Abstract
Bacteria associated with eight field-collected and five cultured soft corals of Briareum sp., Sinularia sp., Sarcophyton sp., Nephtheidae sp., and Lobophytum sp. were screened for their abilities in producing antimicrobial metabolites. Field-collected coral samples were collected from Nanwan Bay in southern Taiwan. Cultured corals were collected from the cultivating tank at National Museum of Marine Biology and Aquarium. A total of 1,526 and 1,138 culturable, heterotrophic bacteria were isolated from wild and cultured corals, respectively; seawater requirement and antimicrobial activity were then assessed. There is no significant difference between the ratio of seawater-requiring bacteria on the wild and cultured corals. The ratio of antibiotic-producing bacteria within the seawater-requiring bacteria did not differ between the corals. Nineteen bacterial strains that showed high antimicrobial activity were selected for 16S rDNA sequencing. Three strains could be assigned at the family level (Rhodobacteraceae). The remaining 16 strains belong to eight genera: Marinobacterium (2 strains), Pseudoalteromonas (1), Vibrio (5), Enterovibrio (1), Tateyamaria (1), Labrenzia (2), and Pseudovibrio (4). The crude extract from bacteria strains CGH2XX was found to have high cytotoxicity against the cancer cell line HL-60 (IC50 = 0.94 μg/ml) and CCRF-CEM (IC50 = 1.19 μg/ml). Our results demonstrate that the marine bacteria from corals have great potential in the discovery of useful medical molecules.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
An increase of bacterial resistance to existing antibiotics has led to the search for new drugs, especially antibiotics. Bioactive compounds are traditionally screened from terrestrial microorganisms. However, the opportunity of finding novel antibiotics from terrestrial microorganisms has diminished (Hong et al. 2009a). The environments between oceanic and terrestrial systems are very different. As such, marine microorganisms may produce bioactive compounds not found in terrestrial habitats (Penesyan et al. 2009). It has been reported that marine invertebrates harbor a higher population of bacteria able to produce novel bioactive compounds (Burgess et al. 1999; Nithyanand et al. 2011). Previous studies on microbes associated with sponges have revealed that some bacteria have a great potential for producing useful natural products (Li 2009). However, coral-associated microbes and their produced metabolites have received little attention (Li 2009). Thus, coral reef ecosystems could be a largely unexplored source for marine microorganisms producing bioactive compounds.
Like sponges, many microbes, including bacteria (Rohwer et al. 2002; Chiou et al. 2010), fungi (Bentis et al. 2000; Wegley et al. 2007), and archaea (Kellogg 2004; Ferris et al. 1996), live in the skeleton, tissues, and surface mucus layer of corals. Some of the associations between corals and microbes are stable as well as species-specific and often distinct from the surrounding planktonic microorganisms (Rohwer et al. 2001; Rohwer et al. 2002). The functions of these coral-associated microorganisms are mostly unknown. Yet, it is suspected that they could play an important role in coral health, nutrition, and disease (Hong et al. 2009b).
Recently, several attempts have been made to culture and assay coral-associated isolates for the production of antimicrobial substances (Nissimov et al. 2009; Shnit-Orland and Kushmaro 2009; Radjasa et al. 2008; Lampert et al. 2006; Ritchie 2006). Nissimov et al. (2009) surveyed the antimicrobial activities of bacterial isolates from the mucus of coral Oculina patagonica and found that several bacterial isolates displayed antagonistic activities toward coral pathogens Vibrio shilonii, V. coralliilyticus, and Serratia marcescens. Another study has shown that corals also harbor diverse species of actinomycetes (Lampert et al. 2008; Lampert et al. 2006; Lombo et al. 2006) and some of these actinomycetes can inhibit the biofilm formation of Streptococcus pyogenes (Nithyanand et al. 2010). Reshef et al. (2006) suggested that these antibiotic-producing bacteria may play an important role in the first-line defense for coral against invading pathogens.
In this study, field-collected and cultured soft corals were used as a source for screening marine bacteria that exhibit antimicrobial activity using a culture-based method. The antimicrobial potentials of these bacteria were assessed and the difference of bacterial flora with antimicrobial activities between field-collected and cultured coral was compared. We hypothesized that the bacterial flora with antimicrobial activities would differ between the field-collected and cultured soft corals. The original objective of this study was primarily to isolate marine actinomycetes. However, only few actinomycetes were isolated and none of them was seawater-dependent. We, therefore, decided to study the antimicrobial activity of marine bacteria.
Materials and methods
Collection of coral samples
Thirteen coral samples, including five cultured corals and eight field-collected corals, were collected in this research (Table 1). The cultured soft corals were collected in January and March 2009 from an 80 ton cultivating tank located in the National Museum of Marine Biology and Aquarium (NMMBA) at 33 ‰ salinity, 25–26 °C. They included four soft coral genera (Briareum sp., Lobophytum sp., Nephtheidae sp., and Sarcophyton sp.). The field-collected soft corals were obtained in January, April, October, and November 2009 from the coast of Nanwan Bay (21°57′10.69″N, 120°46′8.29″E), Kenting National Park, southern Taiwan, at depths of about 10 m. The field-collected corals were composed of four soft coral genera (Sinularia sp., Sarcophyton sp., Briareum sp., and Nephtheidae sp.). All corals appeared to be healthy at the time of sampling except a cultured Briareum sp. The field-collected and cultured coral samples were kept at 4 °C after collection, transported to the laboratory, and processed within 4 h.
Culture of coral-associated bacteria
Approximately 2 g of soft coral was thoroughly washed twice with filtered (0.22 μm) seawater, ground with a sterilized mortar and pestle, and transferred to a 50-ml centrifuge tube. The soft coral was then vigorously vortexed for 3 min after adding 10 ml filtered seawater. The sample was serially diluted with seawater and spread on three different actinomycete-selective media: the M1 agar (M1): 10 g/l starch, 4 g/l yeast extract, 2 g/l peptone, and 15 g/l agar (Jensen et al. 2005); the Gause mineral agar (GH): 20 g/l starch, 1 g/l KNO2, 0.5 g/l K2HPO4, 0.5 g/l MgSO4·7H2O, 0.01 g/l FeSO4·7H2O, 0.05 g/l nystatin, 0.05 g/l cycloheximide, and 15 g/l agar (Ivanitskaia et al. 1978); and the glycerol asparagine agar (GA): 12.5 g/l glycerol, 1 g/l asparagine, 1 g/l K2HPO4, 1 g/l NaCl, 0.5 g/l MgSO4·7H2O, 0.01 g/l FeSO4·7H2O, 0.001 g/l CuSO4, 0.001 g/l MnSO4, 0.001 g/l ZnSO4, 0.05 g/l nystatin, 0.05 g/l cycloheximide, and 15 g/l agar (Dharmaraj and Sumantha 2009). All the above media were supplemented with 80 % of sea water. Plates were incubated at 25 °C for 5 days. Individual colonies were picked and streaked onto fresh M1 agar until pure cultures were obtained. For long-term preservation, cultures were grown at 25 °C in M1 broth and stored at −80 °C in 20 % glycerol solutions.
Detection of seawater requirement
All the isolated strains were streaked on M1 agar supplemented with 0 and 80 % (v/v) of seawater. After streaking the plates with test strains, the plates were incubated at 25 °C for 7 days. Isolates that only grow in M1 agar with 80 % seawater were designated as obligate marine bacteria and all the following biological activity screenings were conducted using these obligate marine bacteria.
Indicator microorganisms
Bacterial strains Vibrio parahaemolyticus (BCRC 10806) and Pseudomonas aeruginosa (BCRC 10303), as well as the fungal strain Candida albicans (BCRC 22903), were obtained from the Bioresource Collection and Research Center (BCRC; Hsinchu, Taiwan). Bacterial strain Vibrio shilonii (ATCC BAA-91) was obtained from American Type Culture Collection (ATCC; Manassas, VA, USA). Bacterial strains Staphylococcus aureus (ATCC 12600) and Escherichia coli (ATCC 11775) were provided by Dr. J. K. Liu, Department of Biological Sciences, National Sun Yat-sen University, Taiwan.
Indicator microbes were cultured as follows: V. parahaemolyticus, P. aeruginosa and V. shilonii were cultured in marine broth at 25 °C for 24 h and maintained on the marine agar. C. albicans was cultured in YM broth at 30 °C for 48 h and maintained on the YM agar. S. aureus and E. coli was cultured in LB broth at 37 °C for 18 h and maintained on the LB agar.
Determination of antimicrobial activity
The screening method consisted of two steps: primary and secondary screening. The primary screening was used to select the antimicrobial isolates against a Gram-negative strain (Vibrio shilonii), a Gram-positive strain (Staphylococcus aureus), and a eukaryotic fungus (Candida albicans). Isolates that exhibited an inhibition zone >1 mm in diameter against at least one test strain in the primary screening were selected for secondary screening against six pathogens E. coli, V. parahaemolyticus, P. aeruginosa, V. shilonii, S. aureus, and C. albicans.
An agar block method (Stern et al. 2006) was used for the primary and secondary screening: each coral isolate was spread evenly on the surface of M1 agar plate, and incubated for 3–5 days at 25 °C. Cylindrical agar blocks (8-mm diameter) were excised from well-growth agar. The agar blocks were placed directly on the top of a new agar plate freshly spread with a fixed amount of indicator microorganisms (108 cells/ml). The agar plate containing the agar blocks and newly inoculated indicator microorganism was incubated for 24 h at the proper temperature for the indicator microorganism. The antibacterial activity was measured in terms of diameter of the inhibition zone with triplication.
Crude extract preparation
Marine isolates were cultured in 500-ml flasks containing 100 ml M1 medium with 80 % seawater. Flasks were incubated at 25 °C on a rotatory shaker at 150 rpm. After 5 days of incubation, the fermented broths were extracted twice with ethyl acetate (100 ml × 2). The solvent extracts were combined and evaporated to dryness under vacuum. The extracts obtained were weighed and stored at −20 °C. The extracts were then used for cytotoxic activity assays.
Detection of cytotoxic activity
Cell lines CCRF-CEM (human T lymphoblastic leukemia), HL-60 (human promyelocytic leukemia) and normal PBMC (normal peripheral blood mononuclear cells) were purchased from the ATCC. Cells were maintained in RPMI 1640 medium supplemented with 10 % fetal calf serum, 2 mM glutamine, and antibiotics (100 units/ml penicillin and 100 μg/ml streptomycin) at 37 °C in a humidified atmosphere of 5 % CO2.
The cytotoxic activity of the crude extract was assessed by a colorimetric method using 3-(4, 5-dimethyl-thiazol-2-yl)-2, 5-diphenyltetra-zolium bromide (MTT; Lu et al. 2009).
Bacterial growth in different media
About 2 g of cultured Briareum sp. was washed and crushed as mentioned before. The coral sample was then serially diluted with seawater and spread on four different media: marine agar, M1, GH, and GA. After incubation for 5 days at 25 °C, the bacterial colonies that developed on the plates were counted and numbers were expressed in colony-forming unit (CFU).
16S rRNA gene sequencing
Nineteen bacterial isolates that showed significant antimicrobial activity were identified by their 16S rRNA gene sequence. 16S rRNA fragment of each strain was amplified by using the universal 16S rRNA bacterial primer pair 27F (5′-GAGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-CTACGGCTACCTTGTTACGA-3′). Sequencing was performed by Tri-I Biotech (Taipei, Taiwan).
Phylogenetic analysis
The nucleotide sequences of the 16S rRNA gene fragments were first compared with those in GenBank databases using the basic local alignment search tool (BLAST; Altschul et al. 1990) algorithm to identify the closest bacterial neighbors. To build the phylogenetic tree, sequences were aligned using the multiple sequence alignment program from MEGA 4 program (Tamura et al. 2007) using the ClustalW method (Thompson et al. 1994). A neighbor-joining tree (Saitou and Nei 1987) was constructed with Kimura’s two-parameter model (Kimura 1980) using the MEGA 4 program. The statistical significance of tree branches was evaluated by bootstrap analysis involving the construction of 1,000 trees from randomly resampled data.
Statistics
Student’s t tests and Mann–Whitney U tests were performed with the software SPSS version 12.0. In order to make the data set a better approximation of normal distribution, the percentage of bacterial isolates with seawater requirement and antimicrobial activity was converted into an arcsin value before performing the Student’s t test and Mann–Whitney U test.
All results from biological activity assays were expressed as mean ± standard error.
Nucleotide sequence accession numbers
The nucleotide sequences obtained in this study have been deposited in the GenBank database under the accession numbers from JQ342679 to JQ342697.
Results
Sample collection, bacteria isolation and seawater requirement assay
It is important to ensure that the isolates were truly coral-associated microbes and are not due to contamination from the land. We estimated the degree of terrestrial microorganism contamination in the marine corals by calculating the percentage of seawater tolerated isolated out of the total isolated. We assumed that a low percentage of seawater-sensitive bacteria in a sample could be indicative of low contamination from terrestrial bacteria.
Using three different culture media, a total of 2,664 heterotrophic bacteria were isolated from thirteen coral samples and of these 1,138 and 1,526 bacterial strains were from cultured (sample A-E) and field-collected (sample F-M) coral samples, respectively (Table 1).
The number and percentage of isolates which showed seawater requirement for each species of coral are also presented in Table 1. Among all the isolated strains (2,664), 76.2 % (2,030) isolates had a seawater requirement. Isolates from field-collected Briareum sp. (sample F) had the highest percentage of seawater-requiring bacteria (96.81 %), while isolates from diseased cultured Briareum sp. (sample B.) had the lowest percentage of seawater-requiring bacteria (33.43 %). Among isolates from field-collected corals, isolates from Sinularia sp. (sample L) had the lowest percentage of seawater-requiring bacteria (62.75 %), while isolates from Nephtheidae sp. (sample D) had the highest percentage of seawater-requiring bacteria (82.81 %) among isolates from cultured corals.
The mean values and standard errors of the percentage of seawater-requiring isolates for each coral sample observed from cultured and field-collected coral sample were 66.27 ± 8.51 % and 77.94 ± 4.60 %, respectively. There were no differences in the percentage of seawater-requiring bacteria between cultured (n = 4, excluding sample B) and field-collected coral isolates (Student’s t test; P > 0.05). However, the total average of the percentage of seawater-requiring isolates for field-collected corals (88.47 %) was higher than for cultured corals (72.02 %).
Antimicrobial activity assay
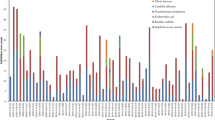
All isolates that showed a seawater requirement were subsequently screened for their antimicrobial activity. The antimicrobial test was carried out in primary screening by agar block methods against three pathogens Vibrio shilonii, Staphylococcus aureus, and Candida albicans (Fig. 1). For each coral sample, the percentage of isolates displayed antimicrobial activity and the percentage of isolates against the three indictor bacteria are shown in Table 2. Among all the tested isolates (2,030), 6.55 % (133) of the isolates showed antimicrobial activity against one or more of the indicator microorganisms. 4.68 % (95) strains showed activity against V. shilonii, 3.82 % (78) against S. aureus, and 1.97 % (40) against both V. shilonii as well as S. aureus. No antimicrobial activity against fugal strain C. albicans was observed in any of the coral isolates. The highest and the second highest percentage of isolates displaying antimicrobial activity were observed in Sarcophyton sp. (sample J, 84.62 %) and Nephtheidae sp. (sample G, 20.29 %). Among the cultured corals, isolates from sample A (Briareum sp., 3.28 %) had the maximum percentage of isolates showing antimicrobial activity. No evidence of antimicrobial activity was found in the cultured corals Nephtheidae sp. (sample D) and Sarcophyton sp. (sample E), as well as the field-collected corals Sarcophyton sp. (sample H) and Sinularia sp. (sample L).
The mean values and standard error of the percentage of antimicrobial isolates for each coral sample observed from each cultured and field-collected coral sample were 1.53 ± 0.79 % and 15.20 ± 10.20 %, respectively. A Mann–Whitney U test indicated that there was no difference in the percentage of antimicrobial bacteria between cultured (n = 4, excluding sample B) and field-collected isolates (n = 8; P > 0.05). However, the total average of the percentage of antimicrobial isolates for field-collected corals (8.6 %) was higher than for cultured corals (2.50 %).
To further confirm and analyze the distribution of anti-bacteria activity, 66 isolates that showed an inhibition zone > 1-mm diameter against at least one test strain were selected for secondary screening using agar block methods against six pathogens E. coli, V. parahaemolyticus, P. aeruginosa, V. shilonii, S. aureus, and C. albicans and the results were shown in Fig. 2 as well as Fig. 3. The results indicated that 80.30 % of these isolates possessed inhibitory activities against more than one indicator organisms (Fig. 2). 21.21 % inhibited Gram-positive bacteria (S. aureus), 92.42 % inhibited Gram-negative bacteria (E. coli, V. parahaemolyticus, V. shilonii and/or P. aeruginosa), and 13.64 % inhibited both Gram-positive and Gram-negative bacteria. None of the isolates exhibited anti-C. albicans activity. Four isolates (6.06 %) were able to inhibit more than three indicator organisms. Most of antibiotic producer were isolated from the field-collected corals (87.88 %), such as Briareum sp. (50.00 %, sample L), Nephtheidae sp. (18.18 %, sample G), and Sarcophyton sp. (16.67 %, sample J).
Distribution of the diameter of inhibition zone size for bacterial strains isolated from soft corals against five indicator bacteria using agar block method. The first letter of isolate indicates the coral sample (as in Table 1)
The percentage of susceptibility of E. coli, S. aureus, V. parahaemolyticus, V. shilonii, and P. aeruginosa to the 66 coral isolates were 7.58, 21.21, 81.82, 71.21, and 59.09 % (Fig. 3), respectively. The percentage of susceptibility to the 66 coral isolates with inhibition zone diameters >4 mm were 3.03, 4.55, 12.12, 16.67, and 10.61 % for the aforesaid order of indicator bacteria. 19 isolates that showed an inhibition zone >4-mm diameter against at least one test strain or were able to inhibit at least four indicator organisms in secondary screening were selected for cytotoxic activity assays.
Cytotoxic activity assay
The cytotoxic activities of the crude extracts of all 19 isolates against two human cancer cell lines were tested (Table 3). Six isolates (31.6 %) were found to exhibit cytotoxic activity against the HL-60 tumor cell line, among which three of them also exhibited cytotoxic activity on CCRF-CEM cells.
The crude extract of a potent strain CGH2XX revealed high cytotoxic activity with IC50 value 0.94 ± 0.59 and 1.19 ± 1.08 μg/ml against HL-60 and CCRF-CEM cell, respectively. Given that the extract of this isolate also inhibited the growth of a broad spectrum of bacteria (Fig. 2), the extract of this isolate was further tested for cytotoxic activity against normal PBMC. The IC50 value of CGH2XX extract against normal PBMC was 243.7 ± 87.7 μg/ml and the stimulation index (SI) against HL-60 and CCRF-CEM cell was 259 and 204, respectively. The high stimulation index suggests that crude extract of CGH2XX has potential for antitumor treatment.
Comparison of bacterial growth in different media
We also compared the number of bacteria grown on marine agar, M1, GH, and GA, using cultured Briareum sp. as test sample. The results were expressed in CFU/g Briareum sp. (means ± standard errors of the means): 2.38 × 104 ± 0.18 × 104, 1.94 × 104 ± 0.44 × 104, 1.89 × 104 ± 0.41 × 103, and 3.09 × 103 ± 0.41 × 103 for the aforesaid order of media. The ratio of CFU/g Briareum sp. among the above media was 1: 0.815: 0.795: 0.130, respectively.
Identification of antibiotic producing bioactive bacterial isolates
16S rRNA genes sequences of the 19 isolates that exhibited high antimicrobial activities were sequenced and subsequently aligned to construct a phylogenetic tree (Fig. 4), based on around 650 bp sequences. These 19 selected isolates are, therefore, neither a representative sample of culturable bacteria nor a representative sample of culturable bacteria with antimicrobial activities of the coral. 19 isolates are belonging to the phylum Proteobacteria, among which 10 of them are of class Alphaproteobacteria whereas the remaining nine are of class Gammaproteobacteria (Fig. 4).
Phylogenetic tree of 19 selected bacterial strains isolated from the soft corals. The tree was constructed from a comparison of an approximately 650-bp region of the 16S rRNA gene sequence using the neighbor-joining analysis of a distance matrix with Kimura’s two-parameter model. Bootstrap values (expressed as percentages of 100 replications) more than 75 % are shown at branch points. The 16S rRNA sequence of Methanocaldococcus jannaschii DSM 2661 were used as outgroup. The scale bar represents 0.05 substitutions per nucleotide position
In addition, we also used Ribosomal Data Project (RDP) classifier tool (Cole et al. 2009) to classify bacterial based on their 16S rRNA sequences (Fig. 5). Among these 19 isolates, three isolates could not be classified to a known genus, but they are likely belonging to the family of Rhodobacteraceae (Fig. 4). The other 16 isolates belong to eight genera: Vibrio (5 isolates), Pseudovibrio (4), Marinobacterium (2), Labrenzia (2), Pseudoalteromonas (1), Enterovibrio (1), and Tateyamaria (1). Two genera, Vibrio and Pseudoalteromonas, were only isolated from cultured corals, whereas four genera, Enterovibrio, Marinobacterium, Labrenzia, and Tateyamaria, as well as three Rhodobacteraceae species were just isolated from field-collected corals. It is also interesting to note that all 10 Alphaproteobacteria species were all within family Rhodobacteraceae.
Discussion
Environmental factors, such as geographic and seasonal factors, have been reported to be some of the most important factors affecting the coral-associated bacterial community (Littman et al. 2009; Bourne et al. 2008). In this study, the culturable, heterotrophic bacteria inhabiting cultured and field-collected corals were compared using both the ratio of antibiotic and seawater-requiring isolates from cultured and field-collected corals as indices. Statistical analyses showed that there was no difference in the ratio of antibiotic and seawater-requiring isolates between cultured and field-collected corals, indicating that the difference in microbial communities associated with cultured and field-collected corals were negligible. Therefore, we conclude that both cultured and field-collected soft corals could be used as a source for screening microorganisms with bioactive natural products. However, this result is based only on two indirect index of microbial flora. As such, further studies are required to analyze culturable bacterial flora between cultured and field-collected corals.
Several marine microbes can produce bioactive compounds, and in this study the cultured and field-collected soft corals were successfully used as a source to screen for antibiotic-producing bacteria. Of a total of 2,030 isolates, 133 strains (6.55 %) showed antibiotic activity toward at least one indicator microbe used in the primary screening using agar block method. This observation is consistent with a previous study that 5.77 % of culturable bacteria isolated from the mucus of Oculina patagonica were antibiotic-producing strains (Nissimov et al. 2009). In contrast, the percentage of antibiotic-producing bacteria is lower than the observation by Shnit-Orland and Kushmaro (2009), in which about 25–27 % and 13 % of the bacterial isolates from the mucus of stony and soft coral possessed antimicrobial activity, respectively. Ritchie (2006) also showed that 20 % isolates from coral Acropora palmata were antibiotic producers. It was suggested that the percentage of antibiotic-producing strains can be affected by both the assay method as well as the species and the number of indicator microbes used in the screening (Long and Azam 2001; Shnit-Orland and Kushmaro 2009). For example, Gram-positive bacteria have been known to be more sensitive to antibiotic substances than Gram-negative bacteria (Nikaido 1996). Therefore, using more Gram-positive bacteria as indicator bacteria may lead to more antibiotic-producing bacteria. Nevertheless, our results suggest that soft coral can be a source for isolating marine bacteria producing antibiotics.
Several previous studies have supported the view that coral can be an untapped source of microbial diversity of economic importance (Shnit-Orland and Kushmaro 2009; Ritchie 2006). Therefore, evaluation of screening strategy, such as screening seawater dependent bacteria, may enhance the recovery rate of novel marine bacteria and improve our current understanding of antibiotic-producing coral bacteria (Mincer et al. 2002). A large fraction (total average 76.2 %) of our bacterial isolates displayed an obligate requirement of seawater for growth (Table 1). The requirement of seawater for bacterial growth suggests a high degree of marine adaptation or marine origin, and possibly little contamination from terrestrial sources. A previous study found that the percentage of seawater requiring bacteria were 57.7 % for isolates from the sponge Haliclona simulans (Kennedy et al. 2008), which is lower than our observations. However, it is interesting to note that our results agree with previous studies of seawater requirement of Gram-positive bacteria (about 82 %) in near-shore tropical marine samples (Jensen and Fenical 1995). Interestingly, the percentage of seawater-requiring bacteria (33.43 %) from cultured Briareum sp. (sample B.) was found to be significantly lower than all the other coral (in Table 1). This result may have been the result of this coral species being diseased and suggests that there may be a large disparity concerning culturable bacterial flora between healthy and diseased coral, which agree with several previous studies that bacterial community of diseased or bleaching coral was significantly different from healthy coral (Koren and Rosenberg 2008; Luna et al. 2007).
The original objective of this study was to isolate marine bacteria with a focus on marine actinomycetes with antimicrobial activity from soft coral. Therefore, three actinomycete-selective media, M1, GH, and GA, were used to culture actinomycetes. However, we found that only few actinomycetal strains were cultured from soft coral, and none of them were seawater-requiring strains. Therefore, we decide to isolate bacteria from soft coral while still using these three media and compared them to the marine agar that is often used for marine bacteria isolation. Using cultured Briareum sp. as the sample, the ratios of CFU/g was 1: 0.815: 0.795: 0.130 among marine agar, M1, GH and GA, respective. The higher number of bacteria grown on M1 and GH indicates that they are both good media for isolation of bacteria with activity. However, it is possible that some species may behave differently among various media.
In Fig. 2, we found that the coral bacteria displayed different strength and spectra of activity against various indicator bacteria. The distribution of inhibition zone diameters for 66 antimicrobial bacterial strains from secondary screening (Fig. 3) showed a high percentage of inhibitive effects against pathogenic indicator bacteria from the ocean, namely Vibrio parahaemolyticus, Pseudomonas aeruginosa and Vibrio shilonii, indicating the production of activities to inhibit the growth of specific marine microbial competitors. It is interesting to note that Vibrio shilonii has been shown to be a coral bleaching pathogen (Kushmaro et al. 1998) and are found in association with different coral species (Chimetto et al. 2008). All the above results suggest that the interactions between coral bacteria as well as host and coral bacteria could be diverse and complicated and different coral bacteria may contribute differently to the protection of the coral from marine pathogens (Nissimov et al. 2009). As pointed out by Shnit-Orland and Kushmaro (2009), these antibiotic-producing bacteria may play an important role in the first-line defense for coral against invading pathogens.
16S rRNA-based phylogenetic analysis was used to examine whether members of certain phylogenetic groups were more likely to possess antimicrobial activities. Our results showed that all of the antimicrobial isolates obtained from soft coral belonged to the phylum Proteobacteria, namely Alphaproteobacteria (53.6 %) and Gammaproteobacteria (47.4 %). Several previous studies have displayed the predominance of Alphaproteobacteria and Gammaproteobacteria antimicrobial isolates from corals. For example, Nissimov et al. (2009) reported that production of antibiotics by marine isolates from the mucus of stony coral Oculina patagonica is due to Alphaproteobacteria (55.6 %) and Gammaproteobacteria (44.4 %). Comparing with several earlier works (Radjasa et al. 2008; Shnit-Orland and Kushmaro 2009), the bacterial diversity was low in this study. We think this is very possible the cost of screening antimicrobial bacteria that meet seawater requirement (Jensen and Fenical 1995). Another possible reason of this is that we only selected bacteria with strong antimicrobial activity for phylogenetic analysis.
Among all the samples, isolates from field-collected Sarcophyton sp. (sample L) displayed the highest percentage (82.81 %) of antimicrobial activity in the primary screening (Table 2). However, high variations in percentage of isolates displaying antimicrobial activity were observed for Sarcophyton sp. (sample E, H, I, and J) suggesting more test is need to confirm the percentage of antimicrobial isolates cultured from Sarcophyton sp. On the other hand, isolates from Briareum samples (except sample B) displayed high percentage of antimicrobial activity revealing that Briareum is a good source of antibiotic-producing bacteria.
For the isolate CGH2XX, its 16S rRNA gene BLAST results showed 98.3 % identity with Pseudoalteromonas sp. H02P24-23 (Genbank accession no. HQ161380). The phylogenetic tree also exhibited a similar relationship. The crude extracts from this red-pigmented isolate were found to have antimicrobial activity against three indicator microbes and cytotoxic activity against two cancer cells, plus low cytotoxic to normal PBMC. Pseudoalteromonas species have been known to develop metabolic pathways that produce biological compounds and antifouling agents (Wilson et al. 2010). In previous culture-based studies, Nissimov et al. (2009) as well as Shnit-Orland and Kushmaro (2009) isolated a number of Pseudoalteromonas species exhibiting antimicrobial activity from the mucus of coral species Oculina patagonica as well as Pocillopora sp., Platygyra sp., Stylophora sp., and Porites sp., respectively. Therefore, this bacterium is a good candidate for further natural product isolation and characterization research. In fact, we already isolated a new compound from the culture broth of CGH2XX (Chen et al. 2012).
Based on these results, we conclude that both cultured and field-collected soft coral could be used for discovery of marine bacteria producing new antimicrobial agents. Also, our study demonstrated that several bacterial strains isolated from soft coral demonstrated antimicrobial activity, suggesting that soft coral can be a source of natural antimicrobial agents; and these antibiotic-producing bacteria may also play an important role in the ecological interaction between bacteria and their eukaryotic host. Following studies with these bioactive isolates will focus on natural product isolation and chemical structure identification in order to unravel their biotechnological potential.
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Bentis C, Kaufman L, Golubic S (2000) Endolithic fungi in reef-building corals (Order: Scleractinia) are common, cosmopolitan, and potentially pathogenic. Biol Bull 198:254–260
Bourne D, Iida Y, Uthicke S, Smith-Keune C (2008) Changes in coral-associated microbial communities during a bleaching event. ISME J 2:350–363
Burgess JG, Jordan EM, Bregu M, Mearns-Spragg A, Boyd KG (1999) Microbial antagonism: a neglected avenue of natural products research. J Biotechnol 70:27–32
Chen YH, Lu MC, Chang YC, Hwang TL, Wang WH, Weng CF, Kuo J, Sung PJ (2012) Pseudoalteromone A: a novel bioactive ubiquinone from a marine bacterium Pseudoalteromonas sp. CGH2XX (Pseudoalteromonadaceae). Tetrahedron Lett 53:1675–1677
Chimetto LA, Brocchi M, Thompson CC, Martins RC, Ramos HR, Thompson FL (2008) Vibrios dominate as culturable nitrogen-fixing bacteria of the Brazilian coral Mussismilia hispida. Syst Appl Microbiol 31:312–319
Chiou SF, Kuo J, Wongd TY, Fan TY, Tew KS, Liu JK (2010) Analysis of the coral associated bacterial community structures in healthy and diseased corals from off-shore of southern Taiwan. J Environ Sci Health B 45:408–415
Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM (2009) The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37:D141–D145
Dharmaraj S, Sumantha A (2009) Bioactive potential of Streptomyces associated with marine sponges. World J Microbiol Biotechnol 25:1971–1979
Ferris MJ, Muyzer G, Ward DM (1996) Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl Environ Microbiol 62:340–346
Hong K, Gao A-H, Xie Q-Y, Gao HG, Zhuang L, Lin H-P, Yu H-P, Li J, Yao X-S, Goodfellow M, Ruan J-S (2009a) Actinomycetes for marine drug discovery isolated from mangrove soils and plants in China. Mar Drugs 7:24–44
Hong MJ, Yu YT, Chen CA, Chiang PW, Tang SL (2009b) Influence of species specificity and other factors on bacteria associated with the coral Stylophora pistillata in Taiwan. Appl Environ Microbiol 75:7797–7806
Ivanitskaia LP, Singal EM, Bibikova MV, Vostrov SN (1978) Directed isolation of Micromonospora generic cultures on a selective medium with gentamycin. Antibiotiki 23:690–692
Jensen PR, Fenical W (1995) The relative abundance and seawater requirements of gram-positive bacteria in near-shore tropical marine samples. Microb Ecol 29:249–257
Jensen PR, Gontang E, Mafnas C, Mincer TJ, Fenical W (2005) Culturable marine actinomycete diversity from tropical Pacific Ocean sediments. Environ Microbiol 7:1039–1048
Kellogg CA (2004) Tropical archaea: diversity associated with the surface microlayer of corals. Mar Ecol Prog Ser 273:81–88
Kennedy J, Baker P, Piper C, Cotter PD, Walsh M, Mooij MJ, Bourke MB, Rea MC, O’Connor PM, Ross RP, Hill C, O’Gara F, Marchesi JR, Dobson ADW (2008) Isolation and analysis of bacteria with antimicrobial activities from the marine sponge Haliclona simulans collected from Irish waters. Mar Biotechnol 11:384–396
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Koren O, Rosenberg E (2008) Bacteria associated with the bleached and cave coral Oculina patagonica. Microb Ecol 55:523–529
Kushmaro A, Rosenberg E, Fine F, Ben-Haim Y, Loya Y (1998) Effect of temperature on bleaching of the coral Oculina patagonica by Vibrio AK-1. Mar Ecol Prog Ser 171:131–137
Lampert Y, Kelman D, Dubinsky Z, Nitzan Y, Hill RT (2006) Diversity of culturable bacteria in the mucus of the Red Sea coral Fungia scutaria. FEMS Microbiol Ecol 58:99–108
Lampert Y, Kelman D, Nitzan Y, Dubinsky Z, Behar A, Hill RT (2008) Phylogenetic diversity of bacteria associated with the mucus of Red Sea corals. FEMS Microbiol Ecol 64:187–198
Li Z (2009) Advances in narine microbial symbionts in the China Sea and related pharmaceutical metabolites. Mar Drugs 7:113–129
Littman RA, Willis BL, Pfeffer C, Bourne DG (2009) Diversities of coral-associated bacteria differ with location, but not species, for three Acroporid corals on the Great Barrier Reef. FEMS Microbiol Ecol 68:152–163
Lombo F, Velasco A, Castro A, de la Calle F, Brana AF, Sanchez-Puelles JM, Mendez C, Salas JA (2006) Deciphering the biosynthesis pathway of the antitumor thiocoraline from a marine actinomycete and its expression in two Streptomyces species. Chem Bio Chem 7:366–376
Long RA, Azam F (2001) Antagonistic interactions among marine pelagic bacteria. Appl Environ Microbiol 67:4975–4983
Lu M-C, Du Y-C, Chuu J–J, Hwang S-L, Hsieh P-C, Hung C-S, Chang F-R, Wu Y-C (2009) Active extracts of wild fruiting bodies of Antrodia camphorata (EEAC) induce leukemia HL 60 cells apoptosis partially through histone hypoacetylation and synergistically promote anticancer effect of trichostatin A. Arch Toxicol 83:121–129
Luna GM, Biavasco F, Danovaro R (2007) Bacteria associated with the rapid tissue necrosis of stony corals. Environ Microbiol 9:1851–1857
Mincer TJ, Jensen PR, Kauffman CA, Fenical W (2002) Widespread and persistent populations of a major new marine actinomycete taxon in ocean sediments. Appl Environ Microbiol 68:5005–5011
Nikaido H (1996) Multidrug efflux pumps of gram-negative bacteria. J Bacteriol 178:5853–5859
Nissimov J, Rosenberg E, Munn CB (2009) Antimicrobial properties of resident coral mucus bacteria of Oculina patagonica. FEMS Microbiol Lett 292:210–215
Nithyanand P, Thenmozhi R, Rathna J, Pandian SK (2010) Inhibition of Streptococcus pyogenes biofilm formation by coral-associated actinomycetes. Curr Microbiol 60:454–460
Nithyanand P, Manju S, Karutha Pandian S (2011) Phylogenetic characterization of culturable actinomycetes associated with the mucus of the coral Acropora digitifera from Gulf of Mannar. FEMS Microbiol Lett 314:112–118
Penesyan A, Marshall-Jones Z, Holmstrom C, Kjelleberg S, Egan S (2009) Antimicrobial activity observed among cultured marine epiphytic bacteria reflects their potential as a source of new drugs. FEMS Microbiol Ecol 69:113–124
Radjasa OK, Wises J, Sabdono A, Imhoff JF (2008) Corals as source of bacteria with antimicrobial activity. J Coast Dev 11:121–130
Reshef L, Koren O, Loya Y, Zilber-Rosenberg I, Rosenberg E (2006) The coral probiotic hypothesis. Environ Microbiol 8:2068–2073
Ritchie KB (2006) Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar Ecol Prog Ser 322:1–14
Rohwer F, Breitbart M, Jara J, Azam F, Knowlton N (2001) Diversity of bacteria associated with the Caribbean coral Montastraea franksi. Coral Reefs 20:85–91
Rohwer F, Seguritan V, Azam F, Knowlton N (2002) Diversity and distribution of coral-associated bacteria. Mar Ecol Prog Ser 243:1–10
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Shnit-Orland M, Kushmaro A (2009) Coral mucus-associated bacteria: a possible first line of defense. FEMS Microbiol Ecol 67:371–380
Stern NJ, Svetoch EA, Eruslanov BV, Perelygin VV, Mitsevich EV, Mitsevich IP, Pokhilenko VD, Levchuk VP, Svetoch OE, Seal BS (2006) Isolation of a Lactobacillus salivarius strain and purification of Its bacteriocin, which is inhibitory to Campylobacter jejuni in the chicken gastrointestinal system. Antimicrob Agents Chemother 50:3111–3116
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Thompson JD, Higgins DG, Gibson TJ (1994) Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Wegley L, Edwards R, Rodriguez-Brito B, Liu H, Rohwer F (2007) Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environ Microbiol 9:2707–2719
Wilson GS, Raftos DA, Corrigan SL, Nair SV (2010) Diversity and antimicrobial activities of surface-attached marine bacteria from Sydney Harbour, Australia. Microbiol Res 165:300–311
Acknowledgments
This work was supported by intramural funding from the National Museum of Marine Biology and Aquarium.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chen, YH., Kuo, J., Sung, PJ. et al. Isolation of marine bacteria with antimicrobial activities from cultured and field-collected soft corals. World J Microbiol Biotechnol 28, 3269–3279 (2012). https://doi.org/10.1007/s11274-012-1138-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11274-012-1138-7