Abstract
Gene for organic solvent stable lipase was overexpressed from soil metagenomic library. The clone with maximum activity was selected, and enzyme was purified by gel-permeation chromatography with a molecular mass of approx. 40 kDa. The deduced aminoacid sequence indicated that the protein belongs to the lipase family I.3 and containing a C-terminal secretion signal for ABC dependent transport together with possible motifs for Ca2+ binding sites. The enzyme expressed maximum activity at 30 °C and pH 7.0 and found to be stable in pH and temperature ranging from 6.0–9.0 and 20–60 °C, respectively. Furthermore, the enzyme was found highly resistant to many organic solvents, especially isopropanol, DMSO, methanol, xylene and hexane. The enzyme showed enhanced activity in the presence of divalent cations (Mg2+, Mn2+, Ca2+, Hg2+, Cu2+), whereas the presence of trivalent cation (Fe3+) inhibited the activity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Soil harbors enormously diverse microbial communities and is the major reservoir of genomic and taxonomic diversity. The microbial diversity in soil exceeds that of other environments and by far, that of eukaryotic organisms. Soil microorganisms have been the major source for many enzymes and other biomolecules of industrial importance (Strohl 2000). However, of late, the discovery rate of novel biomolecules is decreasing by applying traditional cultivation techniques, because most of the soil microorganisms cannot be cultured under standard laboratory conditions and only a small fraction of soil microbial diversity is assessed in this way (Amann et al. 1995). Therefore, culture-dependent methods have been complemented by culture-independent approaches, which theoretically provide access to the collective nucleic acids of all indigenous microorganisms present in the studied environment. Metagenomics is the field combining molecular biology and genetics in an attempt to identify, and to characterize the genetic material from environmental samples and apply that knowledge for isolation and characterization of biomolecules with unique properties. Many enzymes such as, esterases (Rhee et al. 2005), amylases (Handelsman et al. 1998) and chitinase (Cottrell et al. 1999) have been identified and characterized by culture independent methods.
Lipases are one of the most versatile and multifunctional enzymes used in various industries. In addition to their natural function of hydrolyzing carboxylic ester bonds, lipases can catalyze esterification, interesterification, and transesterification reactions in nonaqueous media. Several genes, encoding metagenomic lipases have been identified in metagenomic libraries over the last decade (Lee et al. 2004; Ranjan et al. 2005; Lee et al. 2006). Kim et al. (2009) reported a novel cold-adapted alkaline lipase, LipEH166, from an intertidal flat metagenome. LipEH166 and its homologues showed similarity only in the consensus region to known lipolytic enzyme and classified as a new family of lipolytic enzymes. Lipase diversity in glacier soil was also assessed by metagenomic approach and five divergent lipase family clusters were reported based on phylogenetic analysis (Zhang et al. 2009). A new family of bacterial lipases has also been reported from a Korean flat met library, an important feature of this family is an Arg-Ala sequence that can serve as oxyanion hole, which is known to be a unique signature sequence for filamentous fungal lipases (Lee et al. 2006). Glogauer et al. (2011) constructed a fosmid metagenomic library with the “prokaryotic-enriched” DNA from a fat contaminated soil and identified a novel, organic solvent stable lipase gene.
Lipases catalyze the hydrolysis of a wide range of carboxyl esters at the water–lipid interface and reversing the reaction in non-aqueous media (Jaeger et al. 1999; Villeneuve et al. 2000). This versatility makes lipases the enzymes of choice for potential applications in the food, dairy, pharmaceutical, detergents, textile, biodiesel, cosmetic and paper industry, in synthesis of fine chemicals and new polymeric materials (Hasan et al. 2006). Moreover, these enzymes can be used as chiral catalysts for stereo-selective conversion of a variety of amines and primary and secondary alcohols for production of cosmetics and pharmaceuticals (Jaeger and Eggert 2002).
Substrates of the lipase are often insoluble in aqueous solutions, and organic solvents or organic-aqueous two-phase media are favorable to some reactions. The use of organic solvents instead of water promises many advantages: (a) the relative high solubility of substrates, (b) the relative ease of recovery of products in organic phase, (c) the possibility of reducing the degree of undesirable substrate and/or product inhibition in organic solvent–water two-phase system and (d) the ability to shift the reaction equilibrium in the synthetic direction by continuously removing the products with organic solvent water two-phase systems (Hun et al. 2003). Therefore, organic solvents as reaction media could greatly expand the repertoire of enzyme-catalyzed transformations. In recent years, some organic solvent tolerant lipases have been characterized (Ogino et al. 1994; Hun et al. 2003; Glogauer et al. 2011), but only a few enzymes have been demonstrated to have adequate stability to allow routine use in organic solvents. Most of these enzymes are stable only in non-polar solvents. The non polar solvents are often less harmful to enzyme activity than their hydrophilic counterparts. Hydrophobic solvents could not easily strip essential water required for enzymatic function, thereby maintain the active conformation necessary for the reaction. Lipase from Bacillus sphaericus 205y showed enhanced activity in presence of n-hexane and p-xylene whereas, activity was completely inhibited in n-hexadecene (Hun et al. 2003). Eltaweel et al. (2005) reported that lipase from Bacillus was stable in non-polar solvents like hexane and hexadecane but enzyme activity was lost in polar solvents with low log P value such as propanol, ethylacetate and propylacetate. In other studies also it was reported that lipase activity was increased only in the presence of n-hexane while decreased with the increase in concentration of acetone, chloroform, ethanol and isopropanol (Ogino et al. 1994; Ghori et al. 2011). The availability of lipases that were stable in both polar as well as non-polar solvents would favour new applications in biotechnological processes.
In the view of above facts this work was planned to over express, purify and characterize solvent stable lipase from soil metagenomic library. The constructed library was subjected to activity-based screening for genes encoding lipolytic enzymes. This study demonstrated that function-driven screening of large soil libraries usually results in identification of full-length genes (and therefore functional gene products) with novel properties and this will further accelerate the speed of discovery and the diversity of the recovered biomolecules.
Materials and methods
Construction and screening of soil metagenomic library
Big Easy linear cloning kit, Lucigen was used for construction of gene library. Purified soil DNA was partially digested with Sau3AI. Fractions containing DNA fragments 1–3 kb were end repaired with DNA terminator kit for blunt cloning, ligated with pJAZZ vector and transformed into E. coli JM109. The positive white clones were randomly selected from the LB agar plates containing 0.8 mg/ml X-gal, 3 mM IPTG, and 100 μg/ml ampicillin. For screening two types of indicator plates were employed. Chromogenic substrate plates were prepared by using phenol red (0.01 %) along with 1 % olive oil as substrate as described by Singh et al. (2006). Lipolytic activity was further confirmed on Luria–Bertani agar containing olive oil (1 %, v/v) and the fluorescent dye rhodamine B (0.001 %, wt/vol).
Subcloning and screening of the secondary library
For the overexpression and purification of lipase enzyme, lipase gene from soil metagenomic library was subcloned into QIAexpress Cloning Kit from QIAGEN. Plasmid was isolated by alkaline lysis method and amplified by four different sets of lipase specific primers (Table 1). The 50-μl reactions included 1× PCR buffer (Hi Media, India), 0.4 mM dNTPs, and 0.25 U Taq polymerase (Hi Media, India). PCR was carried out under the following conditions: 95 °C for 5 min, followed by 94 °C for 30 s, 55 °C for 2 min and 72 °C for 30 s. The final extension step was at 72 °C for 5 min. PCR products were visualized on an agarose gel and purified using a gel extraction kit, ligated to QIAexpress vectors. The ligation mixtures were transformed into E. coli, and the transformants were spread onto LB plates supplemented with ampicillin and tributyrin to select subclones showing clear zones around the colonies.
Lipase sequence analysis
DNA sequencing was performed using ABI PRISM, DNA sequencer. Nucleotide and deduced amino acid sequence analyses, open reading frame search, multiple alignment, molecular-mass and isoelectric-point calculations were performed using expasy bioinformatics resource portal [http://expasy.Org/tools/]. Prediction of transmembrane regions and signal peptide sequence were performed using the TMHMM 2.0 (Krogh et al. 2001) and SignalP 3.0 servers (Bendtsen et al. 2004), respectively.
Purification of lipase
The LB broth containing 1.0 % olive oil was inoculated with lipolytic clone SMlipA and incubated for 24 h at 37 °C. The culture was then supplemented with 1 mM (final concentration) isopropyl-β-D-thiogalactopyranoside (IPTG) and incubated for another 4 h at 37 °C for overexpression of the lipase gene. The supernatant obtained by centrifugation was used as crude enzyme and then precipitated with 40 % saturation of ammonium sulphate. The precipitate was recovered by centrifugation (10,000 rpm × 10 min at 4 °C), dissolved in phosphate buffer saline (pH 7.6) and the solution was dialyzed against PBS for overnight. After centrifugation the supernatant was applied into a Sephadex G-75 column, which was previously equilibrated with 50 mM PBS, pH 7.0 at 4 °C and the proteins were eluted from the column with the same buffer. The fraction containing the lipase activity was pooled and dialyzed. The dialyzed enzyme solution was used for further studies. Purified enzyme was analyzed on 12 % SDS-PAGE. A medium range of protein standard markers (Bangalore Genei) were used as the molecular weight markers. The proteins in the gels were stained with Coomassie brilliant blue and compared with the molecular weight markers.
Quantitative lipase assay
The lipase activity was estimated using a spectrophotometric assay with p-nitrophenol acetic acid (100 mM) as a substrate. The activity measurement was carried out at 30 °C; pH 7.5. One unit of lipase activity was defined as the amount of enzyme releasing 1.0 μmol of p-nitrophenol per minute under the assay conditions (Jaeger and Eggert 2002). All the assays were performed in triplicate, and the average values were taken. Relative activity was defined as the percentage of the maximum enzyme activity value.
Characterization of lipase
Effect of temperature and pH on enzyme stability and activity
The optimum temperature and temperature range for the lipase activity were evaluated at pH 7.0, by measuring the hydrolytic activity at different temperatures ranging from 20 to 60 °C. The stability of the lipase to temperature was investigated by measuring the residual activity after incubating the purified lipase at 20–60 °C for 1 h.
The optimum pH and pH range for the lipase were measured by incubating the reaction mixture in 0.1 M phosphate buffer over a pH range from 5.0 to 9.0 at 30 °C. The effect of pH on lipase stability was determined by analyzing the residual activity after incubating the purified lipase in the buffers with different pH values (pH 4.0–10.0) at 4 °C for 1 h.
Effect of metal ions on enzyme stability and activity
The effect of metal ions on lipase activity was obtained by pre-incubation with 1 mM of metal ion at room temperature for 2.5 h. The metal ions used were CaCl2, MgCl2, MnSO4, CuSo4, HgCl2 and FeCl3. The lipase activity of the enzyme solution without any metal was used as control (100 % of relative activity).
Effect of solvents on enzyme stability and activity
The effect of different solvent on enzyme activity was determined by adding equal volume organic solvent to the purified enzyme and incubated at room temperature, with an agitation at 150 rpm for 12 h to ensure the continuous mixing of the enzyme and solvent. The enzyme stability was expressed as the remaining activity assayed relative to the control value. Phosphate buffer was used instead of solvent as the control.
Result and discussion
Cloning and nucleotide sequence of SMlipA lipase gene
The lipase gene was successfully cloned and expressed in QIAexpress pQE, an expression vector carrying a powerful T5 phage promoter, using E. coli JM109 as the host. To screen the lipolytic clones from the soil metagenomic libraries, a function-driven approach was chosen. This strategy bears the potential to discover novel genes without any sequence information. In addition it is selective for full-length genes and functional gene products. The screening for genes exhibiting lipolytic activity was done by the ability of clones to form halos when grown on agar medium containing tributyrine. Halo formation is caused by the hydrolysis of tributyrine. The lipase activity was further confirmed on Rhodamine B agar Plates. Lipase production was monitored by irradiating plates with UV light at 350 nm. After 16 h of incubation positive clones began to show an orange fluorescence around the discs, with continuing incubation time orange fluorescent halos were formed around the colonies of Lipase producing strains. Fifteen lipase positive clones were obtained after screening, and the clone SMlipA that showed maximum activity was selected for further studies. The function-based screen has been used to identify the lipolytic activity of individual microorganisms (Hantsis-Zacharov and Halpern 2007), and recombinant E. coli strains that harbor gene libraries from single microorganisms (Hotta et al. 2002) or metagenomic libraries (Hu et al. 2010). In case of metagenomic libraries, genes conferring lipolytic activity have also been recovered earlier from diverse environments such as mangrove sediment (Couto et al. 2010), marine sediment (Hårdeman and Sjöling 2007), water samples (Wu and Sun 2009).
The entire 1,295 bp containing lipase gene was sequenced and gene sequence was submitted to Gene bank (Accession No. AEW90242.1). The open reading frame (ORFs) was identified with the ORF Finder tool. SMlipA sequence contained an ORF comprising 1,119 bp extending from 169 to 1,290, which encoded 373 amino acids. The ProtParam tool was used to calculate the theoretical parameters of the protein and the deduced molecular mass based on ORF was calculated to be 39.8 kDa. The domain analysis carried out using Pfam showed that the enzyme probably has a type I α/β-hydrolase fold between residues 103 and 201. The amino acid sequences were compared with the non-redundant sequence database deposited at the NCBI using BLAST. SMlipA showed 70, 82 and 70 % of identity with LipA gene from uncultured bacterium [Gene Bank: ACT99860.1], LipB from uncultured Pseudomonas [Gene Bank: AAU13930.1] and cold-active lipase from uncultured bacterium [Gene Bank: ABI94371.1], respectively. Among the lipase with known 3D structure it possess 58 and 51 % of identity with extracellular lipase of Pseudomonas [PDB: 2Z8X] and Serretia marcescens [PDB: 2QUA], respectively (Fig. 1).
Aminoacid sequence alignment of SMlipA (AEW90242.1) lipase with other lipase from unculturable bacterium. Active site (as), which is important for lipase activity, is highlighted in yellow colour, calcium binding domain (cs) green in colour and C terminal signal sequence pink in colour (colour figure online)
The GXSXG motif, which includes the active-site serine residue, was found from residues 125 to 129. The amino acid sequence analysis revealed that the SMlipA lipase lacked a typical N-terminal signal peptide for its secretion, transmembrane domains, and cys residues. Similar findings were reported in case of the lipase from P. fluorescens KB700A, for which it has been shown that secretion is dependent on the C-terminal region of the protein. Interpro Scan tool (Zdobnov and Apweiler 2001) for the signature-recognition of protein indicated that C-terminal region of SMlipA contained hemolysin-type calcium-binding region and Serralysin-like metalloprotease belongs to peptidase M10 family, from amino acid residues 355–390 and 348–408, respectively. It has been shown that family I.3 lipases are involved in the binding of calcium ions in a parallel beta roll structure. The peptidase unit is found at the C terminus forms a corkscrew. It is thought to be important for secretion of the protein through the bacterial cell wall. The secretion of these proteins occurs in one step through a three-component ATP-binding-cassette transporter system (Angkawidjaja et al. 2007). The C-terminal domain of SMlipA was highly similar to that of P. fluorescens KB700A, suggesting similar mechanisms for secretion of SMlipA (Rashid et al. 2001). In addition, the GXXGXD motifs, which are supposed to be involved in Ca2+ binding in proteases and lipases, an extreme C-terminal motif consisting of a negatively charged amino acid followed by four hydrophobic residues (Baumann 1994), proven necessary for the secretion of metalloprotease was also found in SMlipA. Thus it is possible to conclude that SMlipA belongs to lipase Subfamily I.3.
Purification and characterization of lipase
Hydrolysis of p-nitrophenyl ester of acetic acid was used to determine the lipolytic and 23.4 U/ml enzyme activity was measured in crude enzyme.
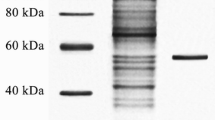
The enzyme from the culture supernatant was partially purified using ammonium sulphate precipitation and the maximum lipase activity was recorded in the fraction precipitated, by 40 % saturation. The partially purified lipase after centrifugation, ammonium sulfate precipitation and dialysis was chromatographed through the Sephadex G-75 with the phosphate buffer saline; in the preceding gradient, the active fraction was appeared. After the three-step procedure, the purified lipase was purified was analyzed on SDS-PAGE for homogeneity and showed a single band of approx. 40.0 kDa on SDS-PAGE (Fig. 2). The specific activity of purified lipase measure was 513 U/mg.
Effect of pH and temperature on enzyme activity
The enzyme exhibited the highest activity at pH 7.0. The enzyme activity increased gradually from pH 4.0 to 6.0 and reached to the maximum at pH 7.0. It was also found that enzyme retained its activity significantly in all the pH range from 5 to 8 and showed 85, 91, 90 and 82 % relative activity at pH 5, 6, 8 and 9 respectively (Fig. 3). Therefore from the results it can be concluded that lipase was active in broad range of pH.
The optimal temperature of the lipase enzyme was found to be 30 °C. Notably the enzyme retained 62 % of the maximal activity at 20 °C, 84 and 70 % at 40 and 50 °C respectively (Fig. 4). The results showed that the optimal temperature of SMlipA was lower than those of the other members of lipase subfamily I.3 and SMlipA retained the stability throughout the temperature range of 20–60 °C.
Most of the lipases that belongs to subfamily I.3 showed temperature optima only at narrow temperature range. Lipase from Pseudomonas strain SIK W1, and strain MIS38 had an optimal temperature in the range of 45–55 °C (Lee et al. 1993; Amada et al. 2000) and KB700A another strain of Pseudomonas sp. showed temperature optima at 35 °C but found to be highly heat labile and lost over 70 % of its activity after only 5 min of incubation at 60 °C (Rashid et al. 2001). It is quite interesting that although SMlipA showed high similarity (80 %) with other lipases of subfamily I.3 but there are considerable differences in optimal temperatures and thermostability characteristics of these proteins from SMlipA lipase. This property of SMLipA may be helpful in elucidation of mechanism of cold and thermostability of lipases from unculturable bacterium.
Effect of metal ions
All the divalent anions such as Mg2+, Mn2+, and Ca2+, Cu2+ and Hg2+ were found to enhance the enzyme activity, while trivalent anion Fe3+ reduced the enzyme activity. The maximum activity was achieved in presence of Mg2+ salts and showed a relative activity of about 176 %. The relative activity of enzyme in presence of various metal ions is summarized in Table 2. The presence of Ca2+-binding motifs, GXXGXD, in the C terminus of the enzyme also confirmed that the SMlipA activity was dependent on divalent cations. Sharma et al. (2009), reported that the increase in catalytic activity in the presence of Mg2+, Mn2+, and Ca2+ was due to the formation of insoluble salts of fatty acids released in the hydrolysis, thus avoiding product inhibition.
Makhzoum et al. (1995), observed approximately a 360 % increase in the enzyme activity in presence of Ca2+ ions to an extracellular lipase of P. fluorescens 2D but Hg2+ and Co2+ strongly inhibited the enzyme activity. Sharon et al. (1998) reported a lipase from P. aeruginosa KKA-5 retained its activity in presence of Ca2+ and Mg2+ whereas salts of heavy metals (Fe2+, Zn2+, Hg2+, Fe3+) strongly inhibited the lipase, suggesting that they were able to alter the enzyme conformation.
Stability of SMlipA lipase in various organic solvents
Lipases catalyzed reactions involved triglyceride hydrolysis, ester synthesis, transesterification of fats etc. Since substrates of such reactions are often insoluble in aqueous solution, organic solvents or organic aqueous two-phase media are required. But most of enzymes are not stable in the presence of organic solvents and are susceptible to denaturation (Ogino et al. 1994).
In this study, activity and stability of SMlipA in various organic solvents with different polarities Log P values from −0.76 to 3.6 was determined. Here, Log P was used as the quantitative measure of the solvent polarity. From the results it was observed that, interestingly the purified lipase activity was greatly activated by 50 % (v/v) polar solvents including isopropanol, DMSO, methanol and non-polar solvent such as xylene and hexane more than ~50 % increment. The relative activity of enzyme in presence of different solvents is shown as Table 3. This is the unique property of the lipase isolated from the metagenomic library. Purified recombinant lipase from Staphylococcus epidermis AT2 was also reported to be activated in presence of 30 % (v/v) DMSO (Rahman et al. 2010). Solvents having a Log P < 2 are not suitable in biocatalytic systems, since they strongly distort the essential water-biocatalyst interaction, thereby inactivating the biocatalyst (Laane et al. 1987). In the present study, results showed that, SMlipA lipase had the maximum activity in the solvent Xylene which is having a Log P value of 3.1. The enzyme activity is enhanced in presence of organic solvents, due to the interaction of hydrophobic solvent and hydrophobic amino acid residues present in the lid of the enzyme that maintain the lid in its open confirmation (Doukyu and Ogino 2010).
Conclusion
SMlipA lipase, over expressed and characterized from soil metagenomic library however, belongs to lipase subfamily 1.3, but it showed considerable difference in biochemical characteristics from other members of the family. The unique feature of SMlipA is its high stability in organic solvents. Since this enzyme function well in broad temperature and stable in presence of organic solvents, it is a novel lipase and promising candidate for biotechnological applications. It can be used for most of the industrial applications especially in the reverse mode, i.e. for ester synthesis, inter-esterification and resolution of racemic mixture to produce optically active compounds.
References
Amada K, Haruki M, Imanaka T, Morikawa M, Kanaya S (2000) Overproduction in Escherichia coli, purification and characterization of a family I.3 lipase from Pseudomonas sp. MIS38. Biochim Biophys Acta 1478:201–210
Amann RI, Ludwig W, Schleifer KH (1995) Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59:143–169
Angkawidjaja C, Dong-ju Y, Matsumura H, Kuwahara K, Koga Y, Takano K, Kanaya S (2007) Crystal structure of a family I.3 lipase from Pseudomonas sp. MIS38 in a closed conformation. FEBS Lett 581:5060–5064
Baumann U (1994) Crystal structure of the 50 kDa metallo protease from Serratia marcescens. J Mol Biol 242:244–251
Bendtsen JD, Nielsen H, Von Heijne G, Brunak S (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340:783–795
Cottrell MT, Moore JA, Kirchman DL (1999) Chitinases from uncultured marine microorganisms. Appl Environ Microbiol 65:2553–2557
Couto G, Glogauer A, Faoro H, Chubatsu L, Souza E, Pedrosa F (2010) Isolation of a novel lipase from a metagenomic library derived from mangrove sediment from the south Brazilian coast. Genet Mol Res 9:514–523
Doukyu N, Ogino H (2010) Organic solvent-tolerant enzymes. Biochem Eng J 48:270–282
Eltaweel MA, Rahman NR, Salleh A, Basri M (2005) An organic solvent-stable lipase from Bacillus sp. strain. Ann Microbiol 55:187–192
Ghori MI, Iqbal MJ, Hameed A (2011) Characterization of a novel lipase from Bacillus sp. isolated from tannery wastes. Braz J Microbiol 42(1):22–29
Glogauer A, Martini VP, Helisson F, Couto GH, Müller-Santos M, Rose AM, Mitchell DA, de Souza EM, Fabio OP, Krieger N (2011) Identification and characterization of a new true lipase isolated through metagenomic approach. Microb Cell Factories 10:54–60
Handelsman J, Rondon MR, Brady SF, Clardy J, Goodman RM (1998) Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem Biol 5:245–249
Hantsis-Zacharov E, Halpern M (2007) Culturable psychrotrophic bacterial communities in raw milk and their proteolytic and lipolytic traits. Appl Environ Microb 73:7162–7168
Hårdeman F, Sjöling S (2007) Metagenomic approach for the isolation of a novel low-temperature-active lipase from uncultured bacteria of marine sediment. FEMS Microbiol Ecol 59:524–534
Hasan F, Shah AA, Hameed A (2006) Industrial applications of microbial lipases. Enzyme Microbial Technol 39:235–251
Hotta Y, Ezaki S, Atomi H, Imanaka T (2002) Extremely stable and versatile carboxylesterase from a hyperthermophilic archaeon. Appl Environ Microb 68:3925–3931
Hu Y, Fu C, Huang Y, Yin Y, Cheng G, Lei F, Lu N, Li J, Ashforth E, Zhang L, Zhu B (2010) Novel lipolytic genes from the microbial metagenomic library of the South China sea marine sediment. FEMS Microbiol Ecol 72:228–237
Hun CJ, Rahman RNZ, Salleh AB, Basri M (2003) A newly isolated organic solvent tolerant Bacillus sphaericus 205y producing organic solvent–stable lipase. Biochem Eng J 15:147–151
Jaeger KE, Eggert T (2002) Lipases for biotechnology. Curr Opin Biotechnol 13:390–397
Jaeger KE, Dijkstra BW, Reetz MT (1999) Bacterial biocatalysts: molecular biology, three-dimensional structures, and biotechnological applications of lipases. Annu Rev Microbiol 53:315–351
Kim E-Y, Oh K-H Lee M-H, Kang C-H, Oh T-K, Jung-Hoon Y (2009) Novel cold-adapted alkaline lipase from an intertidal flat metagenome and proposal for a new family of bacterial lipases. Appl Environ Microbiol 75(1):257–260
Krogh A, Larsson B, Von Heijne G, Sonnhammer EL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580
Laane C, Boeren S, Vos K, Veeger C (1987) Rules for optimization of biocatalysis in organic solvents. Biotechnol Bioeng 30:81–87
Lee YP, Chung GH, Rhee JS (1993) Purification and characterization of Pseudomonas fluorescens SIK W1 lipase expressed in Escherichia coli. Biochim Biophys Acta 1169:156–164
Lee SW, Won K, Lim HK, Kim JC, Choi GJ, Cho KY (2004) Screening for novel lipolytic enzymes from uncultured soil microorganisms. Appl Microbiol Biotechnol 65(6):720–726
Lee MH, Lee CH, Oh TK, Song JK, Yoon JH (2006) Isolation and characterization of a novel lipase from a metagenomic library of tidal flat sediments: evidence for a new family of bacterial lipases. Appl Environ Microbiol 72(11):7406–7409
Makhzoum A, Knappm JS, Owusu RK (1995) Factor affecting growth and lipase 595 production by Pseudomonas fluorescens 2D. Food Microbiol 12:277–290
Ogino H, Miyamoto K, Ishikawa H (1994) Organic-solvent-tolerant bacterium which secretes organic-solvent-stable lipolytic enzyme. Appl Environ Microbiol 60:3884–3886
Rahman RNR, Kamarudin NA, Yunus J, Salleh A, Basri M (2010) Expression of an organic solvent stable lipase from Staphylococcus epidermidis AT2. Int J Mol Sci 11(9):3195–3208
Ranjan R, Grover A, Kapardar RK, Sharma R (2005) Isolation of novel lipolytic genes from uncultured bacteria of pond water. Biochem Biophys Res Commun 335:57–65
Rashid N, Yuji S, Satoshi E, Haruyuki A, Tadayuki I (2001) Low temperature lipase from psychrotrophic Pseudomonas sp. Strain KB700A. Appl Environ Microbiol 67:4064–4069
Rhee J, Ahn D, Kim Y, Oh J (2005) New thermophilic and thermostable esterase with sequence similarity to the hormone-sensitive lipase family, cloned from a metagenomic library. Appl Environ Microbiol 71:817–825
Sharma A, Bardhan D, Patel R (2009) Optimization of physical parameters for lipase production from Arthrobacter sp. BGCC#490. Indian J Biochem Biophys 46(2):178–183
Sharon C, Furugoh S, Yamakido T, Ogawa H, Kato Y (1998) Purification and characterization of a lipase from Pseudomonas aeruginosa KKA-5 and its role in castor oil hydrolysis. J Ind Microbiol Biotechnol 20:304–307
Singh R, Gupta N, Goswami VK, Gupta R (2006) A simple activity staining protocol for lipases and esterases. Appl Microbiol Biotechnol 70:679–682
Strohl WR (2000) The role of natural products in modern drug discovery. Drug Discov Today 5:39–41
Villeneuve P, Muderhwa JM, Graille J, Haas MJ (2000) Customizing lipases for biocatalysis: a survey of chemical, physical and molecular biological approaches. J Mol Catal B Enzym 9:113–148
Wu C, Sun B (2009) Identification of novel esterase from metagenomic library of Yangtze river. J Microbiol Biotechnol 19:187–193
Zdobnov EM, Apweiler R (2001) InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17:847–848
Zhang Y, Shi P, Liu W, Meng K, Bai Y, Wang G, Zhan Z, Bin Y (2009) Lipase diversity in glacier soil based on analysis of metagenomic DNA. J Microbiol Biotechnol 19(9):888–897
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Khan, M., Jithesh, K. Expression and purification of organic solvent stable lipase from soil metagenomic library. World J Microbiol Biotechnol 28, 2417–2424 (2012). https://doi.org/10.1007/s11274-012-1051-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11274-012-1051-0