Abstract
Samples of the marine sponge Haliclona simulans were collected from Irish coastal waters, and bacteria were isolated from these samples. Phylogenetic analyses of the cultured isolates showed that four different bacterial phyla were represented; Bacteriodetes, Actinobacteria, Proteobacteria, and Firmicutes. The sponge bacterial isolates were assayed for the production of antimicrobial substances, and biological activities against Gram-positive and Gram-negative bacteria and fungi were demonstrated, with 50% of isolates showing antimicrobial activity against at least one of the test strains. Further testing showed that the antimicrobial activities extended to the important pathogens Pseudomonas aeruginosa, Clostridium difficile, multi-drug-resistant Staphylococcus aureus, and pathogenic yeast strains. The Actinomycetes were numerically the most abundant producers of antimicrobial activities, although activities were also noted from Bacilli and Pseudovibrio isolates. Surveys for the presence of potential antibiotic encoding polyketide synthase and nonribosomal peptide synthetase genes also revealed that genes for the biosynthesis of these secondary metabolites were present in most bacterial phyla but were particularly prevalent among the Actinobacteria and Proteobacteria. This study demonstrates that the culturable fraction of bacteria from the sponge H. simulans is diverse and appears to possess much potential as a source for the discovery of new medically relevant biological active agents.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Marine sponges are host to a wide range of microbes. The role of these diverse microbes in sponge biology varies from the digestion of microbes as a food source to mutualistic symbioses with the sponge. For mutualistic associations, the sponge is believed to provide shelter from predators, a substrate for colonization, access to sunlight for photosynthetic microbes, and a supply of nutrients. Proposed benefits to the sponge of microbial symbiotic associations include the provision to the sponge of photosynthates and the elimination of toxic metabolic by-products (Taylor et al. 2007). Bioactive metabolite-producing microbes are also believed to have a mutualistic symbiotic relationship with the sponge, with microbes producing metabolites that protect the sponge from microbial disease and predation.
The marine environment and sponges in particular are a rich source of new bioactive metabolites—over 200 novel metabolites were isolated from marine sponges in 2005 (Blunt et al. 2007). In Haliclona species, over 190 metabolites, with wide-ranging biological activities, have been isolated (Yu et al. 2006). These include compounds with anti-fouling (Devi et al. 1998; Sera et al. 2002), anti-microbial (Clark et al. 2001; Richelle-Maurer et al. 2001), anti-fungal (Barrett et al. 1996; Clark et al. 2001), anti-malarial (Sakai et al. 1986), and cytotoxic activities (Erickson et al. 1997; Rashid et al. 2000). The range of compounds isolated from Haliclona sp. include steroids, terpenoids, polyacetylenes, alakaloids, cyclic peptides, unsaturated fatty acids, and polyketides (Yu et al. 2006). The widespread isolation of typical microbial metabolites, such as polyketides, from sponges led to the hypothesis that these metabolites are in fact the products of microbial metabolism. The isolation of secondary metabolite-producing bacteria from sponges and of microbial secondary metabolism gene clusters from the metagenome of sponges has led to the general acceptance that these metabolites are, in many cases, the product of microbial symbionts and are not derived from the microbial diet of sponges (Fortman and Sherman 2005).
The microbiota of sponges is very diverse, and representatives from seven bacterial phyla have been isolated from sponges (Taylor et al. 2007). However, from culture-independent techniques, it is known that at least 16 bacterial phyla are known to be present in sponges (Taylor et al. 2007). For the isolation of secondary metabolite-producing bacteria from sponges, the focus of many researchers has been on genera known to be producers of secondary metabolites, in particular, the Actinobacteria (Jiang et al. 2007; Kim et al. 2005; Zhang et al. 2006). Culture-independent approaches have shown that Actinobacteria were abundant within the sponges Rhopaloeides odorabile and Xestospongia spp. (Webster et al. 2001; Montalvo et al. 2005), and culture-dependent approaches have likewise shown that Actinobacteria can be isolated from sponges but that the abundance and diversity of the cultivable actinobacterial community varies significantly among sponge species (Zhang et al. 2008). The sponges Hymeniacidon perleve (Zhang et al. 2006), Stelletta tenuis, and Halichondria rugosa (Zhang et al. 2008), all collected from the Yellow Sea, were found to contain abundant cultivable Actinobacteria, while sponges Reniochalina sp. and Craniella australiensis, collected from the same location, yielded much fewer Actinobacteria (Zhang et al. 2008). Similarly, a study of cultivable bacteria from Mediterranean sponges (Muscholl-Silberhorn et al. 2008) found significant numbers of cultivable Actinobacteria from two of the sponge species analyzed but few from the remaining eight sponge species.
Analyses of antibiotic activity and secondary metabolite production from sponge isolates have led to the isolation of bacteria that produce known bioactive metabolites such as manzamine (Hill et al. 2005) and rifamycin (Kim et al. 2006). Surveys of antimicrobial activities from sponge isolates have also shown that, while antimicrobial activities have been observed from Actinobacteria (Kim et al. 2005; Muscholl-Silberhorn et al. 2008), these activities can also be present in α- and γ-Proteobacteria and Bacilli (Thakur et al. 2005; Hentschel et al. 2001; Pabel et al. 2003). The microbiological richness of the sponge environment indicates that cultivation studies are highly relevant and are likely to result in the discovery of new microbes and novel metabolic pathways. The aims of this study were to determine both the diversity of the culturable bacterial component of Haliclona simulans and which of the isolated bacteria produced potentially useful antimicrobial substances.
Drug-resistant bacterial infections are growing in importance in both hospital and community settings; in 2005, there were over 19,000 methicillin-resistant Staphylococcus aureus (MRSA)-related deaths in the USA (US Center for Disease Control), and in the UK, MRSA-related deaths have risen sharply from 51 in 1993 to 1,652 in 2006 (UK Office for National Statistics). Similarly, deaths related to Clostridium difficile infections have also risen yearly from 1999 to 2006, with C. difficile being a factor in 6,480 deaths in the UK (UK Office for National Statistics). The alarming growth in C. difficile and MRSA infections and the spread of MRSA into community settings makes the search for new antibiotics increasingly relevant. Thus, this study also focused on the isolation and identification of bacteria from H. simulans with activity against both C. difficile and MRSA with the aim of discovering new clinically relevant biologically active agents.
Experimental Procedures
Collection of Sponges
Samples of H. simulans (class Demospongiae, order Haplosclerida, family Chalinidae) were collected by hand during scuba diving in Gurraig Sound Kilkieran Bay, Galway, on the west coast of Ireland, at a depth of 15 m (N 53°18.944′, W 09°40.140′). Whole sponge samples were stored in seawater at 4°C and transported to the laboratory.
Culture of Sponge-Associated Bacteria
Approximately 1 cm3 samples of sponge tissue were rinsed extensively with sterile artificial seawater, and the washed sponge tissue was cut into small pieces <1 mm3 with a sterile scalpel. The sponge tissue was further processed by grinding with 3 ml of sterile artificial seawater in a Potter-Elvehjem tissue grinder. The sponge extract was serially diluted in artificial seawater to 10−6, and 100 μl aliquots were plated out on to isolation media. Isolation media used were starch-yeast extract-peptone–seawater (SYP-SW) agar [1% (w/v) starch, 0.4% (w/v) yeast extract, 0.2% (w/v) peptone, 3.33% (w/v) artificial sea salts—Instant Ocean Brand, 1.5% (w/v) agar], Marine Agar [0.1% (w/v) yeast extract, 0.5% (w/v) tryptone, 3.33% (w/v) artificial sea salts—Instant Ocean Brand, 1.5% (w/v) agar), Modified Marine Agar (0.005% (w/v) yeast extract, 0.05% (w/v) tryptone, 0.01% (w/v) β-glycerol phosphate disodium salt, pentahydrate, 3.33% (w/v) artificial sea salts—Instant Ocean Brand, 1.5% (w/v) agar], Actinomycete Isolation Seawater Agar [2.2% (w/v) Difco Actinomycete Isolation Agar, 0.5% (v/v) glycerol, 3.33% (w/v) artificial sea salts—Instant Ocean Brand], and NaST21Cx (Magarvey et al. 2004). Amphotericin B (30 μg/mL) was added to inhibit the growth of fungi, and plates were made with and without nalidixic acid (25 μg/mL). Plates were incubated at 28°C for up to 8 weeks.
Colonies were picked from the isolation plates and re-streaked onto SYP-SW agar; pure cultures were obtained by re-streaking from single colonies, at least twice from plates that were visually judged to be from single cultures. For long-term preservation, cultures were grown at 28°C in SYP-SW broth, glycerol was added to 15%, and samples were frozen at −80°C. Spores from sporulating bacteria were also preserved by resuspension in 20% (v/v) glycerol and storage at −20°C.
Preparation of Culture Extracts and Antibiotic Discs
Seed cultures were prepared by inoculating colonies of sponge bacterial isolates into 5 ml SYP-SW broth and incubating at 28°C with shaking for 3 days. One millilitre of these seed cultures was used to inoculate 50 ml of SYP-SW broth in a 250-ml conical flask. These were incubated at 28°C with shaking for 10 days. For the preparation of extracts, bacterial cells were removed by centrifugation at 3,000×g for 15 min or by filtration if the centrifugation step did not result in the formation of a solid pellet. An aliquot (5 ml) of clarified culture broth was removed for direct analysis. To the remainder of the broth, 2 g of XAD-16 resin was added, and this suspension was shaken for 2 h to allow binding of hydrophobic compounds to the resins. The beads of resin were collected and washed with water. Adsorbed compounds were eluted from the resin sequentially with 50% methanol (5 ml), 100% methanol (5 ml), and 100% ethylacetate (5 ml). Cell extracts were prepared by re-suspension of bacterial cell pellets in 10 ml 50% methanol and shaking for 1 h with cell debris removed by centrifugation as above. All extracts were kept at room temperature in the dark. This procedure was scaled up to process 800 ml of culture broth. In this case, 2 × 400 ml cultures (in 1 l Erlenmeyer flasks) were each inoculated with 10 ml of seed culture and incubated at 28°C with shaking for 12 days. Bacterial cells were removed by filtration, and 30 g of XAD-16 resin was added to the culture broth. This suspension was shaken for 2 h to allow binding to the resin. The resin was collected in a column and washed with 100 ml of water. The extract was eluted sequentially with 50 ml 50% methanol, 50 ml 100% methanol, and 50 ml ethylacetate.
For the preparation of antibiotic disks, 100 or 200 μl aliquots of extract were spotted onto 13 mm diameter AA Discs (Whatman), and the discs were allowed to fully dry in the fume hood before applying to culture media.
Isolation of Genomic DNA from Isolates
Two procedures were used for the isolation of genomic DNA from bacterial isolates; bacterial cells from a 3-ml culture grown for 24–72 h in SYP-SW were processed using the Qiagen DNeasy blood and tissue kit, while for suspected Actinobacteria, a small-scale version of the cetyltrimethylammonium bromide procedure for the isolation of genomic DNA from Streptomycetes was used (Kieser et al. 2000).
Amplification of Partial 16S Ribosomal RNA, Polyketide Synthase, and Nonribosomal Peptide Synthase Gene Products
Genomic DNA was used as a template for polymerase chain reaction (PCR) amplification. For 16S ribosomal RNA (rRNA) gene amplification, primers 27f (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492r (5′-TACGGYTACCTTGTTACGACTT-3′) were used (Lane 1991). PCR was carried out under the following conditions: initial denaturation at 94°C for 3 min, followed by 30 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 2 min, with a final extension 72°C for 7 min. For polyketide synthase (PKS) PCR amplification, primers MDPQQRf (5′-RTRGAYCCNCAGCAICG-3′) and HGTGTr (5′-VGTNCCNGTGCCRTG-3′) (Kim et al. 2005) were used. For nonribosomal peptide synthase (NRPS) PCR amplification, primers DKF (5′GTGCCGGTNCCRTGNGYYTC-3′) and MTR (5′-GCNGGYGGYGCNTAYGTNCC-3′) were used (Neilan et al. 1999). Two conditions were used for both PKS and NRPS PCRs. For amplification of PKS and NRPS gene fragments from high GC content DNA (e.g., from Actinomycetes), 10% dimethyl sulfoxide was included in the reaction mixture, and the following conditions were used: initial denaturation at 95°C for 5 min, followed by ten cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 1.5 min, with the annealing temperature reduced by 2°C per cycle, followed by 30 cycles of 95°C for 30 s, 40°C for 30 s, and 72°C for 1.5 min, with a final extension 72°C for 7 min. For all other amplifications, the following conditions were used: initial denaturation at 95°C for 5 min, followed by ten cycles of 95°C for 30 s, 65°C for 30 s, and 72°C for 1.5 min, with the annealing temperature reduced by 2°C per cycle, followed by 30 cycles of 95°C for 30 s, 45°C for 30 s, and 72°C for 1.5 min, with a final extension 72°C for 7 min.
In all cases, the reaction mixture (50 μl) contained deoxyribonucleotide triphosphates (0.2 mM each), 1× reaction buffer (20 mM Tris pH 8.4, KCl 50 mM), MgCl2 (1.7 mM), primers (0.1 μM each), and Taq DNA polymerase (1.25 units; all PCR reagents were purchased from Bioline). PCR products were analyzed by agarose gel electrophoresis.
Sequencing and Phylogenetic Analysis of Partial 16S rRNA Genes
PCR products were purified using Qiaquick gel extraction kit (Qiagen) and were sequenced directly using primer 27f (sequencing was carried out by GATC Biotech, Konstanz, Germany). The 16S rRNA gene sequences were analyzed using BLAST (Altschul et al. 1997) to identify the closest bacterial neighbors. 16S rRNA gene sequences were aligned and a neighbor-joining phylogenetic tree constructed using MEGA 4 (Tamura et al. 2007). Bootstrap tests were performed 1,000 times using MEGA 4.
Cloning, Sequencing, and Phylogenetic Analysis of Partial PKS Genes
PCR products were purified using Qiaquick Gel Extraction Kit (Qiagen) and were subcloned using the Qiagen PCR Cloning Kit. Plasmids containing the PCR products were purified using the Qiaprep Spin Miniprep Kit and sequenced using the T7 promoter primer by GATC Biotech. DNA sequences were analyzed using BLAST and BLASTX analysis (Altschul et al. 1997).
Maintenance of Indicator Strains
Indicator strains for antimicrobial activity were Escherichia coli NCIMB12210; Bacillus subtilis 1A40; Candida glabrata CBS138; Listeria monocytogenes strains F2365 and EGDe; methicillin-resistant S. aureus strains ST544, ST528, ST295, and ST299; vancomycin-insensitive S. aureus (VISA) strains 35403, 35197, 32681, 32679, 32647, and 32682; heterogeneous vancomycin-insensitive S. aureus (hVISA) 22900; Streptococcus pneumoniae Tigr; Bacillus cereus FLP1; vancomycin-resistant enterococci (VRE) strains EC289, EC292, EC295, EC533, EC571, and EC618; C. difficile strains ATCC43593, DPC6357, DPC6360, DPC6361, DPC6356, DPC6353, DPC6355, DPC6507, DPC6537, DPC6538, DPC6539, and DPC6535.
Indicator strains were cultured as follows: E. coli and B. subtilis strains were grown in Luria–Bertani (LB) at 37°C. C. glabrata was grown in yeast-peptone-d-glucose agar (YPD). C. difficile isolates were maintained on Fastidious Anaerobic Agar (Lab M) containing 7.5% defibrinated horse blood and Reinforced Clostridium Medium (Merck) at 37°C. L. monocytogenes strains were grown in brain heart infusion (BHI) broth at 37°C. MRSA, VISA, and hVISA strains were incubated at 37°C in Mueller Hinton Broth aerobically. VRE strains were grown in BHI at 37°C. S. pneumoniae Tigr was cultured in BHI and grown anaerobically at 37°C. B. cereus was cultured in BHI and grown at 30°C. MRSA and VRE strains were provided by the British Society of Antimicrobial Chemotherapy. VISA and hVISA strains were sourced from the Bristol Centre for Antimicrobial Research and Evaluation. C. difficile strains were from the collection at Teagasc, Moorepark and are coded DPC XXXX. All other strains were from the University College Cork culture collection.
Deferred Antagonism Assays
Antimicrobial activity was assessed initially by deferred antagonism assays followed by disc diffusion assays in specific cases. For deferred antagonism assays, 5 μl volumes of the cultures to be tested were spotted on SYP-SW agar plates at 28°C until colonies were ∼0.5–1 cm in diameter (this varied from 2 to 6 days depending on the strain being assayed). For E. coli and B. subtilis, these were overlayed with 10 ml of LB soft agar (1.5% w/v) seeded with 50 μl of an overnight culture of the relevant target strain. For C. glabrata, these were overlayed with 10 ml of YPD soft agar (1.5% w/v) seeded with 50 μl of an overnight culture. For B. cereus, L. monocytogenes, S. pneumoniae, MRSA, and VISA strains, these were overlayed with 20 ml of BHI or Mueller–Hinton soft agar (1.5% w/v) seeded with 50 μl of an overnight culture of the relevant target strain. For Pseudomonas aeruginosa, 100 μl of overnight cultures of sponge bacteria were spotted on SYP-SW agar plates and incubated at 30°C for 3 days. Then the plates were overlaid with 5 ml of LB soft agar 0.5% (w/v) containing P. aeruginosa diluted to a final OD600 of 0.1. The plates were incubated overnight under optimal conditions and examined to identify instances where zones of inhibition were apparent.
Disc Diffusion Assays
Disc diffusion assays were performed with discs prepared as described above. The test strains were grown up overnight in the appropriate media before being diluted 1:100 in liquid broth to achieve a final concentration of ∼107 CFU ml−1. The culture was spread on the surface of the agar of choice using sterile glass beads, and the plate was left to air dry in a laminar flow cabinet for 20 min. Potential antimicrobial-containing discs were aseptically placed onto the surface of the inoculated agar plate. The plates were incubated for 18 to 36 h, and zones of growth inhibition were noted. Discs impregnated with methanol and ethyl acetate were also used for control purposes.
Detection of Surface-Attachment Inhibitors
Sponge isolates were inoculated into 10 ml SYP-SW broth and incubated at 30°C for 11 days with shaking. The broths were clarified by centrifugation (10 min at 4,500×g) to prepared clarified broth. An overnight culture of P. aeruginosa strain PAO1 was diluted to an OD600 of 0.25 in LB broth, and 100 μl of the diluted culture was used to inoculate wells of a 96 well plate (Sarstedt, USA). Clarified broths (100 μl) prepared from the different sponge bacteria were also added to the 96-well plate. Cells were allowed to adhere to the well statically at 37°C for 1 h. Subsequently, the wells were stained by the addition of 100 μl 0.5% (w/v) crystal violet for 15 min at room temperature. The plates were rinsed with water to remove unattached cells and allowed to dry, after which 250 μl of 96% ethanol was added to each well to dissolve the crystal violet. The optical density at 570 nm was measured to detect the relative amount of attached cells.
Results
Isolation of Bacteria
The total colony-forming units for each isolation media were compared 4 days after initial inoculation. Colonies were most abundant on SYP-SW media (∼2.4 × 106 g−1 sponge) with lowest titers on Actinomycete Isolation Seawater Agar (AIA-SW) (∼6 × 105 g−1 sponge). In general, the addition of nalidixic acid to the media resulted in a 50% reduction in the colony-forming units (CFU; CFUs could not be accurately counted on NaST21Cx). The maximum bacterial count of 2.4 × 106 g−1 sponge tissue is not inconsistent with bacterial estimates of >108 g−1 for high-microbial-abundance sponges or bacteriosponges, assuming that the culturable microbial component is <1% as is widely estimated for many environmental niches (Amann et al. 1995).
Phylogeny
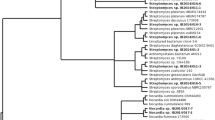
Bacteria were picked, and 52 isolates were selected for further analysis on the bases of diversity of colony color, size, and morphology, with particular emphasis on the isolation of sporulating or potentially filamentous bacteria for the isolation of Actinobacteria. The bacteria selected (Table 1) are not therefore a representative sample of culturable microbiota of the sponge. Phylogenetic analysis (Fig. 1) showed the isolated bacteria to be members of four bacterial phyla (γ-and α-Proteobacteria, Firmicutes, Actinobacteria, and Bacteroidetes) and 12 bacterial genera. Most common among the isolates were those grouping with the Streptomycetes (19), Pseudoalteromonads (12), Halomonads (5), Pseudovibrio (4), and Bacilli (3). The prevalence of actinobacterial Streptomycetes among the isolates is solely due to the selection of sporulating bacteria and does not imply that they are a dominant culturable group of bacteria. The γ-proteobacterial Pseudoalteromonads, however, would be less likely to be enriched by the selection process and are probably the single most dominant group of culturable bacteria from the sponge. Pseudoalteromonads were also found to be the most common cultivable group in a recent study of sponge-associated bacteria from the Norwegian coast (Dieckmann et al. 2005).
Evolutionary relationships of cultured sponge bacteria. The evolutionary history was inferred using the neighbor-joining method (Saitou and Nei 1987). The optimal tree with the sum of branch length = 0.97256429 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown next to the branches (Felsenstein 1985). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the maximum composite likelihood method (Tamura et al. 2004) and are in the units of the number of base substitutions per site. All positions containing gaps and missing data were eliminated from the dataset (complete deletion option). There were a total of 274 positions in the final dataset. Phylogenetic analyses were conducted in MEGA4 (Tamura et al. 2007)
Phylogenetic analysis, based on 16S rRNA gene alignments, showed that most of the bacterial isolates shared 99% identity with known species; however, one group of Streptomycete-related bacteria, SM15, and another Bacillus-related bacteria, BC3, shared only 97% identity with their closest neighbor by BLAST search.
Salt Preference
All the strains analyzed were isolated on media containing 100% artificial seawater. The seawater preference of all strains was analyzed, and it was found that 30 of the 52 strains had an absolute requirement for seawater. This requirement included all of the α-Proteobacteria and 17 out of 21 γ-Proteobacteria. Among the Actinobacteria, only four out of 20 had an absolute salt requirement, although an additional eight strains exhibited a strong preference for salt, taking into account effects on growth and sporulation (Table 1).
Bioactivity
All the isolated strains were initially tested for antimicrobial activity using agar plate overlay assays. The results (Table 2) showed that over 50% of the isolated strains showed antimicrobial activity against one or more of the test organisms. Analysis of individual bacterial phyla showed that the Actinobacteria and Firmicutes were the most likely to exhibit antimicrobial activity, with the Proteobacteria also a significant source. Fourteen of the strains tested displayed significant activity against P. aeruginosa, MRSA, VISA, or hVISA strains.
To allow further analysis of the biological activities, strains exhibiting strong or broad-spectrum antimicrobial activities were grown in liquid culture, and both broth and organic extracts were analyzed for biological activities (Table 3). Of 16 strains tested in this way, six exhibited antimicrobial activities in these assays with extracts from strains SM2 and SM4 showing high activity against a wide range of organisms. SM2 displayed activity against a number of different C. difficile (C. difficile DPC 6357, DPC6360, DPC6361, DPC 6356, DPC 6355, DPC6507, DPC6537, DPC6538, DPC6539, and DPC6535), VISA (VISA 35197, VISA35403, VISA 32681, and VISA32647), MRSA (ST528 and ST295), and VRE (EC292, EC, 295, EC571, and EC618) strains. Similarly, extracts from SM4 displayed activity against VISA (VISA 35197, VISA35403, and VISA 32647), MRSA (ST528 and ST295), and VRE (EC292, EC, 295, EC571, and EC618) strains.
The strains were also assayed for their ability to disrupt biofilm formation. Although most cleared supernatants of marine isolates increased surface attachment of P. aeruginosa, one strain, PM1 (genus Pseudomonas), was found to inhibit the surface attachment of P. aeruginosa strain PAO1.
Secondary Metabolism Genes
The presence of putative polyketide synthase and nonribosomal peptide synthase genes in the genomes of the cultured bacteria was analyzed using degenerate PCR. PCR products indicative of the presence of PKS (∼0.7 kb) and NRPS (∼1.0 kb) genes were detected in 29 and 15 of the cultured bacteria, respectively, a selection of which were subsequently cloned and sequenced. Both of these PCR products were amplified from 11 of the strains analyzed, implying that the genomes of these strains encoded both PKS and NRPS metabolic pathways. Of the 29 putative PKS-containing strains, 17 were Actinobacteria, and the remaining 12 were Proteobacteria. Of the 15 NRPS-containing strains, seven were Actinobacteria, four were Proteobacteria, three were Firmicutes (Bacilli), and one was a Bacteroidetes. Of the strains found to contain both putative NRPS and PKS genes, seven were Actinobacteria, and four were Proteobacteria. Of the four Pseudovibrio strain cultures, three were found to contain both putative NRPS and PKS genes.
To further investigate the apparent abundance of putative secondary metabolism genes among the Pseudovibrio, the PKS gene products from strains PV1, PV2, and PV4 were cloned and sequenced. Upon phylogenetic analysis, the DNA sequences formed two clusters. One of these clusters, represented by sequences FAS-PV1 and FAS-PV2, had close similarities to a group of sequences derived from bacteria cultured from unknown marine sponges (Fig. 2). The closest characterized sequences were a group of fatty acid synthases involved in capsular polysaccharide (KPS) production in the plant symbiotic and nitrogen-fixing bacteria α-Proteobacteria, Rhizobia (Parada et al. 2006).
Phylogenetic analysis of PKS sequences. The evolutionary history was inferred using the neighbor-joining method (Saitou and Nei 1987). The optimal tree with the sum of branch length = 3.84519292 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown next to the branches (Felsenstein 1985). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method (Zuckerkandl and Pauling 1965) and are in the units of the number of amino acid substitutions per site. All positions containing gaps and missing data were eliminated from the dataset (complete deletion option). There were a total of 224 positions in the final dataset. Phylogenetic analyses were conducted in MEGA4 (Tamura et al. 2007). (u. indicates uncultured)
The second group of DNA sequences from the Pseudovibrio, PKS-PV1, PKS-PV2, and PKS-PV4, was again found to be most similar to a group of PKS sequences derived from bacteria previously cultured from unknown marine sponges (Fig. 2). Among PKS genes of known function, the closest match was with the hybrid PKS/NRPS genes for the biosynthesis of the neurotoxic metabolite jamaicamide from Lyngbya majuscula (47% amino acid identity; Edwards et al. 2004).
PKS amplicons from the antibiotic-producing strains SM2 and SM4 were also cloned, sequenced, and analyzed. The PKS sequences from these strains formed two groups (Fig. 2). Both these groups, represented by PKS-SM2 and PKS-SM2/SM4, were found to have higher similarities to PKS genes from Cyanobacteria and Myxobacteria than to PKS genes from Streptomyces. The closest characterized PKS genes to both these clusters were involved in biosynthesis of the myxobacterial metabolite and electron-transport inhibitor, stigmatellin (Gaitatzis et al. 2002), with the closest characterized Streptomyces PKS being involved in the biosynthesis of chlorothricin (Jia et al. 2006).
Discussion
It is estimated that over 99% of marine sponge-associated microbes have yet to be cultivated, and previous metagenomic analysis of the H. simulans microbiota shows that the vast majority of the sponge-associated microbes were not detected by the culture methods described in this paper (Kennedy et al. 2008). Nevertheless, the bacteria isolated from the sponge were diverse; from a survey of 50 isolated strains, representatives from four bacterial phyla and 12 genera were found. Most of the isolated strains were closely related to other cultured bacteria, with a group of Streptomycetes (SM15) and a Bacillus species (BC3) being most divergent (at 97% identity) from isolated bacteria present in the GenBank database. Most bacterial groups found had previously been isolated from other marine sponges, with the two Bacteroidetes isolates (FB1 and CP1) being most distant from other microbes cultured from sponges, at 89% and 87% identity, respectively.
Comparison of the 16S rRNA gene sequences of the isolated bacteria with those found in a metagenomic analysis of the same sponge species sampled from the same site (Kennedy et al. 2008) revealed little overlap. The dominant γ-Proteobacteria operational taxonomical units (OTUs) found in the sponge metagenome were not among the cultured bacteria, and only two of the metagenomic OTUs had a cultured relative within the same genus (>95% identity of 16S rRNA gene). The Pseudovibrio 16S rRNA gene sequence, HAL4, found within the sponge metagenome was closely related (98–99% identity) to 16S rRNA genes from both the PV1 and PV4 Pseudovibrio isolates. Likewise the Pseudoalteromonas OTU, HAL-T-32, from the sponge metagenome was 99% identical to the PA1 and PA5 groups of Pseudoalteromonas isolates. The general lack of overlap between the cultured bacteria and those detected by metagenomic techniques is consistent with studies from other sponges (Webster and Hill 2001).
Bioactive Bacterial Isolates
The isolated bacteria were a very rich source of biological activities with over 50% of the microbes exhibiting anti-microbial activities in the initial plate overlay assays. It is also highly likely that this figure could be increased by increasing the number of test strains and conditions used; for instance, while this study found little activity from the Pseudoalteromonas isolates, a study of bacteria isolated from Mediterranean sponges found that all ten Pseudoalteromonas strains tested exhibited antimicrobial activity against a Gram-negative marine bacterium (Hentschel et al. 2001). Interestingly, none of these Pseudoalteromonas isolates exhibited activity against either E. coli or S. aureus, perhaps indicating the production of activities to inhibit the growth of specific marine microbial competitors.
The high proportion of Bacilli and Streptomycete isolates that demonstrated antimicrobial activity (19 out of 22) validated the approach taken to specifically target sporulating bacterial species for isolation. Both these Gram-positive genera are well-known producers of biologically active secondary metabolites, and genome surveys have revealed that bacteria from these genera can devote over 3% of their genome to secondary metabolite biosynthesis genes such as polyketide synthases and nonribosomal peptide synthases (Donadio et al. 2007). Twelve Streptomycete and one Bacillus strains were found to produce substances active against drug-resistant pathogenic bacteria.
The discovery that the little-known Pseudovibrio isolates (three out of four) were also a potent source of biologically active substances, including activities against MRSA, was more unexpected. This bacterial genus was first described in 2004 and has since been discovered associated with marine sponges, sea squirts, and tunicates (Fukunaga et al. 2006; Shieh et al. 2004; Sertan-de Guzman et al. 2007; Thiel and Imhoff 2003; Webster and Hill 2001; Hentschel et al. 2001). A Pseudovibrio sp. has also been found associated with sponge larvae, and it is thus one of the few verified vertically transmitted sponge symbionts (Enticknap et al. 2006). There is also a single published example of bioactive product formation from this genus with Pseudovibrio denitrificans strain Z143-1 producing the antibacterial and antimalarial red pigment heptylprodigiosin (Sertan-de Guzman et al. 2007). The antibiotic activities of the Pseudovibrio strains isolated in this study were not detected from strains grown in liquid culture, and conditions to optimize the production of these metabolites are currently under investigation.
A Pseudomonas species that inhibits surface attachment of P. aeruginosa was also identified. Inhibition of surface attachment and biofilm formation are important strategies for the development of new antibacterial therapeutics (Cegelski et al. 2008). Pseudomonas species are known to produce surface attachment inhibitors; for example, Pseudomonas putida produces the NRPS-derived cyclic lipopeptides, putisolvin I and II, which reduce surface tension, inhibit biofilm formation, and break down biofilms (Dubern et al. 2006). The identification and characterization of the surface-attachment inhibitor produced by this marine Pseudomonas isolate is currently being pursued.
PKS and NRPS Genes
Polyketides, nonribosomal peptides, and PK/NRP hybrid compounds are important classes of natural products and include many important drugs (e.g., the antibacterial drugs erythromycin, penicillin, and daptomycin; the hypocholesterol drug lovastatin; the anticancer drug doxorubicin; and the immunosuppressants cyclosporine, FK506, and rapamycin). The conserved domain structure of the polyketide synthase and nonribosomal peptide synthetase gene families enables their detection using a PCR-based approach. Indeed this approach has been used as a technique to pre-screen Streptomyces for their potential for secondary metabolite production (Ayuso et al. 2005; Ayuso-Sacido and Genilloud 2005). Sponges are also important sources of these classes of compounds, and while it is widely believed that microbial symbionts are the true producers of these compounds in marine sponges, it is unclear which microbes among the multitude of microorganisms associated with the sponges are responsible for their biosynthesis. We sought to survey the microbial isolates using degenerate PCR to determine which of the isolates had the potential to produce polyketide and nonribosomal peptide secondary metabolites. The results of this survey showed that, as expected, most Actinobacteria were found to contain either PKS or NRPS genes or both. Similarly, all three Bacilli were found to contain NRPS genes. More surprisingly, we found that five out of six α-Proteobacteria from the Genera Sulfitobacter and Pseudovibrio contained both NRPS and PKS genes, suggesting a high potential for secondary metabolite production among these organisms. Genomic analyses of α-Proteobacteria have detected relatively low levels of secondary metabolism genes in these bacteria with only ten of 23 genomes analyzed having PKS or NRPS genes present (Donadio et al. 2007). Bioactivity was detected from three of the four Pseudovibrio, but no activity was detected when these strains were grown in liquid culture. It has been reported previously that an antimicrobial activity associated with a Pseudovibrio strain was unstable (Muscholl-Silberhorn et al. 2008), possibly suggesting an extrachromosomal origin for the antimicrobial activity. Interestingly, large plasmids have been detected in Pseudovibrio isolated from sponge larvae (Enticknap et al. 2006). It is also interesting to note that the one Pseudovibrio isolate that lacked detectable biological activity, PV3, also tested negative for the presence of either PKS or NRPS genes. This strain was indistinguishable from bioactive producing PV4 by 16S rRNA gene analysis.
As the presence of PKS genes has previously not been reported from Pseudovibrio strains, the PKS amplicons were sequences to confirm their identity. This resulted in the characterization of one group of amplicons as PKS gene fragments belonging to the hybrid NRPS/PKS family, thus confirming Pseudovibrio as a novel source of PKS and NRPS genes. The second group of amplicons were related to a fatty acid synthase involved in the production of capsular polysaccharide. In Rhizobia, the production of capsular polysaccharide is linked to the ability to form a symbiotic relationship with the plant (Parada et al. 2006), and a similar mechanism could potentially be involved in a symbiotic relationship between the Pseudovibrio and the sponge. This genus has been consistently found associated with marine sponges and their larvae, often abundantly (Enticknap et al. 2006), and is believed to be a sponge true symbiont. The discovery of PKS and NRPS genes within this genus also suggests an important role for this organism in the production of sponge bioactive metabolites.
The extraction of wide-spectrum antibiotic activities from Streptomyces isolates SM2 and SM4 is further evidence that the culturable sponge microbiota is an important source of biologically active compounds. The compound(s) present within this extract have been shown to have activity against C. difficile, C. glabrata, and drug-resistant Enterococci and S. aureus strains among others. These strains were also found to contain novel PKS genes, of a type more closely related to those from Cyanobacteria and Myxobacteria than from Streptomyces. The identity of the active component(s) of this extract is currently being investigated. Thus, this study demonstrates that the culturable fraction of bacteria from the sponge H. simulans is quite diverse and appears to posses the potential to be a good source for the discovery of bacteria which produce novel clinically relevant biological active agents.
References
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Amann RI, Ludwig W, Schleifer KH (1995) Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59:143–169
Ayuso A, Clark D, Gonzalez I, Salazar O, Anderson A, Genilloud O (2005) A novel actinomycete strain de-replication approach based on the diversity of polyketide synthase and nonribosomal peptide synthetase biosynthetic pathways. Appl Microbiol Biotechnol 67:795–806
Ayuso-Sacido A, Genilloud O (2005) New PCR primers for the screening of NRPS and PKS-I systems in actinomycetes: detection and distribution of these biosynthetic gene sequences in major taxonomic groups. Microb Ecol 49:10–24
Barrett AG, Boys ML, Boehm TL (1996) Total synthesis of (+)-papuamine: an antifungal pentacyclic alkaloid from a marine sponge, Haliclona sp. J Org Chem 61:685–699
Blunt JW, Copp BR, Hu WP, Munro MH, Northcote PT, Prinsep MR (2007) Marine natural products. Nat Prod Rep 24:31–86
Cegelski L, Marshall GR, Eldridge GR, Hultgren SJ (2008) The biology and future prospects of antivirulence therapies. Nat Rev Microbiol 6:17–27
Clark RJ, Garson MJ, Hooper JN (2001) Antifungal alkyl amino alcohols from the tropical marine sponge Haliclona n. sp. J Nat Prod 64:1568–1571
Devi P, Vennam J, Naik CG, Parameshwaran PS, Raveendran TV, Yeshwant KS (1998) Antifouling activity of Indian marine invertebrates against the green mussel Perna viridis L. J Mar Biotechnol 6:229–232
Dieckmann R, Graeber I, Kaesler I, Szewzyk U, Von Dohren H (2005) Rapid screening and dereplication of bacterial isolates from marine sponges of the sula ridge by intact-cell-MALDI-TOF mass spectrometry (ICM-MS). Appl Microbiol Biotechnol 67:539–548
Donadio S, Monciardini P, Sosio M (2007) Polyketide synthases and nonribosomal peptide synthetases: the emerging view from bacterial genomics. Nat Prod Rep 24:1073–109
Dubern JF, Lugtenberg BJ, Bloemberg GV (2006) The ppuI-rsaL-ppuR quorum-sensing system regulates biofilm formation of Pseudomonas putida PCL1445 by controlling biosynthesis of the cyclic lipopeptides putisolvins I and II. J Bacteriol 188:2898–2906
Edwards DJ, Marquez BL, Nogle LM, Mcphail K, Goeger DE, Roberts MA, Gerwick WH (2004) Structure and biosynthesis of the jamaicamides, new mixed polyketide-peptide neurotoxins from the marine cyanobacterium Lyngbya majuscula. Chem Biol 11:817–833
Enticknap JJ, Kelly M, Peraud O, Hill RT (2006) Characterization of a culturable alphaproteobacterial symbiont common to many marine sponges and evidence for vertical transmission via sponge larvae. Appl Environ Microbiol 72:3724–3732
Erickson KL, Beutler JA, Cardellina IJ, Boyd MR (1997) Salicylihalamides A and B, novel cytotoxic macrolides from the marine sponge Haliclona sp. J Org Chem 62:8188–8192
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fortman JL, Sherman DH (2005) Utilizing the power of microbial genetics to bridge the gap between the promise and the application of marine natural products. Chembiochem 6:960–978
Fukunaga Y, Kurahashi M, Tanaka K, Yanagi K, Yokota A, Harayama S (2006) Pseudovibrio ascidiaceicola sp. nov., isolated from ascidians (sea squirts). Int J Syst Evol Microbiol 56:343–347
Gaitatzis N, Silakowski B, Kunze B, Nordsiek G, Blocker H, Hofle G, Muller R (2002) The biosynthesis of the aromatic myxobacterial electron transport inhibitor stigmatellin is directed by a novel type of modular polyketide synthase. J Biol Chem 277:13082–13090
Hentschel U, Schmid M, Wagner M, Fieseler L, Gernert C, Hacker J (2001) Isolation and phylogenetic analysis of bacteria with antimicrobial activities from the Mediterranean sponges Aplysina aerophoba and Aplysina cavernicola. FEMS Microbiol Ecol 35:305–312
Hill RT, Peraud O, Hamann MT, Noer K (2005) Manzamine producing actinomycetes. United States Patent Application
Jia XY, Tian ZH, Shao L, Qu XD, Zhao QF, Tang J, Tang GL, Liu W (2006) Genetic characterization of the chlorothricin gene cluster as a model for spirotetronate antibiotic biosynthesis. Chem Biol 13:575–585
Jiang S, Sun W, Chen M, Dai S, Zhang L, Liu Y, Lee KJ, Li X (2007) Diversity of culturable actinobacteria isolated from marine sponge Haliclona sp. Antonie van Leeuwenhoek 92:405–416
Kennedy J, Codling CE, Jones BV, Dobson AD, Marchesi JR (2008) Diversity of microbes associated with the marine sponge, Haliclona simulans, isolated from Irish waters and identification of polyketide synthase genes from the sponge metagenome. Environ Microbiol 10(7):1888–1902
Kieser T, Bibb M, Buttner MJ, Chater K, Hopwood DA (2000) Practical streptomyces genetics. The John Innes Foundation, Norwich
Kim TK, Garson MJ, Fuerst JA (2005) Marine actinomycetes related to the “Salinospora” group from the Great Barrier Reef sponge Pseudoceratina clavata. Environ Microbiol 7:509–518
Kim TK, Hewavitharana AK, Shaw PN, Fuerst JA (2006) Discovery of a new source of rifamycin antibiotics in marine sponge actinobacteria by phylogenetic prediction. Appl Environ Microbiol 72:2118–2125
Lane DJ (1991) 16S/23S rRNA squencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acids techniques in bacterial systematics. Wiley, Chichester
Magarvey NA, Keller JM, Bernan V, Dworkin M, Sherman DH (2004) Isolation and characterization of novel marine-derived actinomycete taxa rich in bioactive metabolites. Appl Environ Microbiol 70:7520–7529
Montalvo NF, Mohamed NM, Enticknap JJ, Hill RT (2005) Novel actinobacteria from marine sponges. Antonie van Leeuwenhoek 87:29–36
Muscholl-Silberhorn A, Thiel V, Imhoff JF (2008) Abundance and bioactivity of cultured sponge-associated bacteria from the Mediterranean sea. Microb Ecol 55:94–106
Neilan BA, Dittmann E, Rouhiainen L, Bass RA, Schaub V, Sivonen K, Borner T (1999) Nonribosomal peptide synthesis and toxigenicity of cyanobacteria. J Bacteriol 181:4089–4097
Pabel CT, Vater J, Wilde C, Franke P, Hofemeister J, Adler B, Bringmann G, Hacker J, Hentschel U (2003) Antimicrobial activities and matrix-assisted laser desorption/ionization mass spectrometry of Bacillus isolates from the marine sponge Aplysina aerophoba. Mar Biotechnol 5:424–434
Parada M, Vinardell JM, Ollero FJ, Hidalgo A, Gutierrez R, Buendia-Claveria AM, Lei W, Margaret I, Lopez-Baena FJ, Gil-Serrano AM, Rodriguez-Carvajal MA, Moreno J, Ruiz-Sainz JE (2006) Sinorhizobium fredii HH103 mutants affected in capsular polysaccharide (KPS) are impaired for nodulation with soybean and Cajanus cajan. Mol Plant Microbe Interact 19:43–52
Rashid MA, Gustafson KR, Boswell JL, Boyd MR (2000) Haligramides A and B, two new cytotoxic hexapeptides from the marine sponge Haliclona nigra. J Nat Prod 63:956–959
Richelle-Maurer E, Braekman JC, De Kluijver MJ, Gomez R, Van De Vyver G, Van Soest RW, Devijver C (2001) Cellular location of (2R, 3R, 7Z)-2-aminotetradec-7-ene-1, 3-diol, a potent antimicrobial metabolite produced by the Caribbean sponge Haliclona vansoesti. Cell Tissue Res 306:157–165
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sakai R, Higa T, Jefford CW, Bernardinelli G (1986) Manzamine A, a novel antitumor alkaloid from a sponge. J Am Chem Soc 108:6404–6405
Sera Y, Adachi K, Fujii K, Shizuri Y (2002) Isolation of Haliclonamides: new peptides as antifouling substances from a marine sponge species, Haliclona. Mar Biotechnol (NY) 4:441–446
Sertan-De Guzman AA, Predicala RZ, Bernardo EB, Neilan BA, Elardo SP, Mangalindan GC, Tasdemir D, Ireland CM, Barraquio WL, Concepcion GP (2007) Pseudovibrio denitrificans strain Z143–1, a heptylprodigiosin-producing bacterium isolated from a Philippine tunicate. FEMS Microbiol Lett 277:188–196
Shieh WY, Lin YT, Jean WD (2004) Pseudovibrio denitrificans gen. nov., sp. nov., a marine, facultatively anaerobic, fermentative bacterium capable of denitrification. Int J Syst Evol Microbiol 54:2307–2312
Tamura K, Nei M, Kumar S (2004) Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A 101:11030–11035
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Taylor MW, Radax R, Steger D, Wagner M (2007) Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol Mol Biol Rev 71:295–347
Thakur AN, Thakur NL, Indap MM, Pandit RA, Datar VV, Muller WE (2005) Antiangiogenic, antimicrobial, and cytotoxic potential of sponge-associated bacteria. Mar Biotechnol 7:245–252
Thiel V, Imhoff JF (2003) Phylogenetic identification of bacteria with antimicrobial activities isolated from Mediterranean sponges. Biomolecular Engineering 20:421–423
Webster NS, Hill RT (2001) The culturable microbial community of the Great Barrier Reef sponge Rhopaloeides odorabile is dominated by an alpha-proteobacterium. Mar Biol 138:843–851
Webster NS, Wilson KJ, Blackall LL, Hill RT (2001) Phylogenetic diversity of bacteria associated with the marine sponge Rhopaloeides odorabile. Appl Environ Microbiol 67:434–44
Yu S, Deng Z, Proksch P, Lin W (2006) Oculatol, oculatolide, and A-nor sterols from the sponge Haliclona oculata. J Nat Prod 69:1330–1334
Zhang H, Lee YK, Zhang W, Lee HK (2006) Culturable actinobacteria from the marine sponge Hymeniacidon perleve: isolation and phylogenetic diversity by 16S rRNA gene-RFLP analysis. Antonie Van Leeuwenhoek 90:159–169
Zhang H, Zhang W, Jin Y, Jin M, Yu X (2008) A comparative study on the phylogenetic diversity of culturable actinobacteria isolated from five marine sponge species. Antonie Van Leeuwenhoek 93:241–248
Zuckerkandl E, Pauling L (1965) Molecules as documents of evolutionary history. J Theor Biol 8:357–366
Acknowledgments
JK, PB, and MJM are in receipt of Marie Curie Transfer of Knowledge Host Fellowships; [grant no. MTKD-CT-2006-042062]. This project was funded by the Irish Marine Institute under the Strategic Marine Biodiscovery RTDI Programme and by the Marine Biodiscovery Research Award funded by the Irish Government under the National Development Plan (2007–2013). We thank Dr Grace McCormack from the National University of Ireland, Galway, for the H. simulans sponge samples.
Author information
Authors and Affiliations
Corresponding author
Additional information
DNA sequences have been deposited at GenBank and have been given the following accession numbers EU768811–EU768843.
Rights and permissions
About this article
Cite this article
Kennedy, J., Baker, P., Piper, C. et al. Isolation and Analysis of Bacteria with Antimicrobial Activities from the Marine Sponge Haliclona simulans Collected from Irish Waters. Mar Biotechnol 11, 384–396 (2009). https://doi.org/10.1007/s10126-008-9154-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10126-008-9154-1