Abstract
Six diazotrophic bacteria were isolated from surface-sterilized roots of rice variety HUR-36, which is grown with very low or no inputs of nitrogen fertilizer. Out of six bacteria one isolate, RREM25, showed appreciable level of nitrogenase activity, IAA production, and Phosphate solubilization ability, and was further characterized with a view to exploiting its plant growth promoting activity. Based on 16S rRNA gene sequence analysis, this isolate was identified as Burkholderia cepacia. Diazotrophic nature of this particular isolate was confirmed by Western blot analysis of dinitrogenase reductase and amplification of nifH. Microscopic observation confirmed colonization of gfp/gusA-tagged RREM25 in the intercellular spaces of cortical as well as vascular zones of roots. Inoculation of RREM25 to rice plants resulted in significant increase in plant height, dry shoot and root weight, chlorophyll content, nitrogen content and nitrogenase activity. Plant growth promoting features suggest that this endophytic bacterium may be exploited in rice cultivation after a thorough and critical pathogenicity test.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Biological nitrogen fixation (BNF), generally found in legumes, is a valuable nature’s gift through which atmospheric nitrogen is fixed by symbiotically associated rhizobia that make it available to plants. Among cereals, rice (Oryza sativa L.) is an important staple food crop that feeds a large proportion of the world population (Ladha et al. 1997). There is tremendous pressure on agriculturists worldwide to increase the rice yield to meet growing consumption to feed a world population increasing day by day. However, unlike symbiotic plant–microbe interactions commonly observed in legumes, endophytic associations in several cereal crop plants including rice have been observed (Yanni et al. 1997, 2001; Singh et al. 2006). Extensive research on endophytic bacteria and its beneficial effects on plant growth started with the isolation of endophytic Gluconoacetobacter diazotrophicus from Brazilian sugarcane (James and Olivares 1998). Burkholderia, a phylogenetically well-defined genus which is remarkably diverse in view of its wide environmental distribution and its capabilities for promotion of plant growth (Chen et al. 2006). Endophytic Burkholderia reside inside the plant tissue without doing substantive harm and 42–64% increases in growth of rice plants was observed when Burkholderia brasilensis and Burkholderia vietnamiensis were inoculated under gnotobiotic conditions (Govindarajan et al. 2008). Nitrogen is the major limiting factor for plant growth, the application of N2-fixing endophytic bacteria as biofertilizer has emerged as one of the most efficient and environmentally sustainable methods for increasing growth and yield of crop plants. Diazotrophic endophytic bacteria provide more of fixed nitrogen as compared to rhizospheric bacteria because the interior of plants is a more suitable niche for nitrogen fixation in view of low partial oxygen pressure (pO2) and direct accessibility of the fixed nitrogen to the plants (James and Olivares 1998). Besides nitrogen fixation, endophytic bacteria may also have other plant-growth promoting activities such as production of phytohormones, inhibition of ethylene biosynthesis, and Phosphate solubilization (Govindarajan et al. 2008; Son et al. 2005). Nitrogen balance studies suggest that sufficient supply of biologically fixed nitrogen (as much as 150 kg N ha−1 year−1) by Acetobacter diazotrophicus to Brazilian variety of sugarcane, and Azoarcus to Kallar grass (Leptochloa fusca (20–40 t ha−1 year−1) is made available to the plants under natural condition (Boddey et al. 1995; Reinhold-Hurek et al. 1993; Sandhu et al. 1981). These investigations point to the potential of endophytic diazotrophs to increase the productivity of non legumes including important crop plants (Sturz et al. 2000). In present study, we report the colonization of endophytic diazotrophic Burkholderia cepacia in rice followed by subsequent plant growth promotion.
Materials and methods
Isolation and enumeration of diazotrophic endophytic bacteria
Healthy plants of four varieties of rice, viz., NDR97, HUR-36, Sarjoo52, and Pusa Basmati 1, were randomly collected from different sampling locations i.e. Varanasi, Balia, Mirzapur, Ghorakhpur, and Chandouli during the growing season from farmers field of eastern Uttar Pradesh, India. Plants of each variety from four plots were uprooted and brought immediately to the laboratory. These varieties of rice grown in fields getting minimal application of synthetic chemical N fertilizer. For screening of diazotrophic bacteria, initially, the plant roots/shoots were thoroughly washed, surface-sterilized, and subjected to Acetylene Reduction Assay (ARA). Only those plants that showed positive ARA were selected for the isolation of endophytic bacteria. Putative endophytic diazotrophic bacteria were isolated and cultivated on BAz/BAc and JNFb− media (Estrada-de los Santos et al. 2001, Singh 2008) while Nutrient agar (NA) was used for efficiency of sterilization and bacterial enumeration. Surface-sterilization of rice roots and pure endophytic bacterial colonies were isolated following standard protocol (Singh et al. 2006). In brief, roots were cleaned, surface sterilized in 95% ethanol and 0.1% HgCl2, and macerated in a sterile pestle and mortar. One milliliter of each root homogenate was aseptically inoculated to different ‘‘legume trap’’ hosts, i.e., Vigna radiata, V. mungo, V. unguiculata, Pisum sativum, Glycine max, Cajanus cajan, Cicer arietinum, Trifolium alexandrinum, Phaseolus vulgaris, and Sesbania aculeata grown in test tubes. These tubes were filled with nitrogen-free Fahraeus medium (NFM) (Fahraeus 1957) solidified with 1.5% agar. Seedlings were grown for 35 days in a plant growth chamber programmed for a 14-h photoperiod, 26–28/22–24°C day/night cycle of temperature and 70% relative humidity. Uninoculated plants served as control. Established reference strains of B. cepacia strain RRE3, RRE5 (Singh et al. 2006) and B. vietnamiensis strain LMG10929, were used as positive control to compare the symbiotic performance of the rice-borne Burkholderial isolates. All six isolates grown in King’s B medium were tested for fluorescence by observing under UV transilluminator at 365 nm.
Production of Indole Acetic Acid (IAA), phosphate solubilization and nitrogenase activity of endophytic isolates
IAA was extracted from culture supernatants/cultures by a modified method of Manulis et al. (1994). High-performance liquid chromatography (HPLC) of extracted IAA was performed in a Shimadzu system (Shimadzu Corporation, Kyoto, Japan) equipped with two LC-10 ATVP reciprocating pumps, a variable Shimadzu SPD-10 AVP UV–VIS detector and a Rheodyne (Model 7725) injector with a loop size of 20 μl. IAA present in the samples was identified by comparing retention time (Rt) with an individual reference standard and further by co-injection. Screening of phosphate solubilization and quantitative estimation of solubilized P and Nitrogenase activity were done following standard protocol (Singh 2008). Three replicates were used for analysis.
16S rRNA and nifH gene amplification, cloning, and sequencing
Genomic DNA was extracted by D Neasy Tissue kit (Qiagen GmbH, Hilden, Germany) as per the instructions of manufacturer. Nearly full-length rRNA gene of RREM25 isolate was amplified using the universal primers (PA 5′-AGAGTTTGATCCTGGCTCAG-3′ and PH 5′-AAGGAGGTGATCCAGCCGCA-3′), as per the standard protocol Singh et al. (2006). In case of nifH gene, the DNA of RREM25 isolate was used as test while Escherichia coli that of as negative control and the PCR amplification using the primers 19F (5′-GCIWTYTAYGGIAARGGIGG-3′) and 407R (5′-AAICCRCCRCAIACIACRTC-3′) was carried out as described Ueda et al. (1995). PCR amplified products of 16S rRNA and partial nifH gene were purified using QIAquick PCR purification kit (Qiagen GmbH, Hilden, Germany), cloned into pGEM®T vector (Promega Corp., Madison, WI, USA) and sequenced with ABI automated DNA Sequencer using ABI Big Dye termination cycle sequencing ready reaction kit (Applied Biosystems, USA) as per the protocol of manufacturer. The 16S rRNA and nifH gene sequences were compared to similar gene sequences available at GenBank database using BLASTn program (Altschul et al. 1997) and were aligned separately with the sequences of closely related species of Burkholdaria genus. The phylogenetic trees based on 16S rRNA and nifH partial gene sequences were constructed by neighbor-joining method using MEGA 4.0 (Saitou and Nei 1987; Tamura et al. 2007). All bootstrap replications (1,000 replications) indicating more than 80% support were placed at the nodes in the phylogenetic tree.
Immunological detection of dinitrogenase reductase protein
Western blot hybridization was used to detect the presence of nitrogenase enzyme in the isolate RREM25. Culture was grown in N-free minimal liquid medium (JNFb−) for 96 h at 30°C, and whole-cell proteins were extracted from the culture. The immunological detection was made by using mixture of polyclonal antibody raised against dinitrogenase reductase protein of Rhodospirillum rubrum and Azotobacter vinelandii according to Schloter et al. (1995). The antibody was a generous gift from P.W. Ludden, University of Wisconsin, Madison, USA. Western blot hybridization was used to detect the presence of nitrogenase enzyme in the isolate RREM25.
Tagging of bacterial strains with gfp/gusA
With a view to identify native of RREM25, it was marked with gfp/gusA reporter gene using a Plasmid pHRGFPGUS (Ramos et al. 2002). B. cepacia (RREM25) resistant to Rifampicin (125 μg ml−1) were chosen as recipients for genetic tagging with gfp/gusA reporter. This strain was sensitive to Kanamycin (50 μg ml−1). Plasmid pHRGFPGUS containing the gfp and gusA genes expressed under the control of a gentamycin promoter was introduced by biparental mating using donor strain E. coli S17-1 (Simon et al. 1983; Bhatia et al. 2002; Sharma et al. 2005). Plasmid pHRGFPGUS is a derivative of plasmid pBBR1, which is a small (2.6 kb), broad-host range plasmid and stably maintained in a number of Gram-positive and Gram-negative bacteria (Quanhrani-Bettache et al. 1999). The conjugation mix was incubated for 24 h on Luria agar plates and then spread on Luria agar plates containing appropriate antibiotics. Ex-conjugants showing green fluorescence under UV illumination were selected for further study. Seedlings were inoculated as per the method described by Singh et al. (2009).
Confocal laser scanning microscopy (CLSM)
Seven days after inoculation, rice seedlings (inoculated and uninoculated) were taken out from the culture tubes, washed with sterile water, cut into small pieces, and mounted on bridge slide with 10% (v/v) glycerol (microscopy grade). Optical sections of the root pieces were observed in a Bio-Rad Radiance 2000 Multiphoton CLSM system attached to a Nikon E-300 inverted microscope. GFP-tagged bacterial cells were excited with the 488 nm Argon laser line. Images were collected in a z-series from 10 to 25 optical sections ranging from 1 to 2 μm in thickness.
In planta nitrogenase activity and growth of rice variety HUR-36 in gnotobiotic and green house conditions
The AR (Acetylene Reduction) activity of inoculated plants roots was determined following standard protocol (Govindarajan et al. 2008). For the assessment of plant growth–promoting potential of B. cepacia RREM25 a plant infection test was performed along with RRE3, RRE5 and LMG10929 in gnotobiotic and green house condition. For raising gnotobiotic culture of rice cultivar HUR-36 with Burkholderia, seeds were de-hulled, treated with 96% ethanol followed by 0.1% acidified HgCl2 for 10 min and were washed six times with sterile water. Seeds were incubated in dark at 30°C for 3 days for germination on 0.8% water agar plates. Seedlings were transferred to culture tubes at the rate of one seedling per tube having agar slants containing nitrogen free Fahraeus medium. Simultaneously Burkholderial cells, grown in BAc/BAz medium, were harvested by centrifugation at 10,000g for 10 min at 4°C and resuspended in phosphate buffer (pH 7.0). Each tube was inoculated with 1 ml bacterial suspension ±108 c.f.u.ml−1 (mean inoculation level 3 × 105 cells seed−1) and transferred in plant growth chamber programmed with 14/10 h light/dark period, 28–30/23–25°C day/night cycle of temperature and 70% relative humidity. Uninoculated seedlings served as control. In the green house experiment, the seeds of rice cultivar HUR-36 were surface sterilized as described earlier. Other experimental conditions for growth remained the same. Treated seeds were sown in plastic pots containing 200 g sterilized sand. Pots were inoculated with 1 ml culture of similar bacterial content after 3 days of seedlings emergence. Plants were regularly irrigated with sterile water and the effect on plants was assessed after 35 days of inoculation by measuring the plant height, dry shoot and root weight, chlorophyll content, nitrogen content and nitrogenase activity of the test plants (Singh 2008).
Statistical analysis
The data were analyzed by analysis of variance (ANOVA) and the means were compared following Fishers test of least significant difference (LSD) to assess the effect of inoculation on rice cultivar.
Results
Enumeration of diazotrophic endophytic bacteria
Interestingly, out of four varieties, the macerate of only the rice variety HUR-36 showed the presence of endophytic diazotrophic bacteria when tested for growth on JNFb—solid medium. Based on distinct morphotypes of colonies on JNFb—solid agar medium, four root isolates RREM25, RREM34, RREM42, and RREM17, and two culm isolates, RREM37, and RREM51, were picked up and grown on Burkholderial specific nitrogen-free medium (BAc/BAz). The root macerates routinely showed higher number (3 × 105) of diazotrophic bacterial isolates than those of culm (3 × 104 cfu/g fresh wt). However, bacterial population was much higher (2 × 107 cfu/g fresh wt) when the root/culm macerate was plated on NA medium. Growth of any bacteria neither occurred on plates spread with the last wash nor on sections of surface sterilized roots or culms suggesting the recovery of isolates only from the interior parts of the plants. None of the isolates even showed fluorescence under UV light.
Test of IAA production, phosphate solubilization and nitrogenase activity of endophytic isolates
Initial screening revealed IAA production in six isolates out of which highest production was shown by RREM25 (19.50 μg mg−1 dry wt) followed by LMG 10929 (17.55 ± 3.0 μg mg−1 dry wt) after growth in the medium supplemented with tryptophan (Table 1). Standard peaks identified as IAA were recorded and compared with all the three isolates through the analysis of samples by HPLC (Fig. not shown). In addition to nitrogen-fixing ability and IAA production, the two isolates were found positive for P solubilization. Initial screening for P solubilization activity was based on the appearance of a clearing zone around the bacterial colonies on solid agar medium supplemented with insoluble phosphate. Further analysis showed that the isolate RREM25 had the highest level (31.50 μg mg−1 dry wt) of P solubilization (Table 1). Diazotrophic nature of all the isolates was determined by ARA. Eight isolates exhibited nitrogenase activity that ranged between 0.51 and 1.70 μmol C2H4 mg−1 protein h−1, RREM25 being the highest, while nitrogenase activity could not be detected in the isolate RREM37 (Table 1).
16S rRNA and nifH gene identification of selected endophytic isolate
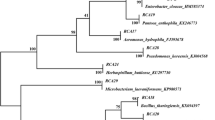
Among all the tested isolates, the isolate RREM25 appeared to be potent in terms of nitrogenase activity, P solubilization, and IAA production (Table 1). These characters of the isolate led to the identification of RREM25 as B. cepacia based on 16S rRNA gene. 16S rRNA gene sequence analysis that showed highest degree of similarity to B. cepacia (99.1%). The phylogenetic tree constructed on the bases of 16S rRNA gene sequences indicated the position of B. cepacia in close relation with other Burkholderia species (Fig. 1a). In case of nifH gene, ~390 bp fragment was obtained in PCR amplification (Fig. 2). The phylogenetic analysis based on partial nifH gene sequence of RREM25 showed significant similarity with other Burkholderia species (Fig. 1b). Amplification of nifH was not observed in negative control. The partial sequences of 16S rRNA and nifH gene of B. cepacia RREM25 have been deposited in NCBI, GenBank under accession numbers EU246850 and HQ699896, respectively.
The phylogeny of B. cepacia RREM25 with different Burkholderia species. a Phylogenetic tree based on 16S rRNA gene sequences. b Phylogenetic tree based on partial NifH DNA sequences. The strain B. cepacia RREM25 used in this study was indicated in Box. Phylogenetic trees were generated by neighbor-joining method using MEGA 4.0. The percentage of bootstrap values (1,000 resampling) that supported more than 80% were indicated at the branches
Detection of dinitrogenase reductase protein
Diazotrophy of B. cepacia RREM25 was investigated by Western blot hybridization of nitrogenase. Applying immunoblotting technique, a ~27-kDa fragment of dinitrogenase reductase was detected after hybridization with specific antibodies (Fig. 3). No positive signal was observed in protein extract of E. coli.
Inoculation of rice seedlings by gfp/gusA-tagged Burkholderia cepacia RREM25
Test of growth rate and plant growth promoting features, viz., nitrogen fixation, IAA production, and P solubilization of the transconjugants, revealed no change and were almost identical to that of parent isolate RREM25. When gfp/gusA-tagged isolate was inoculated on germinated seedlings of rice variety, HUR-36, active colonization occurred and was confirmed by reisolation of tagged strain from the inoculated rice plants. As expected, uninoculated control plants grown under identical conditions did not show the presence of any bacteria in the roots or culms. Moreover, no disease symptoms appeared in control or inoculated plants throughout the study. At 40× resolution, gfp/gusA-tagged cells were apparently localized in intercellular spaces of cortical as well as vascular zones, indicating the entry of gfp/gusA-tagged isolate RREM25 (Fig. 4). Rice roots of uninoculated control plant did not show any fluorescence 7 days after inoculation (DAI).
In planta nitrogenase activity and growth of rice variety HUR-36 in gnotobiotic and green house conditions
Our results indicated that B. cepacia isolate RREM25, after inoculation, resulted in significant plant growth promotion after 35 DAI as assessed on different growth parameters viz. plant height, dry shoot and root weight, chlorophyll and nitrogen content (Tables 2 and 3). Maximum number of bacterial count most probable number (MPN) was recorded in RREM25 and HUR-36 interaction (Table 4). Colonization of rice variety with RREM25 also enhanced nitrogenase activity in both gnotobiotic and green house conditions 20.22 and 21.32 nmol C2H4 mg−1 fresh weight of root, respectively (Table 4).
Discussion
B. cepacia complex is regarded as a potential human pathogen associated with several clinical manifestations such as ciystic fibrosis (Chiarini et al. 2006). We report the presence of endophytic B. cepacia in the roots and culms of healthy rice plant (HUR-36). It is pertinent to mention that HUR-36 is mostly grown in rain fed area and requires low input of chemical N fertilizers, which might favor association with endophytic diazotrophic bacteria such as B. cepacia. Apart from diazotrophy, other beneficial activities such as P solubilization and IAA production is an added feature of this bacterium. All the isolates recovered from the roots or culms are indeed endophytes and are present in the interior tissues of rice plants as apparent from the methods of sterilization employed. Occurrence of higher population of endophytes in roots in comparison to aerial parts has been reported in several plants, and our results are in accordance with its reports (Katherine et al. 2008). In previous study, roots have been shown to be a preferred niche for growth and nitrogen fixation by endophytic Burkholderia (Govindarajan et al. 2008). In the present study, six endophytic diazotrophic isolates from roots and culms were isolated from rice variety, HUR-36. There were significant differences in nitrogenase activity and IAA production and P solubilization activity among the six isolates. Only isolate RREM25 exhibited highest record for all the three growth-promoting characters. This isolate was identified as B. cepacia by rRNA gene sequence analysis, and it is placed in the class β-proteobacteria from which only a few members including B. vietnamensis, B. kururiensis, B. tropica, B. unamae, and a few others are known to show N2-fixing ability (Caballero-Mellado et al. 2004). Phylogenetic analysis suggested significant similarity to other strains of Burkholderia, which otherwise are known to perform different roles in the plant growth-promoting rhizobacteria (Govindarajan et al. 2008).We feel that plant-growth ability in B. cepacia strain RREM25 may have resulted from environmental adaptation. However, RREM25 may not be used as biofertilzer agent until its pathogenicity test is critically evaluated. Knowing that ARA is an indirect method to test the diazotrophic nature of any microorganism, confirmation of diazotrophy in RREM25 was established by localizing the key enzyme nitrogenase using immunoblotting technique as reported in Burkholderia (Estrada-de los Santos et al. 2001). The presence of a protein band specifically bound to anti-dinitrogenase reductase in this isolate clearly demonstrated the presence of active nitrogenase polypeptide in the cells. Result of Western blot hybridization was further supported by the amplification of ~ 390 bp fragment of nifH, which encodes dinitrogenase reductase. Diazotrophy was confirmed by amplification of nifH segment using different PCR primers from various organisms and natural samples (Ueda et al. 1995). In this study, fidelity of the amplified segment of nifH was assured from the sequence information, which displayed similarity with nifH sequence available in database of GenBank. Based on the above physiological and molecular evidences, it may be concluded that the isolate RREM25 is indeed a diazotrophic and active N2-fixing bacterium. One prominent feature of gfp/gusA-marked B. cepacia RREM25 noticed in this study is the active colonization of rice plants that was confirmed by reisolation of this strain from surface-sterilized roots and culms of the inoculated seedlings. Intense gfp activity was noted on intercellular spaces of cortical as well as vascular zones, which suggests that this region may be the possible site for colonization of this bacterium. This finding is in agreement with other reports on diazotrophic endophytic Burkholderia sp. in rice (Katherine et al. 2008; Singh et al. 2009). As such, the apoplastic localization in intercellular spaces is considered to be the preferred site for a few endophytic diazotrophs (James and Olivares 1998). Other hand in situ gus staining has a major drawback, the presence of blue color does not unequivocally confirm the location or even the presence of the gus-marked bacteria because the color can diffuse into bacterium-free plant material (Jefferson et al. 1987). Isolate RREM25 may be used as a potent plant growth bioagent is borne out by the experiments conducted with the variety HUR-36 where significant increase in length of root and stem and enhanced formation of lateral and adventitious roots were observed. It is well known that IAA secreted by a bacterium may promote root growth due to stimulatory effect on plant cell elongation or cell division or indirectly by influencing bacterial 1-aminocyclopropane-1-carboxylate (ACC) deaminase activity (Glick 2005). Further increased plant dry weight, chlorophyll and nitrogen content were observed. That might be due to the supply of fixed nitrogen by the colonized bacteria as a result of significant level of nitrogenase activity in rice plants detected 35 DAI which is also supported earlier finding (Govindarajan et al. 2008).
In conclusion, our results showed that the roots and culms of the rice variety HUR-36 harbor a variety of endophytic diazotrophic bacteria including B. cepacia (RREM25). This strain showed high level of nitrogenase activity, strongly solubilized P, and produced IAA. Despite the fact that B. cepacia did not show any disease symptoms in rice, it is an opportunistic and potential human pathogen, and therefore, it would be necessary to test its pathogenicity before exploiting its beneficial characters for other crop plants.
References
Altschul SF, Madden TL, Schafeer AA et al (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acid Res 25:3389–3402
Bhatia R, Dogra RC, Sharma PK (2002) Construction of green protein (GFP)-marked strains of Bradyrhizobium for ecological studies. J Appl Microbiol 93:835–839
Boddey RM, de Oliveira OC, Urquiaga S et al (1995) Biological nitrogen fixation associated with sugar cane and rice: contributions and prospects for improvement. Plant Soil 174:195–209
Caballero-Mellado J, Martynez-Aguilar L, Paredes-Valdez G et al (2004) Burkholderia unamae sp. nov., a N2-fixing rhizospheric and endophytic species. Int J Syst Evol Microbiol 54:1165–1172
Chen WM, James EK, Coeny ET et al (2006) Burkholderia mimosarum sp. nov., isolated from root nodules of Mimosa spp. from Taiwan and South America. Int J Syst Evol Microbiol 56:1847–1851
Chiarini L, Bevivino A, Dalmastri C et al (2006) Burkholderia cepacia complex species: health hazards and biotechnological potential. TRENDS in Microbiol 14:277–286
Estrada-de los Santos P, Bustillos-Cristales R, Caballero-Mellado J (2001) Burkholderia, a genus rich in plant-associated nitrogen fixers with wide environmental and geographic distribution. Appl Environ Microbiol 67:2790–2798
Fahraeus A (1957) The infection of clover root hairs by nodule bacteria studied by simple glass slide technique. J Gen Microbiol 16:374–381
Glick BR (2005) Modulation of plant ethylene levels by the bacterial enzyme ACC deaminase. FEMS Microbiol Lett 251:1–7
Govindarajan M, Balandreau J, Kwon SW et al (2008) Effects of the inoculation of Burkholderia vietnamensis and related endophytic diazotrophic bacteria on grain yield of rice. Microbial Ecol 55:21–37
James EK, Olivares FL (1998) Infection and colonization of sugarcane and other gramineous plants by endophytic diazotrophs. Crit Rev Plant Sci 17:77–119
Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: β- glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6:3901–3907
Katherine AM, Vania LMP, Alexandre R et al (2008) Endophytic colonization of rice (Oryza sativa L) by diazotrophic bacterium Burkholderia kururiensis and its ability to enhance plant growth. Annal Brazi Acad Sci 80:477–493
Ladha JK, de Bruijn FJ, Malik KA (1997) Assessing opportunities for nitrogen fixation in Oryza sativa: a frontier project. Plant Soil 194:1–10
Manulis S, Shafrir H, Epstein E et al (1994) Biosynthesis of indole-3-acetic acid via the indole-3-acetamide pathway in Streptomyces spp. Microbiol 140:1045–1050
Quanhrani-Bettache F, Porte J, Teyssier J et al (1999) pBBR1-GFP: a broad-host-range vector for prokaryotic promoter studies. Bio Tech 26:620–622
Ramos HJ, Roncato-Maccari LD, Souza EM et al (2002) Monitoring Azospirillum-wheat interactions using the gfp and gusA gene constitutively expressed from a new broad-host range vector. J Biotechnol 97:243–252
Reinhold-Hurek B, Hurek T, Gillis M et al (1993) Azoarcus gen. nov., nitrogen-fixing proteobacteria associated with roots of Kallar grass (Leptochloa fusca (L) Knuth), and descriptions of two species, Azoarcus indigens sp. nov. and Azoarcus communis sp. nov. Int J Syst Bacteriol 43:574–584
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sandhu GR, Aslam Z, Salim M et al (1981) The effect of salinity on the yield and composition of Diplachne fusca (Kallar grass). Plant Cell Environ 4:177–181
Schloter M, Bode W, Hartmann A (1995) Characterization and application of a strain-specific monoclonal antibody against the rhizosphere bacterium Azospirillum brasilense Wa3. Hybridoma 16:183–187
Sharma PK, Sarita S, Prell J (2005) Isolation and characterization of an endophytic bacterium related to Rhizobium/Agrobacterium from wheat (Triticum aestivum L.) roots. Curr Sci 89:608–610
Simon R, Priefer UB, Puehler A (1983) Vector plasmids for in vivo and in vitro manipulations of Gram-negative bacteria. In: Puehler A (ed) Molecular genetics of the bacteria—plant interactions. Springer-Verlag KG, Berlin, pp 98–106
Singh MK (2008) Genetic basis of colonization by endophytic bacteria and their role in plant growth promotion of rice. Ph.D. thesis, Banaras Hindu University, Varanasi, India, pp 95–99
Singh RK, Mishra RPN, Jaiswal HK et al (2006) Isolation and identification of natural endophytic rhizobia from rice (Oryza sativa L.) through rRNA gene PCR-RFLP and sequence analysis. Curr Microbiol 52:345–349
Singh MK, Kushwaha C, Singh RK (2009) Studies on endophytic colonization ability of two upland rice endophytes, Rhizobium sp. and Burkholderia sp., using green protein reporter. Curr Microbiol 59:240–243
Son H, Park G, Cha M et al (2005) Solubilization of insoluble inorganic phosphates by a novel salt and pH tolerant Pantoea agglomerans R-42 isolated from soyabean rhizosphere. Bioresource Technol 97:204–210
Sturz AV, Christie BR, Nowak J (2000) Bacterial endophytes: potential role in developing sustainable system of crop production. Crit Rev Plant Sci 19:1–30
Tamura K, Dudley J, Nei M et al (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Ueda T, Suga Y, Yahiro N et al. (1995) Remarkable N2- fixing bacterial diversity detected in rice roots by molecular evolutionary analysis of nifH gene sequences. J Bacteriol 177: 1414–1417
Yanni YG, Rizk RY, Corich V et al (1997) Natural endophytic association between Rhizobium leguminosarum bv trifolii and Oryza sativa roots and assessment of its potential to promote Oryza sativa growth. Plant Soil 194:99–114
Yanni YG, Rizk RY, Fatah FKA et al (2001) The beneficial plant growth-promoting association Rhizobium leguminosarum bv trifolii with Oryza sativa plants. Aust J Plant Physiol 28:845–870
Acknowledgments
We are thankful to HJO Ramose (Curitiba, PR, Brazil), for providing plasmid pHRGFPGUS as gift. MKS is thankful to UGC, India for the award of Research Fellowship, to carry out this study. Research works were partly supported by the grant received from the DST Gov. of India, New Delhi.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Singh, M.K., Singh, D.P., Mesapogu, S. et al. Concomitant colonization of nifH positive endophytic Burkholderia sp. in rice (Oryza sativa L.) promotes plant growth. World J Microbiol Biotechnol 27, 2023–2031 (2011). https://doi.org/10.1007/s11274-011-0664-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11274-011-0664-z