Abstract
During a survey of endophytic diazotrophic bacteria associated with different rice varieties in Tamilnadu, some “endophytes” were obtained. Thirteen bacterial isolates from surface-sterilized roots and shoots were obtained in pure culture, which produced indole acetic acid (IAA) and reduced acetylene to ethylene. Polymerase chain reaction (PCR) amplification confirmed the presence of nif-H gene in all the isolates. Morphological, biochemical, and molecular characteristics indicated that all of them belonged to the genus Burkholderia One of them, MGK3, was consistently more active in reducing acetylene, and 16S rDNA sequences of isolate MGK3 confirmed its identification as Burkholderia vietnamiensis. Colonization of rice root was confirmed by strain MGK3 marked with gusA gene. The inoculated roots showed a blue color, which was most intense at the points of lateral root emergence and at the root tip. Transverse sections of roots, 15 days after inoculation, revealed beta-glucuronidase (GUS) activity within many of the cortical intercellular spaces next to the stele and within the aerenchyma. Nitrogen fixation was quantified by using 15N isotope dilution method with two different cultivars grown in pot and field experiments. Higher nitrogen fixation was observed in variety Ponni than in ADT-43, where nearly 42% (field) and 40% (pot) of the nitrogen was derived from the atmosphere (% Ndfa). Isolate MGK3 was used to inoculate rice seedlings in a comparison with four other diazotrophs, viz., Gluconacetobacter diazotrophicus LMG7603, Herbaspirillum seropedicae LMG6513, Azospirillum lipoferum 4B LMG4348, and B. vietnamiensis LMG10929. They were used to conduct two pot and four field inoculation experiments. MGK3 alone, and combined with other diazotrophs, performed best under both pot and field conditions: combined inoculation produced yield increases between 9.5 and 23.6%, while MGK3 alone increased yield by 5.6 to 12.16% over the uninoculated control treatment.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The high-yielding rice varieties of the “Green Revolution” have resulted in large increases in rice production but require large amounts of nitrogen fertilizers, which contribute to nitrate contamination of soils and ground water supplies, leading to health hazards and environmental pollution. Moreover, for many farmers, the use of chemicals is too costly, especially nitrogen, which is the most frequent limiting factor of rice production. An alternative is the use of bacterial inoculants in crop plants. Rhizobacteria that establish positive interactions with plant roots have been called plant growth-promoting rhizobacteria (PGPR) and are promising for their potential use in sustainable agriculture [18]. Application of bacterial inoculants as biofertilizers has resulted in improved growth and increased yield of cereal crops [36]. Nitrogen-fixing bacteria belonging to the genera Azospirillum, Gluconacetobacter, Azoarcus, Enterobacter, Herbaspirillum, and Burkholderia appear to be frequent colonizers of important cereal crops and grasses and have been extensively studied [2, 3, 6, 8, 19, 21, 25 and 50].
During the last two decades, molecular techniques have been widely used to study the endophytic diazotrophs in many cereal crops. Previously, methods used to detect and isolate endophytes mostly involve maceration and centrifugation of surface-sterilized plant tissues. These methods can give estimates of bacterial population and extent of colonization but fail to reveal the site of infection of the colonization process. The development in use of reporter genes, such as the gusA gene from Escherichia coli has facilitated investigations of plant–microbe interactions [17]; on the other hand, 15N dinitrogen incorporation assay make it possible to quantify the biologically fixed nitrogen in the plants precisely [11].
The genus Burkholderia comprises over 50 species; of which, nine species viz., Burkholderia vietnamiensis, Burkholderia kururiensis, Burkholderia tuberum, Burkholderia phymatum, Burkholderia unamae, Burkholderia tropica, Burkholderia xenovorans, Burkholderia phytofirmans, and Burkholderia terrae were verified as nitrogen fixers [13, 22, 26, 27, 29, 52, 54 and 64]. Recently, Chen et al. [15 and 16] isolated several Burkholderia strains from legume nodules (including B. phymatum and B. tuberum), and some of these have been confirmed to be N-fixing symbionts of Mimosa spp. Of which, the species B. vietnamiensis TVV75 has been extensively studied in Vietnam, for its PGPR effect on rice, both under pot and field conditions. Inoculation significantly increased several yield components, resulting in a final 13 to 22% increase in grain yield [62]. Baldani et al. [4] observed 42–64% increases in growth of rice plants when Burkholderia “brasilensis” and B. vietnamiensis were inoculated under gnotobiotic conditions. Govindarajan et al. [28] inoculated B. vietnamiensis strain MG43 to micropropagated sugarcane plantlets in a comparison with two other diazoptrophs, viz. Gluconacetobacter diazotrophicus and Herbaspirillum seropedicae. Inoculated plants and uninoculated controls were used in a pot experiment followed by two field experiments under different rates of nitrogen fertilizers. Yield increase due to B. vietnamiensis strain MG43 inoculation reached 20% in the field. Taken together, these examples indicate that these bacteria have an important role in improving plant nutrition, in the context of low input sustainable agriculture.
In this study, we report (1) to isolate endophytic nitrogen-fixing Burkholderia spp. from the different cultivated rice fields of Tamilnadu, India, and (2) to investigate endophytic colonization and in planta nitrogen fixation of a selected diazotroph to verify whether the bacteria function as nitrogen-fixing endophytes, and (3) their performance under pot and field conditions as potential bio-fertilizer inoculants capable of increasing rice grain yield.
Material and Methods
In short, the methods used for identification, growth promoting properties, and studying colonization pattern of Burkholderia sp. are: fatty acid analysis, amplification of nif-H gene, sequencing of 16S rDNA, acetylene reduction (AR) activity, IAA production, 15N dinitrogen incorporation assay, and GUS assay. To determine inoculation effects, two pot experiments followed by four field trials were conducted using with four different cultivars.
Culture Media and Growth Conditions
Isolation media were PCAT and LGIM [12, 23]; BDN [23] was used for growing strains before DNA extraction; BMGM media [23] was used to analysis physiological, biochemical, and acetylene reduction activity. LGI-P [51] and LGIM were used for Gluconacetobacter and Burkholderia recovery; JNFb [40] was used to recover Herbaspirillum and Azospirillum.
Reference Strains
The reference strains used in this study were G. diazotrophicus [67] strain LMG7603 has been isolated from sugarcane root tissues in Brazil as strain PAL 5 of Acetobacter diazotrophicus [25]. H. seropedicae [3] strain LMG6513 has been isolated from surface sterilized rice roots in Brazil. B. vietnamiensis [26] strain LMG10929 has been isolated by Tran Van et al. [61], from rice roots in Vietnam. Azospirillum lipoferum [6] strain 4B (LMG4348) has been isolated from a rice rhizosphere in France.
Sampling, Isolation, and Enumeration
In ten different fields of Tamilnadu, the roots, stems, and leaves of rice plants were sampled. Cultivars were IR20, Ponni, ADT-37, ADT-43, and Co-47. Isolation followed the method described by Estrada et al. [23]. Tenfold serial dilutions were used to inoculate (in triplicate) N-free semisolid LGIM tubes. After 96–120 h of incubation, vials were assayed for AR activity following Mascarua-Esparza et al. [45]. Bacteria growing in nitrogenase-positive vials as a white or yellowish pellicle at a depth of 1 to 4 mm were streaked on LGIM agar plates. All isolates were checked for purity and their ability to grow on modified PCAT agar (0.05 g/l yeast extract powder added) plates [12]. Most Probable Numbers (MPN) of PCAT-growing and AR activity positive isolates were calculated using the McGrady tables. Isolates were maintained in semisolid BMGM for further studies.
Fatty Acid Analysis
The MIDI-FAME technique was used to determine the cellular fatty acid profiles of the isolate MGK3 (most active strain, isolated from surface sterilized stems of cv. Ponni) and the B. vietnamiensis LMG10929. Isolates were grown overnight on trypticase soy agar plates. One loopful of fresh cells was harvested and transferred to a screw cap culture tube. One milliliter of saponification reagent was added. Tubes were tightly sealed with a Teflon-lined screw cap and vortexed for 5–10 s. The tubes were placed in water bath at 100 ± 2°C for 5 min. They were then removed from the boiling water bath and cooled slightly, vortexed for 10 s, and incubated in a water bath for an additional 25 min. After a total of 30 min of saponification in the water bath, the samples were placed under tap water. Each tube received 2.0 ml of methylation reagent, was tightly capped and vortexed for 10 s. The tubes were placed in a water bath at 80 ± 1°C for 10 min. After that, samples were cooled at room temperature. Then, 1.25 ml of extraction reagent was added into the tubes. The tubes were centrifuged, and the bottom phase was removed using a pipette. Finally, 3.0 ml of base reagent was added, and this was placed again for 5 min in a laboratory centrifuge. The upper solvent phase was removed and transferred to vials for fatty acid analysis.
Physiological and Biochemical Characterization
Pigment production was monitored on nutrient agar and King “B” media. Colony morphology was examined on PCAT and BMGM agar plates. Morphology and Gram type were determined using a Trinocular Phase Contrast Fluorescent Microscope (Olympus AX 80T). Bacterial motility was tested by growth in a semisolid 0.3% mannitol motility test medium. Oxidase and catalase tests were determined using commercially available discs (Hi media, Bombay, India). Growth and acid production were tested using the BMGM medium in which the carbon source was replaced by individual carbon substrates (5 g/l) such as d-glucose, sorbitol, meso-inositol, mannose, glycerol, l-rhamnose, lactose, fructose, l-arabinose, trehalose, l-raffinose, meso-erythritol, galactose, mannitol, cellobiose, xylose, sucrose, starch, sodium acetate, maltose; organic acids were also tested including adipic, malonic, succinic, oxalic, valeric, fumaric, hippuric, malic, tartaric, keto glutaric, and citric acids. Growth on the following amino acids was tested (in the presence of sorbitol as a carbon source): cysteine, glutamic acid, proline, trytophane, leucine, threonine, histidine, lysine, tyrosine, and valine.
Isolation of Total DNA
Isolate MGK3 and B. vietnamiensis LMG10929 were grown in BDN medium at 28°C for 24 h and centrifuged at 12,300×g. The pellet was washed with Tris–ethylenediaminetetraacetic acid (EDTA) (TE) buffer, then resuspended in 10 ml of TE (1×) with 3 ml of 5% sodium dodecyl sulfate (SDS) in TE (1×) and 3 ml of proteinase K 2.5 mg/ml. This was then incubated at 37°C for 1 h. The cleaned lysates were extracted with phenol is to chloroform is to isoamylic alcohol (25:24:1). DNA was precipitated by adding one-tenth volume of 3 M sodium acetate (pH 5.2) and 2.5 volume of ethanol to the supernatant. Dried pellets were dissolved in 1× TE buffer.
PCR Amplification of the nif-H Genes
PCR amplification was performed to determine the presence of nif-H gene using specific primers described by Ueda et al [63]. Amplification reactions were performed in a total volume of 25 μl. The reaction mixture contained: 2.5 μl 10× PCR buffer, 2.5 μl of 2 mM each of dATP, dCTP, dTTT, and dGTP; 3 μl of each forward and reverse primer (30 ng), 1 μl of template DNA (10 ng) and 0.3 μl of (3 U/μl) Taq polymerase; final volume was made into 25 μl using milli-Q water. The step-up PCR procedure included denaturation at 95°C for 3 min, 52°C for 1 min, and 72°C for 1 min, followed by 30 cycles of 95°C for 1 min, 54°C for 30 s, and 72°C for 1 min, with a final extension at 72°C for 10 min. Amplification products were electrophoresed on 1.5% agarose gel in 1× Tris–borate–EDTA (TBE) buffer.
PCR Amplification and 16S rDNA Sequencing
`The 16S rDNA gene sequences were determined by PCR amplification [43] and direct sequencing [31]. For the phylogenetic analyses, related 16S rDNA gene sequences within the genus Burkholderia were downloaded from GenBank. 16S rDNA sequences were aligned by using the MEGALIGN program of DNASTAR. An evolutionary distance matrix was generated as described by Jukes and Cantor [38]. The evolutionary tree for the dataset was inferred from the neighbor-joining method of Saitou and Nei [53] using the neighbor-joining program of MEGA version 2.1 [42]. The stability of relationships was assessed by performing bootstrap analyses of the neighbor-joining data based on 1,000 resamplings.
Nucleotide Sequence Accession Numbers
The nucleotide sequences of diazotrophs isolated in this work have been deposited under the accession numbers to NCBI: AY789926-MGK3.
Nitrogenase Activity
Nitrogen fixation by the isolate MGK3 was determined in semisolid BMGM medium by the acetylene reduction method. The vials containing 10 ml of the BMGM semisolid media were inoculated with single colonies and incubated at 28°C for 3 days. Acetylene (10% v / v) was injected into the inoculated vials, and again, they were incubated at 28°C for 24 h. To determine the ability of reduced acetylene on different carbon sources, isolates were incubated on BMGM medium in which the carbon sources was replaced by the individual carbon sources (5 g/l). Eight important carbon sources were used: azelaic acid, fructose, glycerol, succinic acid, sucrose, mannitol, malic acid, and glucose. Acetylene reduction activity was measured using a flame ionization gas chromatograph (Systronic) equipped with a Porapak N column. Uninoculated vials were used as negative controls. The AR activity of inoculated plants roots was determined according to Ladha et al. [44]. Ten seedlings from each treatment were taken at panicle initiation and grain filling stages, and roots were separated and washed twice with sterile water to remove loosely associated bacteria. The roots were then transferred to fresh, N-free, liquid Jensen’s medium [58]. The tubes containing the roots were sealed with a rubber seal, and 10% of the headspace volume was replaced with acetylene. Uninoculated plant roots and tubes not injected with acetylene served as controls. Tubes were maintained at 25 ± 2°C with a relative humidity of 75% and 16 h light (60 m lum m−2) and 8 h dark. AR activity was determined as mentioned above.
IAA Production
To quantify the production of IAA by the isolate MGK3 and B. vietnamiensis LMG10929, bacteria were grown in CCM for 1 week, and the cells were pelleted by centrifugation at 10,000×g for 15 min. The pH of the supernatant was adjusted to 2.8 with HCl and then extracted three times with equal volumes of ethyl acetate [60]. The extract was evaporated to dryness and resuspended in 1 ml of ethanol. The samples were analyzed on high performance liquid chromatography (HPLC) (Shimadzu SPE 10A, 10AD) using an ultraviolet (UV) detector and a Techsphere C-18 column. Pure IAA was used as a standard. Methanol is to acetic acid is to water (30:1:70 v / v / v) was used as a mobile phase at the rate of 1.2 ml/min [49].
GUS Labeling of Burkholderia sp. Strain MGK3
E. coli (S17.1 pir) containing the constitutive transposons (mTn5ssgusA11) in plasmid pCAM111 [66], which was used as the donor, was grown in Luria agar medium containing 100 mg l−1 of ampicillin at 37°C with shaking overnight. pCAM111 was transferred to MGK3 by conjugation using a filter-mating technique [57]. The transconjugants7 were isolated on LGIM medium containing spectinomycin (100 mg l−1) and gusA substrate 5-bromo-4-chloro-3-indolyl-β-d-glucuronate (X-Gluc; Biosynth AG, Switzerland, at 500 mg l−1). The transconjugants were selected based on the phenotypic expression in the presence of substrate containing medium. The marker strains were further analyzed for the presence of marker gusA gene by PCR technique [41].
Plant Inoculation and Gus Staining
Dehulled seeds of rice (Oryza sativa) cultivars, ADT-43, ADT-37, Co-47, and Ponni were surface-sterilized with 95% (v / v) ethanol for 2 min and 2.5% (w / v) NaClO3 for 30 min, followed by washing seven times with sterile water. They were then transferred to plates containing 0.75% (w / v) agar for germination. The germinated seeds were transferred aseptically on to filter paper and placed in 50 ml glass tubes with 15 ml modified Jensen’s N-free medium [58] and maintained at 25 ± 2°C with a relative humidity of 75% and 16 h light (60 m lum m−2) and 8 h dark. The broth culture, 1 ml, of transconjugant (isolate tagged with gusA) was diluted to approximately 108 cells ml−1 and used for inoculation. For GUS staining, three plant samples (inoculated and uninoculated) were collected at intervals, washed with sterile 50 mM PBS, pH 7, and stained with 500 μg 5-bromo-4-chloro-3-indolyl-â-d-glucuronate(X-Gluc) ml−1, 0.1% (w / v) SDS in 1% (v / v) Triton X-100, and incubated overnight at 30°C. Samples were then cut into small pieces and fixed in 4% (v / v) glutaraldehyde in 50 mM phosphate buffer (pH 7) containing 0.1% (v / v) Triton X-100 under vacuum for 15 min and then incubated overnight at atmospheric pressure. Fixed samples were rinsed in 50 mM phosphate buffer and taken up for sectioning and viewing under the light microscopy.
Enumeration of Transconjugant MGK3 Colonizing the Inoculated Plants
The roots were washed in excess sterile water and then immersed in sterile water and vortexed for 30 s. The resulting solution was serially diluted and placed on LGIM agar plates containing spectinomycin (100 μg ml−1) and X-Gluc (40 μg ml−1). Blue colonies were then enumerated. In another set, the roots were surface sterilized by immersion in 95% (v / v) ethanol for 5 min followed by treatment with 2.5% (w / v) NaClO3 for 20 min followed by sterile water wash seven times. It was then macerated in saline and the homogenate was serially diluted and plated on LGIM agar as described above.
15N Dinitrogen Incorporation
Thirty-day-old inoculated and uninoculated rice (cv. Ponni and ADT-43) seedlings were grown in pot and field experiments and were transplanted in Pyrex tubes (25.5 × 2.5 cm) containing sterilized vermiculite. All the plants, including the uninoculated control plants were approximately 10 cm tall. For each treatment, eight plants were used. Half strength of N-free Jensen’s medium was used as nutrient source. 15N-labeled ammonium sulfate (100 mg N/tube) of 10% atom excess was added as a tracer to quantify nitrogen fixation. Plants were kept at 30 ± 2°C during the day and 25 ± 2°C at night for 4 weeks. At the time of harvesting, root area of the plant was measured with the Root Image Analysis Program (Washington State University Research Foundation, USA). The plants were dried in an oven at 70°C until no change in weight was noted. The dried plant samples were ground to a fine powder, and total N in these samples was determined by using a semi micro-Kjeldahl method based on wet combustion in a Rapid Kjeldahl (Labconco, Kansas city, Missouri). The analysis for 15N excess was carried out on a double inlet mass spectrometer (Varian MAT GD 150). Quantification of nitrogen fixation based on isotope dilution was calculated by the formula of Fried and Middleboe [24] which is:
Where fs is fixing system and nfs is non-fixing system.
Pot Experiments
Two pot experiments were conducted using two different rice cultivars (ADT-43 and Ponni) differing in length of growth cycle. Dehulled seeds were germinated as mentioned above. Five-day-old uncontaminated seedlings were treated with a suspension of respective cultures containing 108 cfu ml−1. After 30 min contact, seedlings were transplanted into pots. Pots were 60 cm in diameter, 45 cm high, and contained 30 kg of soil. Experiments comprised seven levels of inoculation viz. (1) control pots treated with an autoclaved mixture of bacterial strains, (2) G. diazotrophicus LMG7603, (3) H. seropedicae, (4) A. lipoferum LMG4348, (5) B. vietnamiensis LMG10929, (6) isolate MGK3, and (7) a combination of all five strains. In each pot, eight hills were maintained, with three seedlings per hill. Irrigation was natural rainfall and borewell water. G. diazotrophicus LMG7603 counts were done on medium LGIP where they form a typical subsurface pellicle; isolates were obtained from this pellicle by streaking on LGI agar plates supplemented with 50 mg/l yeast extract [50]. Burkholderia were counted on LGIM [22] followed by streaking on PCAT [11] for confirmation. H. seropedicae and A. lipoferum LMG4348 were counted on JNFb agar plates [40]. Leaf N was determined by the microKjeldahl method [32]. Shoot height, shoot weight, root weight, and tiller numbers were done at two dates (60th and 110th day for ADT-43 and 60th and 150th day for Ponni), and grain weight was measured at harvest.
Field Experiments
Four commercially important cultivars were selected for conducting field trial experiments at different places of Tamilnadu: ADT-43, ADT-37, Co-47, and Ponni. To germinate, rice seeds were tied in gunny bags, dipped overnight in water tanks, and kept in the shade for germination (36 to 48 h). After germination, the first inoculation was performed by adding approximately 108 cells per seed (based on the optical density at 600 nm); water was then added to keep the seeds completely immersed overnight to allow bacterial cells to adhere to seeds and roots. Treatments were: (1) control pots treated with an autoclaved mixture of bacterial strains, (2) G. diazotrophicus LMG7603, (3) H. seropedicae LMG6513, (4) A. lipoferum LMG4348, (5) B. vietnamiensis LMG10929, (6) isolate MGK3, and (7) a combination of all five strains. After incubation, inoculated pre-germinated seeds were sown in nursery beds, which had a surface representing 10% of the experiment area. The nurseries were flooded to 2 cm water above the soil surface after seedling emergence. At the three-leaf stage (25 to 30 days), rice seedlings were transplanted into the field plots. At transplanting, young plantlets were inoculated again by dipping them in the respective bacterial suspension for 30 min. The total area of each experiment was 5,600 m2, divided into seven 800 m2 areas. Control and inoculated plots were separated by mud levees. Irrigation was natural rainfall and bore well water; sub-canals for individual treatments were maintained. Each hill contained three seedlings. For dwarf varieties (ADT-43, ADT-37, and Co-47), the distance between hills was 5 cm and between rows 7 cm. For the tall variety (Ponni), the distance between hills was 7 cm and between rows, 12 cm. Potassium and phosphorus were applied at the rate of 50 and 100 kg ha−1 for all treatments and no N fertilization was applied. For estimation of yield components, five subplots per treatments were randomly selected, and five hills per subplot were sampled. Each sample contained plants and the adhering clod. All samples were kept in plastic bags and processed in the laboratory to determine the plant height (from soil surface to the tip of the upper leaf), root weight, number of fertile tillers, and the dry weight of grains.
Statistical Analysis
The data for each treatment were subjected to a variance analysis using the Statgraphics software (Release 5.0, Uniware STSC, Inc.). When analysis of variance showed significant treatment effects, the least significant difference (LSD, p < 0.05) test was applied to make comparisons between treatments.
Results
Isolation and Characterization of Nitrogen-Fixing Bacteria from Rice
The inoculation of N-free semisolid LGIM medium with samples from surface sterilized roots, stems, and leaves of rice, followed by subsequent streaking on the PCAT media, allowed the recovery of 13 N2-fixing isolates. Colonies on BMGM agar plates were large yellow with round entire margins. Colonies on nutrient agar and King’s B medium were white, smooth without pigment. After transfer to PCAT agar plates, all colonies were small, white, round with entire margins. The most active strain was MGK3, isolated from surface sterilized stems of tall Ponni variety at Ulundurpet, Tamilnadu.
Identification and Growth Promoting Properties
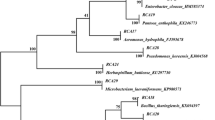
The presence of 16:0 3-OH in fatty acids suggested that isolate MGK3 belonged to the Burkholderia genus. The 16S rDNA gene sequence of isolate MGK3 gave a 100% match with B. vietnamiensis LMG10929 (Fig. 1). PCR amplification of nif-H showed that isolate MGK3 produced the expected 390-bp amplification products (data not shown). Isolate MGK3 produced more indole acetic acid in the presence of tryptophan (100 mg/l) than in its absence. With tryptophan, isolate MGK3 produced the highest amount (16.4 μg/ml); whereas B. vietnamiensis LMG10929 produced 10.5 μg ml−1.
Phylogenetic position of strain MGK3 within the genus Burkholderia on the basis of 16S rDNA gene sequences. The phylogenetic tree was constructed by the neighbor-joining method [50], and the 16S rDNA gene sequence of Pelistega europaea LMG10982 was used as the out group. The numbers at nodes indicate the levels of the bootstrap support based on a neighbor-joining analysis of 1,000 resampled data sets.
Physiological and Biochemical Characterization
Isolate MGK3 was Gram-negative, rod-shaped, aerobic, and motile. Isolate MGK3 showed oxidase and catalase positivity. All grew well at pH 4.0 to pH 7.0 at room temperature (30°C). Isolate MGK3 grew in different carbon sources such as glucose, sorbitol, meso-inositol, mannose, glycerol, fructose, arabinose, trehalose, raffinose, galactose, mannitol, cellobiose, xylose, and sucrose. But isolate MGK3 did not grow on l-rhamnose, meso-erythritol, starch, sodium acetate, and maltose. Isolate MGK3 grew well in the following organic acids: azelaic, succinic, valeric, fumaric, hippuric, malic, tartaric, α-ketoglutaric and citric acids, whereas isolate could grow on the oxalic, malonic, and adipic acids. Isolate MGK3 and type strains grew well in the presence of the following amino acids: l-cysteine, l-threonine, l-glutamic acid, l-proline, l-tryptophane, l-leucine, l-lysine, l-histidine, l-tyrosine, and l-valine with sorbitol as a carbon source.
Acetylene Reduction Activity
Isolate MGK3 was capable of N2 fixation with different carbon sources viz., fructose, mannitol, malate, azelaic, sucrose, glucose, succinic, and glycerol as single carbon sources. However, B. vietnamiensis LMG10929 showed negative AR activity with glucose as single carbon source (Table 1). AR activity also was determined at the rice panicle initiation and grain filling stages. The AR activity was approximately three times higher at the grain-filling stage as compared with the panicle-initiation stage. Roots of cultivars ADT-43 and Ponni produced more ethylene compared to other cultivars of both the panicle initiation and grain filling stages (Table 2). The bacterial populations in roots, as estimated by ARA-based MPN counts, also were higher at the grain-filling stage than at the panicle-initiation stage.
GUS Staining and Plant Colonization
Blue colonies were observed in the transconjugants grown on LGIM agar containing spectinomycin (100 μg ml−1) and X-Gluc (40 μg ml−1) but not in the wild strain because of the absence of gusA gene. Furthermore, specific PCR analysis of the gusA gene revealed a fragment of 1,200 bp in the transconjugant, but no amplification was observed in wild-type isolate MGK3 (data not shown). Inoculated plants roots after 24 h incubation showed that the blue color cells were distributed mainly on the root surface. After 5 days, more intensive colonization was observed in the lateral roots emergence (Fig. 2A). The Ponni rhizosphere populations of MGK3 increased up to 15 days after inoculation with the maximum being after 10 days (Table 3). Although bacteria could be isolated from surface of the roots at the first day, they could be re-isolated from roots as endophytes only after 5 days (2.5 × 107 cells g−1 fresh weight). The same trend was observed in ADT-43 with rhizosphere populations increasing up to 15 days (Table 3), while in the endophytic populations from the roots, re-isolation was possible after 5 days (1.5 × 106 cells g−1 fresh weight) with the maximum at 15 days (1.5 × 108). In Co-47 and ADT-47, the rhizosphere populations were increased for 15 days (Table 3). However, they could be re-isolated from roots as endophytes only after 10 days (1.5 × 107 cells g−1 fresh weight). The occurrence of a blue color at the root tips after MGK3 gusA inoculation also supports the possibility of entry via root tips. Transverse sections of roots 15 days after inoculation revealed GUS activity within many of the cortical intercellular spaces next to the stele and within the aerenchyma.
15N Dinitrogen Incorporation Assay
The acetylene reduction assay is an indirect method to verify nitrogen fixation in planta of endophytes, and the possibility of ethylene emission from plants could not be completely excluded. Therefore, we designed a 15N dinitrogen incorporation experiment to follow 15N2 gas into rice plants through endophytic B. vietnamiensis MGK3. Quantification of nitrogen fixation by 15N isotopic dilution method showed that the effect of B. vietnamiensis MGK3 inoculation was more prominent in cultivar Ponni as compared to ADT-43 in field and in pot trial plants (Table 2). In this rice cultivar, field and pot trial plants showed maximum fixation with 42 and 40% Nfda recorded, respectively. In cultivar ADT-43, field and pot trial plants showed maximum fixation with 39 and 36% Nfda recorded, respectively. Type strain B. vietnamiensis LMG10929 also showed comparable fixation in both cultivars (Table 4).
Rice Inoculation Experiments
In both pot trials, the uninoculated control rice hosted a mixture of diazotrophs in its roots, but in low numbers: 21.2 and 40.9 × 103 CFU/g fresh weight at days 50 and 110, respectively. In both pot experiments, inoculation raised the diazotroph population level up to 1.15 × 105 CFU/g fresh weight at 60 days. It stayed at this level onto 110 days, except for G. diazotrophicus LMG7603, whose level decreased down to 73.5 × 103 CFU/g fresh weight. In the fields, uninoculated control plant roots also contained a mixture of diazotrophs but at a low level, similar to what was observed in the controls of pot experiments (data not shown). Differences between inoculated treatments and the controls were visible in all experiments. In all assays, the mixed inoculum performed best. For all measured parameters, it produced a significant increase over the uninoculated control. Increases of shoot biomass over control was 63.8, 29.0, 60.3, 32.7, 12.2, and 32.3% (mean 38.4%) at 60 days after inoculation followed by 46.8, 21.6, 39.9, 90.4, 19.2, and 20.4% (mean 39.71%) at 110 days after inoculation, and root biomass over control was 130.0, 28.6, 88.2, 34.8, 50.0, and 166.7% (mean 83.05%) at 60 days after inoculation followed by 90.9, 98.6, 69.2, 95.2, 51.0, and 110.5% (mean 85.9%) at 110 days after inoculation. Number of tiller increases over control was 49.7, 51.0, 45.6, 29.6, 47.2, and 93.1 (mean 52.7%) at 60 days after inoculation followed by 39.1, 65.4, 57.9, 35.6, 53.4, and 94.6 (mean 57.7%) at 110 days after inoculation. Yield increase over control was 10.1, 13.2, 9.5, 23.6, 14.4, and 15.80% (mean 14.4%) in the two pot and four field experiments respectively (Tables 5, 6, 7, 8, 9, 10).
Inoculation of B. vietnamiensis MGK3 alone produced a significant increase over the control for all measured parameters. Increases of shoot biomass over control was 39.6, 14.3, 32.4, 25.0, 13.3, and 22.6% (mean 24.53%) at 60 days after inoculation followed by 23.4, 8.4, 21.0, 8.9, 14.5, and 11.8 (mean 14.66%) at 110 days after inoculation, and root biomass over control was 72.8, 12.7, 8.8, 25.8, 36.6, and 133.3% (mean 48.3%) at 60 days after inoculation followed by 36.4, 20.0, 30.7, 9.5, 36.7, and 57.9 (mean 31.86%) at 110 days after inoculation. Tiller increase over control was 36.5, 25.9, 36.5, 19.0, 25.5, and 27.3 (mean 28.5%) at 60 days after inoculation followed by 25.8, 43.8, 44.0, 35.9, 46.0, and 33.8 (mean 38.2%) at 110 days after inoculation. Yield increase over control was 8.2, 12.8, 5.6, 10.7, 6.7, and 12.16% (mean 9.36%) in the two pot and four field experiments, respectively (Tables 5, 6, 7, 8, 9, 10). B. vietnamiensis LMG10929 gave a significant effect on growth parameters as compared to the uninoculated control except in the field trial with cv. Ponni at 150 days (Table 10). It had little or no effect on N leaf content. Yield increase over control was 3.7, 2.8, 3.9, 8.9, 4.2, and 8.9% (mean 5.4%) in the two pot and four field experiments, respectively (Tables 5, 6, 7, 8, 9, 10).
A. lipoferum LMG4348 gave a significant effect on growth parameters as compared to the uninoculated control except for tiller numbers and root weight in the first two field experiments (Tables 7 and 8). It had little effect on N leaf content. Yield increase over control was 3.7, 7.5, 4.4, 11.7, 3.7, and 10.05% (mean 6.8%) in the two pot and four field experiments, respectively (Tables 5, 6, 7, 8, 9, 10). Its overall effect was higher than B. vietnamiensis LMG10929. H. seropedicae LMG6513 sometimes gave a significant effect on growth parameters as compared to the uninoculated control in three field experiments (Tables 8, 9, 10). Nevertheless, yield increase over control was very weak: 3.1, 0.8, 2.2, 0.8, 2.1, and 6.8% (mean 2.6%) in the two pot and four field, experiments respectively. G. diazotrophicus LMG7603 gave a significant effect on growth parameters as compared to the uninoculated control for all vegetative parameters. The effect on N leaf content was inconsistent. Yield increase over control was 6.3, 10.2, 3.3, 7.0, 2.7, and 10.05% (mean 6.6%) in the two pot and four field experiments, respectively (Tables 5, 6, 7, 8, 9, 10). Shoot is to root ratios were significantly varied among the cultivars used in pot and field experiments. Cultivar ADT-43 produced shoot is to root ratio almost 10:1 (Tables 5 and 7), and cultivar Ponni ratio was 3:1 (Tables 6 and 10) in both pot and field experiments. Field experiments with cultivar Co-47 produced shoot is to root ratio nearly 7:1 (Table 8) and cultivar ADT-37 shoot is to root ratio was 3:1 (Table 9).
The weight of 1,000 grains is a late yield component. It is increased by all inoculants (Table 11). The combined inoculum ranks first, followed by B. vietnamiensis MGK3; the controls consistently ranked last, just after H. seropedicae LMG6513, A. lipoferum 4B LMG4348, G. diazotrophicus LMG7603, and B. vietnamiensis LMG10929 rank between MGK3 and H. seropedicae LMG6513. In terms of overall effect on yield, treatments can be ranked using the mean effect on yield in the six experiments: the combined inoculum ranks first (14.4% increase over control), followed by B. vietnamiensis MGK3 (9.36%), A. lipoferum 4B LMG4348 (6.8%), G. diazotrophicus LMG7603 (6.6%), B. vietnamiensis LMG10929 (5.4%), and H. seropedicae LMG6513 (2.6%).
Discussion
The present study was undertaken to isolate and enumerate new nitrogen-fixing Burkholderia endophytes from different rice cultivars of Tamilnadu State, India. This concept of Biological Nitrogen Fixation by endophytes has been introduced by Döbereiner [19] and mostly tested with graminaceous plants. Boddey and Döbereiner [10] suggested that endophytic bacteria better express their nitrogen fixation potential inside plant tissues due to the lower competition for nutrients and protection against high levels of O2 present on the root surface. This habitat has already been identified as an important source of endophytic PGPR that do not induce obvious symptoms of disease. Examples include Azospirillum strains isolated from “inside” host roots (after surface sterilization), which increase yield when inoculated on that homologous host [9], the diazotrophic endophytes of Azoarcus inside Kallar grass [7, 33], and Acetobacter diazotrophicus inside sugar cane [18]. Nature appears to select endophytes that are competitively fit to occupy compatible niches within this nutritionally enriched and protected habitat of the root interior without causing pathological stress on the host plant. Furthermore, Yanni et al [68] reported that field inoculation with the rhizobial endo-colonizer did not qualitatively alter rice grain protein composition and that nutritionally important proteins are all present in treated and control samples in similar ratios.
Identification and Growth-Promoting Properties
From surface-sterilized plant tissues, we obtained 13 isolates able to grow on PCAT. In planta densities were diverse; the most active strain was MGK3. Isolate MGK3 grew well on different carbon sources. In particular, it grew on sorbitol, mannose, and mannitol which differentiate Burkholderia spp. from the genus Ralstonia [26]. Their 16S rDNA sequences and their fatty acid profiles placed them in the species B. vietnamiensis. Isolate MGK3 showed positive for acetylene reduction, and PCR amplification of nif-H confirmed the presence of this structural nitrogenase gene in isolate MGK3.
We have found large differences in nitrogenase activity when isolate grown in different carbon sources (Table 1) and also rice growth stages (Table 2). Similar variation in AR activity with the plant growth stage has been reported by Watanabe et al. [65] in two rice varieties, IR36 and IR26, where maximum AR activity was detected at the grain-filling stage. Higher AR activity at a particular growth stage may be due to a reduction in inhibitory nitrogen concentrations in the soil or overproduction of root exudates that are conducive to diazotroph growth and activity [35].
The root surface of rice was intensively colonized at the end of 24 h of inoculation (Fig. 2A) and colonized many of the cortical intercellular spaces after 15 days of inoculation (Fig. 2B). Primer specific PCR analysis of the gusA gene revealed a fragment of 1,200 bp in the transconjugant [39]. The xylem could be a suitable non-nodular niche for N2 fixation because it could provide the low pO2 required for the expression and function of nitrogenase and also allow the exchange of fixed N2 [37]. Nitrogen fixation by 15N isotopic dilution method showed that the effect of B. vietnamiensis MGK3 inoculation was higher in cultivar Ponni than in ADT-43, where nearly 42% (field) and 40% (pot) of the nitrogen was derived from atmosphere (% Ndfa), respectively. In both cultivars, native isolate B. vietnamiensis MGK3 showed increased fixation than type strain B. vietnamiensis LMG10929 (Table 4).
Rice Inoculation Experiments
Two pot experiments followed by four field experiments were performed at four different places using four commercially important rice cultivars (ADT-43, Co-47, ADT-37, and Ponni) to evaluate and compare isolate MGK3 to: (1) the type strain of B. vietnamiensis LMG10929, (2) other rice diazotrophs (H. seropedicae LMG6513, A. lipoferum LMG4348), (3) a sugarcane diazotroph (G. diazotrophicus LMG7603), and (4) a mixture of all these strains. In terms of overall effect on yield, the ranking is (1) combined inoculum (2) B. vietnamiensis MGK3, (3) A. lipoferum 4B LMG4348, (4) G. diazotrophicus LMG7603, (5) B. vietnamiensis LMG10929, and (6) H. seropedicae LMG6513. Significant increases of shoot and root biomass due to combined inoculation reached 38.4 and 83.0% (mean of two pot and four field experiments), respectively, at 60 days after inoculation. This is indicative of a very early effect of bacteria on their host plant. Jacoud et al. [34] reported that a very short and early contact of maize with PGPR bacteria was enough to ensure a significant effect on subsequent growth. Tillering is the next important step in rice development, and the tiller number per plant was significantly increased (+28%, mean of two pot and four field experiments) 60 days after combined inoculation. Tran Van et al. [62] noticed a similar effect when B. vietnamiensis was inoculated with rice. Root biomass was drastically low compared to shoot biomass among different pot and field experiments; this variation may be difficult to extract all roots in pot and field experiments. The effect on the weight of 1,000 grains somewhat parallels the effect on final yield, suggesting that the effect of inoculated bacteria persists throughout the plant growth cycle.
Some of these strains had already been assayed in the field. B. vietnamiensis LMG10929, for instance, has been inoculated to rice in Vietnam [62] in three pot and four field experiments in three different locations. Yield increases caused by inoculation ranged from 13 to 22% over control, larger than what was observed in Tamilnadu. In our experiment, B. vietnamiensis LMG10929 gave a significant effect on growth parameters as compared to the uninoculated control except in the field trial with cv. Ponni at the 150th day (Table 9). Yield increase over control was the mean of 5.4% in the two pot and four field experiments. This strain is not an Indian but a Vietnamese isolate [59] obtained from an acid sulfate soil, very different from soils used in the present paper. The ability of B. vietnamiensis LMG4348 to internally colonize rice tissues is unknown; however, this species has been recovered from internal tissues of maize [22].
G. diazotrophicus LMG7603 gave a significant effect on growth parameters as compared to the uninoculated control for all vegetative parameters. Yield increase over control was mean of 6.6% in the two pot and four field experiments. G. diazotrophicus LMG7603 has proved beneficial, where the rice seedlings inoculated with the G. diazotrophicus grew to be significantly taller 30 days after inoculation than plants inoculated with the nif- mutant or uninoculated plants under N-deficient conditions [56]. Inoculation of G. diazotrophicus LMG7603 to sugarcane has been proven beneficial, where the plant height [55] and yield [46] of the inoculated plants were higher than the control. Field trials conducted in the sugarcane system revealed the usefulness of G. diazotrophicus LMG7603 with other diazotrophs, which have contributed to the yield equal to that of the control (280 kg N ha−1). Mixed inoculation of vesicular–arbuscular mycorrhizal (VAM) spores and G. diazotrophicus LMG7603 also proved beneficial in improving the yield of different sugarcane varieties. The yield was also not reduced even under 50–100% reduction from the recommended dose of chemical N compared to the control, attributing the role of inoculated G. diazotrophicus LMG7603 in N contribution [46]. It has been reported that inoculation of micropropagated sugarcane seedlings would make the plants not only grow faster, but also ensure efficient N-fixing plants in fields.
A. lipoferum LMG4348 gave a significant effect on growth parameters as compared to the uninoculated control except for tiller numbers and root weight in the first two field experiments (Tables 7 and 8). Yield increase over control was the mean of 6.8% in the two pot and four field experiments. Strain 4B of A. lipoferum LMG4348 has been isolated from a French rice field [59], proved to be a very efficient diazotroph under gnotobiotic conditions [30], and for that reason, it was used for three field inoculation experiments [14]. Yield increases following inoculation were 20.7, 21.3, and 15.9% over uninoculated controls. Its rather poor performance in the Indian context is surprising. H. seropedicae LMG6513 sometimes gave a significant effect on growth parameters as compared to the uninoculated control especially in three experiments (Tables 8, 9, 10). Nevertheless, yield increase over control was the mean of 2.6% in the two pot and four field experiments. The less performance of H. seropedicae LMG6513 is also surprising because this particular strain was isolated from rice and aggressively colonizes rice tissues [35] and stimulates rice growth in gnotobiotic conditions [5]. This bacterial species is often found in rice and its wild counterparts [20]. There is no explanation for its poor efficiency under Indian conditions. Baldani et al. [4] reported inoculation of rice under gnotobiotic conditions with Herbaspirillum seropedicae LMG6513, B. “brasilensis” and B. vietnamiensis LMG10929. Total plant dry weight increases were observed after the inoculation viz: 42% for B. vietnamiensis LMG10929, 64% for B. “brasilensis” and 71.5% for H. seropedicae LMG6513 (59% for strain LMG6513). In pots on 4-month-old plants, these figures were 48 and 27% for H. seropedicae LMG6513 and B. “brasilensis”, respectively, confirming the former as the best inoculum. This difficulty in comparing inoculation experiments is further increased by the use of different cultivars in different parts of the world. The use of endophytic diazotrophs as a substitute for nitrogen fertilizers [1] is still a debatable matter. The beneficial results reported in the present article are only one more argument in favor of this new practice. Other arguments are: low cost, absence of pollution, similarity to the natural process, and foreseeable sustainability.
The mixed inoculant, in the present work, displays an important potential in spite of the poor performance of some of its components. Muthukumarasamy et al. [46] inoculated a mixture of diazotrophs and mycorrhizal fungi to micropropagated sugarcane and obtained an effect equivalent to half the recommended rate of nitrogen fertilizers under pot conditions. Oliveira et al. [47] also reported a pot experiment in which the combined inoculation of five strains (G. diazotrophicus LMG7603, H. seropedicae LMG6513, H. rubrisubalbicans, A. amazonense, and Burkholderia sp.) gave higher contribution to plant growth, followed by the treatment with a mixture of Herbaspirillum spp. However, the contribution was much lower when the plants were inoculated with a mixture of G. diazotrophicus LMG7603 with A. amazonense and Burkholderia sp. Govindarajan et al. [28] reported that individual inoculation B. vietnamiensis MG43 and G. diazotrophicus LMG7603 gave a higher sugarcane yield as compared to combined inoculation. One can speculate that the different bacteria occupy different niches in the plant, replacing expected competition by a cooperative effect, as proposed by Ueda et al. [63]. Next to the mixed inoculum, strain MGK3 ranks first among single inoculum assays and is a good candidate for further inoculation trials on a larger scale.
Nevertheless, due to its relatedness to Burkholderia species (B. cenocepacia, B. multivorans, and B. stabilis) causing severe pneumonia in cystic fibrosis-affected people, a prerequisite [48] will be to demonstrate that MGK3 is devoid of any pathogenesis genes and is unable to acquire them through horizontal transfer. Several human pathogenic Burkholderia species (including B. cenocepacia, B. pseudomallei, and B. mallei) have been completely sequenced and other environmental species sequencing projects are in progress (B. vietnamiensis G4, B. phytofirmans PsJN, and B. phymatum STM815). It can be expected that knowledge derived from these genome sequencing projects will allow us to gain further insights into functional diversity, evolution, and pathogenicity mechanism. This may provide the scientific basis for important decisions regarding the biotechnological use of Burkholderia species.
References
Andrews, MA, James, EKB, Cummings, SPA, Zavalin, AAC, Vinogradova, LVC, McKenzie, BAD (2003) Use of nitrogen fixing bacteria inoculants as a substitute for nitrogen fertiliser for dry land graminaceous crops: progress made, mechanisms of action and future potential. Symbiosis 35: 209–229
Balandreau, J, Viallard, V, Cournoyer, B, Coenye, T, Laevens, S, Vandamme, P (2001) Burkholderia cepacia genomovar III is a common plant-associated bacterium. Appl Environ Microbiol 67: 982–985
Baldani, JI, Baldani, VLD, Seldin, L, Dobereiner, J (1986) Characterisation of Herbaspirillum seropedicae gen. nov. sp. nov., a root associated nitrogen fixing bacterium. Int J Syst Bacteriol 36: 86–93
Baldani, VLD, Baldani, JI, Dobereiner, J (2000) Inoculation of rice plants with the endophytic diazotrophs Herbaspirillum seropedicae and Burkholderia spp. Biol Fertil Soils 30: 485–491
Baldani, JI, Pot, B, Kirchhof, G, Falsen, E, Baldani, VLD, Olivares, FL, Hoste, B, Kersters, K, Hartmann, A, Gillis, M, Döbereiner, J (1996) Emended description of Herbaspirillum; inclusion of [Pseudomonas] rubrisubalbicans, a mild plant pathogen, as Herbaspirillum rubrisubalbicans comb. nov.; and classification of a group of clinical isolates (EF group 1) as Herbaspirillum species 3. Int J Syst Bacteriol 46: 802–810
Bally, I, Thomas-Bauzon, D, Heulin, T, Balandreau, J, Richard, C, De Ley, J (1983) Determination of the most frequent N2-fixing bacteria in a rice rhizosphere. Can J Microbiol 29: 881–887
Bilal, R, Malik, KA (1987) Isolation and identification of an N2-fixing zoogloea-forming bacterium from kallar grass histoplane. J Appl Bacteriol 62: 289–294
Bilal, R, Rasul, G, Qureshi, JA, Malik, KA (1990) Characterisation of Azospirillum and related diazotrophs associated with roots of plants growing in saline soils. World J Microbiol Biotechnol 6: 46–52
Boddey, R, Dobereiner, J (1988) Nitrogen fixation associated with grasses and cereals: recent results and perspectives for future research. Plant Soil 108: 53–65
Boddey, RM, Dobereiner, J (1995) Nitrogen fixation associated with grasses and cereals; recent progress and perspectives for the future. Fertil Res 42: 241–250
Boddey, RM, Polidoro, JC, Resende, AS, Alves, BJR, Urquiaga, S (2001) Use of the 15N natural abundance technique for the quantification of the contribution of N2 fixation to sugarcane and other grasses. Aust J Plant Physiol 28: 889–895
Burbage, DA, Sasser, M (1982) A medium selective for Pseudomonas cepacia. Phytopathol Abstracts 72: 706
Caballero-Mellado, J, Martínez-Aguilar, L, Paredes-Valdez, G, Estrada-de los Santos, P (2004) Burkholderia unamae sp. nov., an N2-fixing rhizospheric and endophytic species. Int J Syst Bacteriol 54: 1165–1172
Charyulu, PBBN, Fourcassie, F, Barbouche, AK, Rondro, Harisoa, L, Omar AMN, Weinhard, P, Marie, R, Balandreau, J (1985) Field inoculation of rice using in vitro selected bacterial and plant genotypes. In: Klingmuller W (Eds.) Azospirillum III. Genetics Physiology Ecology, Springer-Verlag Publications, pp 163–179
Chen, WM, de Fario, SM, Straaliotto, R, Pitard, RM, Simoes-Araujo, JL, Chou, JH, Chou, YJ, Barrios, E, Prescott, AR, Elliott, GN, Sprent, JI, Young, JPW, James, EK (2005a) Proof that Burkholderia forms effective symbioses with legumes: a study of novel mimosa-nodulating strains from South America. Appl Environ Microbiol 71: 7461–7471
Chen, WM, James, EK, Chou, JH, Sheu, SY, Yang, SZ, Sprent, JI (2005b) Beta-rhizobia from Mimosa pigra, a newly discovered invasive plant in Taiwan. New Phytol 168: 661–675
Christiansen-Weniger, C (1997) Ammonium-excreting Azospirillum brasilense C3: gusA inhabiting induced tumors along stem and roots of rice. Soil Biol Biochem 29: 943–950
Di cello, F, Bevivino, A, Chiarini, L, Fani, R, Paffetti, D, Tabachioni, S, Dalmastri, C (1997) Biodiversity of a Burkholderia cepacia population isolated from the maize rhizosphere at different growth stages. Appl Environ Microbiol 63: 4485–4493
Döbereiner, J, Day, JM (1976) Associative symbiosis in tropical grasses. Characterization of microorganisms and dinitrogen fixing sites. In: Newton, WE, Nyman, CJ (Eds.) Proceedings of the first international symposium on nitrogen fixation, Washington State University Press, Pullman, 2, pp 518–538
Dobereiner, J, Reis, V, Paula, M, Olivares, F (1993) Endophytic diazotrophs in sugar cane, cereals, and tuber plants. In: Palacios, R, Mora, J, Newton, WE (Eds.) ‘New Horizons in Nitrogen Fixation’, Kluwer Academic Publishers, Dordrecht, pp 671–679
Elbeltagy, A, Nishioka, K, Sato, T, Suzuki, H, Ye, B, Hamada, T, Isawa, T, Mitsui, H, Minamisawa, K (2001) Endophytic colonization and in planta nitrogen fixation by a Herbaspirillum sp. isolated from wild rice species. Appl Environ Microbiol 67: 5285–5293
Estrada-de-los-Santos, P, Bustillo-Cristalles, R, Caballero-Mellado, J (2001) Burkholderia, a genus rich in plant-associated nitrogen fixers with wide environmental and geographical distribution. Appl Environ Microbiol 67: 2790–2798
Estrada, P, Mavingui, P, Cournoyer, B, Fontaine, F, Balndreau, J, Cabellero-Mellado, J (2002) A N2-fixing endophytic Burkholderia sp. associated with maize plants cultivated in Mexico. Can J Microbiol 48: 285–294
Fried, M, Middleboe, V (1977) Measurement of amount of nitrogen fixed by a legume crop. Plant Soil 40: 713–715
Gillis, M, Kersters, K, Hoste, B, Janssens, D, Kropenstedt, RM, Stephen, MP, Teixeira, KRS, Dobereiner, J, De Ley, J (1989) Acetobacter diazotrophicus sp. nov., a nitrogen-fixing acetic acid bacterium associated with sugarcane. Int J Syst Bacteriol 39: 361–364
Gillis, M, Tran Van, V, Bardin, R, Goor, M, Hebar, P, Willems, A, Segers, P, Kersters, K, Heulin, T, Fernandez, MP (1995) Polyphasic taxonomy in the genus Burkholderia leading to an emended description of the genus Burkholderia and transposition of Burkholderia vietnamiensis sp. nov. for N2-fixing isolates from rice in Vietnam. Int J Syst Bacteriol 45: 274–289
Goris, J, De Vos, P, Caballero-Mellado, J, Park, JH, Falsen, E, James M, Tiedje, Vandamme, P (2004) Classification of the PCB- and biphenyl-degrading strain LB400 and relatives as Burkholderia xenovorans sp. nov. Int J Syst Evol Microbiol 54: 1677–1681
Govindarajan, M, Balandreau, J, Muthukumarasamy, R, Revathi, G, Lakshminarasimhan, C (2006) Improved yield of micropropagated sugarcane following inoculation by endophytic Burkholderia vietnamiensis. Plant Soil 280: 239–252
Yang, H-C, Im, W-T, Kim, KK, An, D-S, Lee, S-T (2006) Burkholderia terrae sp. nov., isolated from a forest soil. Int J Syst Evol Microbiol 56: 453–457
Heulin, T, Rahman, M, Omar, AMN, Rafidison, Z, Pierrat, JC, Balandreau, J (1989) Experimental and mathematical procedures for comparing efficiencies of rhizosphere N2 fixing bacteria. J Microbiol Methods 9: 163–173
Hiraishi, A (1992) Direct automated sequencing of 16S rDNA amplified by polymerase chain reaction from bacterial cultures without DNA purification. Lett Appl Microbiol 15: 210–213
Humphries, EC (1956) Mineral components and ash analysis. In: Peach, K, Tracged, MV (Eds.) Modern Methods of Plant Analysis, Springer-Verlag, Berlin. pp 468–502
Hurek, T, Reinhold-Hurek, B, van Montague, M, Kellenberger, E (1994) Root colonization and systemic spreading of Azoarcus sp. strain BH72 in grasses. J Bacteriol 176: 1913–1923
Jacoud, C, Job, D, Wadoux, P, Bally, R (1999) Initiation of root growth stimulation by Azospirillum lipoferum CRT1 during maize seed germination. Can J Microbiol 45: 339–342
Jagnow, GC (1983) Nitrogenase (C2 H2) activity in non-cultivated and cereal plants; influence of nitrogen fertilizer on population and activity of nitrogen fixing bacteria. Z Pflanzenernaehr Bodenkd 146: 217–227
James, EK, Olivares, FL (1998) Infection and colonization of sugar cane and other graminaceous plants by endophytic diazotrophs. Crit Rev Plant Sci 17: 77–119
James, EK, Gyaneshwar, P, Mathan, N, Barraquio, WL, Reddy, PM, Iannetta, PP, Olivares, FL, Ladha, JK (2002) Infection and colonization of rice seedlings by the plant growth-promoting bacterium Herbaspirillum seropedicae Z67. Mol Plant Microb Interact 15: 894–906
Jukes, TH, Cantor, CR (1969) Evolution of protein molecules. In: Munro, HN (Eds.) Mammalian Protein Metabolism, Academic Press, New York, III, pp 21–132
Kennedy, IR, Tchan, YT (1992) Biological nitrogen fixation in non-leguminous field crops: recent advances. Plant Soil 141: 93–118
Kirchhof, G, Reis, VM, Baldani, JI, Eckert, B, Dobereiner, J, Hartmann, A (1997) Occurence, physiological and molecular analysis of endophytic diazotrophic bacteria in gramineous energy plants. Plant Soil 194: 45–55
Kirchhof, G, Baldani, JI, Reis, VM, Hartmann, A (1998) Molecular assay to identify Acetobacter diazotrophicus and detect its occurrence in plant tissues. Can J Microbiol 44: 12–19
Kumar, S, Tamura, K, Jakobsen, IB, Nei, M (2001) MEGA2: Molecular Evolutionary Genetics Analysis Software. Bioinformatics 17: 1244–1245
Kwon, SW, Kim JS, Park, IC, Yoon, SH, Park, DH, Lim, CK, Go, SJ (2003) Pseudomonas koreensis sp. nov., Pseudomonas umsongensis sp. nov. and Pseudomonas jinjuensis sp. nov., novel species from farm soils in Korea. Int J Syst Evol Microbiol 53: 21–27
Ladha, JK, Triol, AC, Daroy, LG, Caldo, G, Ventura, W, Watanabe, I (1986) Plant associated N2 fixation (C2H2) reduction by five rice varieties, and relationship with plant growth characters as affected by straw incorporation. Soil Sci Plant Nutr 32: 91–106
Mascarua-Esparza, MA, Villa-Gonzallez, R, Caballero-Mellado, J (1988) Acetylene reduction and indolacetic acid production by Azospirillum isolates from Cactaceous plants. Plant Soil 106: 91–95
Muthukumarasamy, R, Revathi, G, Lakshminarasimhan, C (1999) Diazotrophic associations in sugarcane cultivation in South India. Trop Agric (Trinidad) 76: 171–178
Oliveira, ALM, Urquiaga, S, Döbereiner, J, Baldani, JI (2002) The effect of inoculating endophytic N2-fixing bacteria on micropropagated sugarcane plants. Plant Soil 242: 205–215
Parke, JL, Gurian-Sherman, D (2001) Diversity of the Burkholderia cepacia complex and implications for risk assessment of biological control strains. Annu Rev Phytopathol 39: 225–258
Rasul, G, Mirza, MS, Latif, F, Malik, KA (1998) Identification of plant growth hormones produced by bacterial isolates from rice, wheat and kallar grass. In: Malik, KA, Mirza, MS, Ladha, JK (Eds.) Nitrogen Fixation with Non-Legumes, Kluwer Academic Publishers, Netherlands
Reinhold-Hurek, BR, Hurek, T, Gillis, M, Hoste, B, Vancanneyt, M, Kersters, K, De Ley, J (1993) Azoarcus gen. nov. Nitrogen-fixing proteobacteria associated with roots of kallar grass (Leptochoa fusca (L.) Kunth) and description of two species, Azoarcus indigens sp. nov. and Azoarcus communis sp. nov. Int J Syst Bacteriol 43: 574–584
Reis, VM, Olivares, FI, Dobereiner, J (1994) Improved methodology for isolation of Acetobacter diazotrophicus and confirmation of its endophytic habitat. World J Microbial Biotechnol 10: 101–104
Reis, VM, Estrada-de los Santos, P, Tenorio-Salgado, S, Vogel, J, Stoffels M, Guyon, S, Mavingui, P, Baldani, VLD, Schmid, M, Baldani, JI, Balandreau, J, Hartmann, A, Caballero-Mellado, J (2004) Burkholderia tropica sp. nov., a novel nitrogen-fixing, plant-associated bacterium. Int J Syst Evol Microbiol 54: 2155–2162
Saitou, N, Nei, M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425
Sessitsch, Coenye, T, Sturz, AV, Vandamme, P, Ait Barka, E, Salles, JF, Van Elsas, JD, Faure, D, Reiter, B, Glick, BR, Wang-Pruski, G, Nowak, J (2005) Burkholderia phytofirmans sp. nov., a novel plant-associated bacterium with plant-beneficial properties. Int J Syst Evol Microbiol 55: 1187–1192
Sevilla, M, de Olivares, A, Baldani, JI, Kennedy, C (1998) Contribution of the bacterial endophyte Acetobacter diazotrophicus to sugarcane nutrition. Symbiosis 25: 181–191
Sevilla, M, Kennedy, C (2000) Colonisation of rice and other cereals by Acetobacter diazotrophicus, an endophyte of sugarcane. In: Ladha, JK, Reddy, PM (Eds.) The Quest for Nitrogen Fixation, IRRI Press, Manila, pp 151–165
Simon, R, Priefer, U, Puhler, A (1983) A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis of Gram negative bacteria. Bio/Technology 1: 784–791
Somasekaran, P, Huben, HJ (1994) Handbook for Rhizobia. Springer-Verlag, pp 340
Thomas-Bauzon, D, Weinhard, P, Villecourt, P, Balandreau, J (1982) The spermosphere model. Its use in growing counting and isolating N2-fixing bacteria from the rhizosphere of rice. Can J Microbiol 28: 922–928
Tien, TM, Gaskins, MH, Hubbell, DH (1979) Plant growth substances produced by Azospirillum brasiliense and their effect on growth of Pearl millet (Pennisetum americanum L.). Appl Environ Microbiol 37: 1016–1024
Tran Van, V, Berge, O, Balandreau, J, Ngo Kê, S, Heulin, T (1996) Isolement et activité nitrogénasique de Burkholderia vietnamiensis, bactérie fixatrice d’azote associée au riz (Oryza sativa L) cultivé sur un sol sulfaté acide du Viêt-nam. Agronomie 16: 479–491
Tran Van, V, Berge, O, Ngo Ke, S, Balandreau, J, Heulin, T (2000) Repeated beneficial effects of rice inoculation with a strain of Burkholderia vietnamiensis on early and late yield components in low fertility sulphate acid soils of Vietnam. Plant Soil 218: 273–284
Ueda, T, Suga, Y, Yahiro, N, Matsuguchi, T (1995) Remarkable N2-fixing bacterial diversity detected in rice roots by molecular evolutionary analysis of nif-H gene sequence. J Bacteriol 177: 1414–1417
Vandamme, P, Goris, J, Chen, WM, de Vos P, Willems, A (2002) Burkholderia tuberum sp. nov. and Burkholderia phymatum sp. nov., nodulate the roots of tropical legumes. Syst Appl Microbiol 25: 507–512
Watanabe, I, Barraquio, WI, de Guzman, MR, Cabera, DA (1979) Nitrogen fixing (C2 H2 reduction) activity and population of aerobic heterotrophic nitrogen fixing bacteria associated with wetland rice. Appl Environ Microbiol 37: 813–819
Wilson, KJ, Sessittsch, A, Corbo, J, Giller, KEA, Akkermans, DL, Jefferson, RA (1995) β-glucuronidase (Gus) transposons for ecological for ecological and genetic studies of rhizobia and other gram negative bacteria. Microbiology 141: 1691–1705
Yamada, Y, Hoshino, K, Ishikawa, T (1997) The phylogeny of acetic acid bacteria based on the partial sequences of 16S ribosomal RNA: the elevation of the subgenus Gluconoacetobacter to the generic level. Biosci Biotechnol Biochem 61: 1244–1251
Yanni, YG, Rizk, RY, Abd El-Fattah, FK, et al. (2001) The beneficial plant growth-promoting association of Rhizobium leguminosarum bv. trifolii with rice roots. Aust J Plant Physiol 28: 1–26
Acknowledgments
This work is part of the Ph.D. thesis of first author. Thanks are due to Prof. P. Vandamme LMG-BCCM, University of Gent, Belgium for providing B. vietnamiensis and B. cepacia reference cultures. For reprints, preprints, unpublished data, practical assistance, and helpful discussions, we thank our friends and colleagues in throughout the world.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Govindarajan, M., Balandreau, J., Kwon, SW. et al. Effects of the Inoculation of Burkholderia vietnamensis and Related Endophytic Diazotrophic Bacteria on Grain Yield of Rice. Microb Ecol 55, 21–37 (2008). https://doi.org/10.1007/s00248-007-9247-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-007-9247-9