. The methanol-induced yeast transformants solution was 20-fold concentrated by centrifugal filter unit (Millipore). Five milliliters of culture supernatant treated by this way was added to 45 mL PDA at 45 °C, mixed rapidly, and poured into petri dishes. A 4 mm diameter plug of the actively growing mycelium of phytopathogen was placed at the center of the PDA plate and incubated at 28 °C. The discs of phytopathogen on PDA with 5 mL of supernatant obtained from the control yeasts (empty vector) served as a control. There were four replicates per treatment. Colony diameter was observed and measured daily. Growth inhibition rate determined by the following formula:
R is the phytopathogenic colony radius of control and r is the radius of phytopathogenic colony with treatment.
Results
Cloning of the proteinase gene
The genomic DNA of T. asperellum and the cDNA of gene TaAsp were isolated and sequenced. The DNA sequence of proteinase gene TaAsp is 1,323-bp in length, containing three exons and two introns. The lengths of the three exons are 99, 154 and 935-bp, and two introns are 167 and 68-bp in length. Sequence analysis of the target fragment demonstrats the presence of an ORF of 1,185 bp, designated TaAsp, in the insert of recombinant plasmid pPIC9K. This ORF is predicted to encode a polypeptide of 395 amino acids (aa) with a molecular mass of about 42.3 kDa. A band of 40.75 kDa could possibly originate from this predicted ORF after removal of a 17 aa signal peptide. Analysis of the predicted TAASP protein sequence suggests that the first 17 aa constituted a signal peptide and a signal peptidase cutting site is located between the 17th and the 18th residues where a typical AXA motif for signal peptidase I is found. The predicted molecular mass of the 395 aa peptide is about 42 kDa. A aspartic protease conserved domain was found in the enzyme by using Conserved Domain Architecture Retrieval Tool (CDART) program within NCBI website. It was predicted to belong to the aspartic protease superfamily with the InterProScan tool. Analysis of the NCBI’s Blastp showed it contained the inhibitior binding sites of aspartic proteases, fingerprints of catalytic motif and catalytic residues (Fig. 1a).
With the NCBI database search, the deduced amino acid sequence showed 82 % identity with the proteases of Gibberella zeae, XP_390958.1. Aspartic proteases of T. asperellum (ACF20292), Magnaporthe grisea and Neurospora crassa had closest evolutionary relationship in amino acid sequence. At the same time, the proteases of Phaeosphaeria nodorum and Kluyveromyces lactis had further distant evolutionary relationship with the other species. The secondary structure of the protease were predicted by using SSpro bioinformatics analysis software (http://scratch.proteomics.ics.uci.edu/), prediction results are showed in Fig. 1b, in which C, H, E means β-turn, α-helix and β-sheet respectively, the protease active site is located in the shaded area, which mostly are the β-turn and α-helix.
Heterologous expression of TaAsp in Pichia pastoris
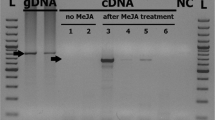
Cloning of the TaAsp ORF into the pPIC9 vector formed a construct that encoded TAASP with the αMF factor prepro sequence, which ensures proper processing of the secreted proteins (Invitrogen, USA, K1710-01). With this design, the product of the subcloned gene incorporated an additional 17 aa at the N-terminal end of the TAASP, following signal cleavage. Sequencing of the construct showed an in-frame positioning of the TAASP cDNA with respect to the initiation codon (ATG) of the vector. The correct structure of the expression vector pPIC9-TaAsp was confirmed by DNA sequencing. Recombinant pPIC9-TaAsp was transformed into P. pastoris GS115 and transformants GS115-TaAsp was selected on MD plates. Northern dot blot analysis provided with high signal intensity (Fig. 2a). SDS-PAGE analysis was conducted to determine whether the TAASP protein had been synthesized in P. pastoris GS115. Compared with the control transformant harboring empty pPIC9 (GS115), the transformant harboring the TaAsp gene produced a clearly visible protein band with a molecular mass of approximately 42 kDa in the SDS-PAGE gel (Fig. 2b). This result indicated that the TAASP protein had been successfully synthesized in the yeast cells GS115, and secreted into the culture.
a Northern blotting analysis of total RNA from the pPIC9K-TaAsp yeast transformant. C: transformants of GS115-pPIC9K (methanol inhibit); 12, 36, 60, 72 shows transformants of GS115-pPIC9K-TaAsp inducing for 12, 36, 60, 72 h. b SDS-PAGE analysis of recombinant protein in GS115 transformant. M: protein marker (DL2000); 1–3: proteins sample of GS115-pPIC9K; 4–6: proteins sample of GS115-pPIC9K-TaAsp
Analysis of property of protease activity in transgenic TaAsp yeast
The protease TAASP activity of the transgenic yeast strain GS115-TaAsp showed a peak value at 4 day with methanol induction (Fig. 3a). Additionally, protease activity was not detected in the culture medium of P. pastoris GS115-pPIC9 after methanol-induction, indicating that protease activity displayed in transgenic yeast cells is due to an expression of the exogenous TaAsp. After induction with methanol for TAASP expression, the transformant GS115-TaAsp that expressed the protease at the highest level was selected for further experimentation.
Activities and properties of recombinant protease TAASP. a Effects of culture time on recombinant protease TAASP activity; b effects of temperature on recombinant protease TAASP stability. The relative activity of TAASP was calculated as the activity in different temperature divided by the activity of TAASP under optimal temperature (as the control); c effect of pH on recombinant protease TAASP. The relative activity of TAASP was calculated as the activity in different pH divided by the activity of TAASP under optimal pH (as the control). The error bars are standard errors
TAASP activity reached its peak at 40 °C and when the temperature reached 60 °C, enzyme activity was just 30 % of the peak. Therefore, the optimal reaction temperature for TAASP was 40 °C (Fig. 3b). At 40 °C, the enzyme activity of recombinant TAASP was stable at pH 3–6, and the peak of activity appeared at pH 4 (Fig. 3c). Under the optimal temperature and pH, the crude enzyme activity of transgenic TaAsp yeast protease was 18.5 U mL−1. The activity of recombinant TAASP was stimulated by all metal ions except for Ba2+ and Na+. But all metal ions did not induce any substantive activation of recombinant TAASP (Fig. 4).
Effects of exposure to metal ions on proteinase TAASP activity. Ions: 1, K+; 2, Ba2+; 3, Ca2+; 4, Na+; 5, Li+; 6, Mg2+; 7, Mn2+; 8, Fe2+; 9, Cu2+;10, Zn2+. The relative activity of TAASP was calculated as the activity in the ion-enhanced buffer divided by the activity of TAASP under normal conditions (as the control). The error bars are standard errors
Assay for biocontrol ability of TAASP to different pathogens
In order to evaluate the antagonistic activity of protease TAASP against pathogenic fungi, in vitro assays for inhibition of mycelial growth of five pathogenic fungi were performed using the culture supernatant of yeast transformants with protease TAASP. During the early stage of experiments, protease TAASP did not show significant inhibition to all of these pathogenic fungi, while, after 24 h, the inhibition effect were becoming more and more detectable but still relative weak. Among the five in vitro assays, the most significant inhibition of mycelial growth by protease TAASP happened with S. sclerotiorum and showed only 22.82 % inhibition rate, while the mycelial growth of R. solani and C. chrysosperma were inhibited with 18.23 and 16.57 % respectively. A weak inhibition of A. alternata was 10.89 %, while no significant inhibition effect on F. oxysporum was found (Table 1).
Discussion
Trichoderma sp. is ubiquitous in soil environment and parasitizes a broad range of phytopathogenic fungi. As the characteristics of active mycoparasite, this filamentous fungus could be used as biocontrol agent. The main mechanism involved in the antagonism to pathogenic fungi by Trichoderma sp. is the release of lytic enzymes, mainly chitinases, glucanases, and proteases, with the presence of a suitable host (Chet and Chernin 2002). In comparison with chitinases and glucanases, proteases is less studied. Recently, the study of the components of the proteolytic system of Trichoderma spp. and their contribution to biocontrol has been paid increasing attention.
In order to obtain enough amount of aspartic protease applied for its functional analysis, P. pastoris as a powerful and versatile heterologous expression system was used in this study. Expression of gene coding for protease from T. asperellum was achieved and the recombined protein product was found in the culture supernatant with activity. The optimal enzyme reaction temperature is 40 °C and the optimal pH is 4.0. This result is similar with what Liu and Yang have been described in the past (Liu and Yang 2007).
Owing to the complexity of the in vitro interaction between the mycoparasitic fungus and plant pathogenic fungi, we have studied the fungicidal activity of recombined aspartic protease by using the culture supernatant of the P. pastoris GS115 transformant. The concentrated culture supernatant was observed to possess different fungicidal activity against five pathogenic fungi, which indicated that aspartic protease TAASP could inhibit against pathogen fungi to some extent. The mechanism of difference on inhibition of mycelial growth against pathogenic fungi is unknown, however, it may be conjectured that although the basic protease plays a fundamental role in mycoparasitism towards different pathogens, the regulation seems to depend on cell wall components of the host (Suarez et al. 2005).
The stability and fungicidal activity of aspartic protease TAASP make it a promising alternative to control pathogenic fungi. In order to construct valuable industrial engineering strain of yeast, further research, based on this study, are proposed as follow: screening high expression transformants of aspartic protease; optimizing the fermentation process and conditions to increase the amount of enzyme production; combining with other enzymes for synergy effect to improve the inhibition on the growth of pathogens.
References
Chet I, Chernin L (2002) Biocontrol, microbial agents in soil. Wiley Online Library
Delgado-Jarana J, Pintor-Toro JA, Benítez T (2000) Overproduction of [beta]-1, 6-glucanase in Trichoderma harzianum is controlled by extracellular acidic proteases and pH. BBA-Protein Struct M 1481(2):289–296
Delgado-Jarana J, Rincon AM, Benitez T (2002) Aspartyl protease from Trichoderma harzianum CECT 2413: cloning and characterization. Microbiol 148:1305–1315
Elad Y, Kapat A (1999) The role of Trichoderma harzianum protease in the biocontrol of Botrytis cinerea. Eur J Plant Pathol 105(2):177–189. doi:10.1023/a:1008753629207
Grinyer J, Hunt S, McKay M, Herbert BR, Nevalainen H (2005) Proteomic response of the biological control fungus Trichoderma atroviride to growth on the cell walls of Rhizoctonia solani. Curr Genet 47(6):381–388
Harman GE, Howell CR, Viterbo A, Chet I, Lorito M (2004) Trichoderma species—opportunistic, avirulent plant symbionts. Nat Rev Microbiol 2(1):43–56
Jones RW, Hancock JG (1987) Conversion of viridin to viridiol by viridin-producing fungi. Can J Microbiol 33(11):963–966
Kapteyn JC, Montijn RC, Vink E, delaCruz J, Llobell A, Douwes JE, Shimoi H, Lipke PN, Klis FM (1996) Retention of Saccharomyces cerevisiae cell wall proteins through a phosphodiester-linked beta-1,3-/beta-1,6-glucan heteropolymer. Glycobiology 6(3):337–345. doi:10.1093/glycob/6.3.337
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227(5259):680–685
Liu Y, Yang Q (2007) Cloning and heterologous expression of aspartic protease SA76 related to biocontrol in Trichoderma harzianum. FEMS Microbiol Lett 277(2):173–181. doi:10.1111/j.1574-6968.2007.00952.x
Liu ZH, Yang XX, Sun DM, Song JZ, Chen G, Juba O, Yang QA (2010) Expressed sequence tags-based identification of genes in a biocontrol strain Trichoderma asperellum. Mol Biol Rep 37(8):3673–3681. doi:10.1007/s11033-010-0019-0
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the folin phenol reagent. J Biol Chem 193:265–275
Maeda S (1989) Expression of foreign genes in insects using baculovirus vectors. Annu Rev Entomol 34(1):351–372
Masuda T, Tamaki S, Kaneko R, Wada R, Fujita Y, Mehta A, Kitabatake N (2004) Cloning, expression and characterization of recombinant sweet-protein thaumatin II using the methylotrophic yeast Pichia pastoris. Biotechnol Bioeng 85(7):761–769
Olmedo-Monfil V, Mendoza–Mendoza A, Gomez I, Cortes C, Herrera-Estrella A (2002) Multiple environmental signals determine the transcriptional activation of the mycoparasitism related gene prb1 in Trichoderma atroviride. Mol Genet Genomics 267(6):703–712. doi:10.1007/s00438-002-0703-4
Pastor F, Pujol X, Blanco A, Vidal T, Torres A, Diaz P (2001) Molecular cloning and characterization of a multidomain endoglucanase from Paenibacillus sp BP-23: evaluation of its performance in pulp refining. Appl Microbiol Biotechnol 55(1):61–68
Pozo MJ, Baek JM, Garcia JM, Kenerley CM (2004) Functional analysis of tvsp1, a serine protease-encoding gene in the biocontrol agent Trichoderma virens. Fungal Genet Biol 41(3):336–348. doi:10.1016/j.fgb.2003.11.002
Schuster A, Schmoll M (2010) Biology and biotechnology of Trichoderma. Appl Microbiol Biotechnol 87(3):787–799. doi:10.1007/s00253-010-2632-1
Suarez B, Rey M, Castillo P, Monte E, Llobell A (2004) Isolation and characterization of PRA1, a trypsin-like protease from the biocontrol agent Trichoderma harzianum CECT 2413 displaying nematicidal activity. Appl Microbiol Biotechnol 65(1):46–55. doi:10.1007/s00253-004-1610-x
Suarez MB, Sanz L, Chamorro MI, Rey M, Gonzalez FJ, Llobell A, Monte E (2005) Proteomic analysis of secreted proteins from Trichoderma harzianum—identification of a fungal cell wall-induced aspartic protease. Fungal Genet Biol 42(11):924–934. doi:10.1016/j.fgb.2005.08.002
Szekeres A, Kredics L, Antal Z, Kevei F, Manczinger L (2004) Isolation and characterization of protease overproducing mutants of Trichoderma harzianum. FEMS Microbiol Lett 233(2):215–222
Viterbo A, Harel M, Chet I (2004) Isolation of two aspartyl proteases from Trichoderma asperellum expressed during colonization of cucumber roots. FEMS Microbiol Lett 238(1):151–158. doi:10.1016/j.femsle.2004.07.028
Waterham HR, Digan ME, Koutz PJ, Lair SV, Cregg JM (1997) Isolation of the Pichia pastoris glyceraldehyde-3-phosphate dehydrogenase gene and regulation and use of its promoter. Gene 186(1):37–44
Wessels JGH (1986) Cell-wall synthesis in apical hyphal growth. Int Rev Cytol 104:37–79
Yan L, Qian Y (2009) Cloning and heterologous expression of SS10, a subtilisin-like protease displaying antifungal activity from Trichoderma harzianum. Fems Microbiol Lett 290(1). doi:10.1111/j.1574-6968.2008.01403.x
Acknowledgments
The technical assistance of Northeast Forestry University by Zhihua Liu, Harbin, is appreciated by the authors. This work is partially sponsored by the Natural Science Foundation of Heilongjiang Province, China (Grant No. C201118) and National High-tech and Program of China (Grant No. 2011AA10A205).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yang, X., Cong, H., Song, J. et al. Heterologous expression of an aspartic protease gene from biocontrol fungus Trichoderma asperellum in Pichia pastoris . World J Microbiol Biotechnol 29, 2087–2094 (2013). https://doi.org/10.1007/s11274-013-1373-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11274-013-1373-6