Abstract
Two hybrid moving bed biofilm reactor-membrane bioreactors were used for the treatment of variable-salinity influent wastewater with maximums of 4.5 and 8.5 mS cm−1 electric conductivity. Operational conditions of the bioreactors were 6 h hydraulic retention time and 2500 mg L−1 total solids. The membrane operated in a cycle of 9 min draw-1 min backwash and at 23.6 L h−1 m−2 flux rate. Membrane biofilm was collected from both systems and cultured in growth media for precipitation of carbonate and phosphate minerals, yielding only Bacillus stratosphericus for the 4.5 mS cm−1 scenario and Bacillus stratosphericus, Bacillus toyonensis, Microbacterium esteraromaticum, Comamonas testosteroni, and Janibacter meloni for the 8.5 mS cm−1 scenario. Scanning electron microscopy and X-ray analysis showed similarities in morphology and composition for the carbonate crystals from both salinity conditions and differences for the phosphate minerals. Study of the bacterial community of membrane biofilm and mixed liquor showed close similarities between them for the same salinity conditions, with both dominated by genera Rhodanobacter, Chujaibacter, and Thermomonas.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
For the purpose of wastewater treatment, one of the best available technologies is the membrane bioreactor (MBR) (Rodriguez-Sanchez et al. 2017a). The MBR joins the biological treatment with a very efficient separation of solids and water that allows for high performances in the removal of solids and pathogens, operation at higher solids concentrations, low sludge production, complete retention of biomass and higher solids retention times in operation, with the drawback of higher energy costs in operation (Rodriguez-Sanchez et al. 2017b).
In spite of their numerous advantages, the MBR systems have to face a major operational problem caused by the clogging of membrane pores, nominated as fouling, which occurs more rapidly at higher suspended solids concentrations during operation. This fouling can be caused by materials of inorganic origin or organic origin (Rodriguez-Sanchez et al. 2018a). To alleviate the fouling problems of MBR systems, the combined system of the moving bed biofilm reactor-membrane bioreactor (MBBR-MBR) has been developed. In this context, the addition of carrier media to the bioreactor allows for the growth of attached biomass, which increases the total solids concentrations in the system without raising the suspended solids, allowing for operation with less risk of fouling. Nevertheless, MBBR-MBR systems still have to face fouling problems (Gonzalez-Martinez et al. 2015).
A major cause of fouling is the colonization of microbial communities of the membrane surface, leading to biofouling, which is mainly caused by the extracellular polymeric substances and the soluble microbial products they produce (Gao et al. 2014). Therefore, it has been proposed that the characterization of the microbial communities in the membrane biofilm is of vital importance in order to understand, predict, and control the process of biofouling (Rodriguez-Sanchez et al. 2018b). Moreover, the serious risk that minerals whose precipitation is caused by bacterial activity have been highlighted before (Gonzalez-Martinez et al. 2017).
It has been proven that bacteria play a role in the formation of some minerals in different natural and engineered environments, and that this is not restricted to a certain taxonomy but widely distributed across many members of the domain Bacteria with different phylogenies (Rivadeneyra et al. 2017). On the other hand, it has been proposed that biomineralization occurs due to changes in the immediate environment of the microorganism, such as changes in pH and ion concentrations, and therefore, it has been proposed that there is a specific association between microbial taxonomy, the environment where it grows, and the mineral formed (Gonzalez-Martinez et al. 2016). In wastewater treatment systems, the most common biominerals found are of carbonate and phosphate nature. Carbonate precipitation in wastewater has been regarded as an effective and economic feasible solution for the sequestration of CO2 in stable forms, such as calcium carbonate, thus reducing the impact of CO2 over the Earth’s biosphere (Uad et al. 2014). On the other hand, phosphate mineral struvite has been associated with operational problems in wastewater treatment systems as far as to develop specific reactors for its precipitation and safeguard the process downstream (Rivadeneyra et al. 2014).
Wastewater can raise its salinity levels by several ways, such as addition of salt for snow-melting activities, addition of salt for cooking, use of seawater in toilets for flushing or by the intrusion of seawater in wastewater treatment plants of coastal areas (Cortes-Lorenzo et al. 2012). Salinity in wastewater can have a dramatic effect over the performance and the bacterial community structure of bioprocesses. It has been proposed that increasing salinity affects microbial metabolism, causes the accumulation of toxic metabolic intermediates, and that increases cell membrane osmotic pressure leading to higher cell death rates (Bassin et al. 2012; Castillo-Carvajal et al. 2014; Cortés-Lorenzo et al. 2016). Moreover, previous research has found these effects precisely in MBR and MBBR-MBR systems (Rodriguez-Sanchez et al. 2017a, b, 2018a). Salinity conditions in MBR and MBBR-MBR systems affect their microbial communities and can successively affect the biofouling of the membrane process within them, a phenomenon that has been unexplored to date.
The MBBR-MBR systems operated under a membrane flux rate of 23.6 L h−1 m−2 in a 9-min draw-1-min backwash cycle for the membrane, and with a hydraulic retention time of 6 h and 2500 mg L−1 total solids. The analysis of the biomineralization phenomenon in the membrane biofilms of systems operating under tidal salinity variations with maximum influent salinities of 4.5 and 8.5 mS cm−1 was done. Bacterial strains with capacity to mediate mineralization of carbonates and phosphates were identified by culture-dependent techniques. The nature of the minerals formed was evaluated by scanning electron microscopy and X-ray analysis. Moreover, the whole bacterial community in the bioreactors operating under such conditions was observed by culture-independent techniques such as next-generation sequencing, with emphasis on the quantification of strains with mineralization capacity.
2 Materials and Methods
2.1 Bioreactor Configuration and Operation
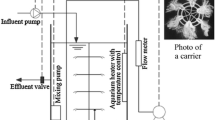
The bioreactor configuration used in the study was a hybrid MBBR-MBR system (Fig. 1). The bioreactor had four chambers of 6 L operational volume and a membrane tank of 4.32 L operational volume. The first, third, and fourth chambers were filled with carriers in 35% of their volumes. The carrier used was the K1 carrier (AnoxKaldnes AS, Norway) (density of 0.92–0.96 g cm−3) accounting for a total of 111.7 m2 m−3 in the whole bioreactor. Fine bubble diffusers AFD 270 (ECOTEC SA, Spain) and air compressors ACO-500 (Hailea, China) were used to aerate the first, third, and fourth chambers, with input air entering at the bottom of the chamber, assuring for a complete mix of the volume of the chamber. The aeration of the chambers was measured by rotameters 2100 Model (Tecfluid SA, Spain) and regulated using a valve. The second chamber was anaerobic and completely mixed by the means of a mechanical stirrer Multi Mixer MM-1000 (Biosan Laboratories Inc., USA). Coarse bubble diffusers CAP 3 (ECOTEC SA, Spain) and air compressors ACO-500 (Hailea, China) were used to aerate and completely mix the membrane tank. The aeration of the membrane tank was measured by rotameters 2100 Model (Tecfluid SA, Spain) and regulated using a valve.
Inside the membrane tanks, a module of 0.04 μm pore diameter hollow fibers of polyvinylidene fluoride with a core reinforcement of polyester was displaced vertically and continuously submerged. The total membrane surface area per bioreactor was of 0.2 m2.
The influent flow was pumped, using a Watson-Marlow peristaltic pump (Watson-Marlow Pumps Group, USA), to the first chamber and forced to pass through the second, third, and fourth, then discharged into the membrane tank. From the membrane tank, a recycling flow of 500% the influent flow was imposed using a Watson-Marlow peristaltic pump. Also, the effluent was withdrawn by the means of pumping, using a Watson-Marlow peristaltic pump through the membrane module. The pump withdrawing the effluent operated in a 10-min cycle consisting of 9 min effluent withdrawal followed by 1 min backwash.
The influent wastewater was a salinity-amended urban wastewater, pumped into the bioreactors from a mixing tank. The amendment was achieved by automatic addition of tap water with NaCl diluted resulting in 50 mS cm−1 electric conductivity, stored in the NaCl tap water tank, to the regular urban wastewater, stored in the urban wastewater tank. The automatic addition of NaCl tap water to the urban wastewater in the mixing tank was done by a TOPAX LF1 conductivity module (Lutz-Jesco GmbH, Germany), which continuously monitored the electric conductivity of the influent wastewater by continuous measuring using a conductivimeter. Two Watson-Marlow peristaltic pumps connected to the conductivity module allowed to automatically increase the proportion of urban wastewater and NaCl tap water in the influent wastewater depending on the specified salinity conditions in the influent. Two different salinity scenarios for operation were chosen, both resembling tidal-like salinity variations: (a) cycle of 6 h maximum electric conductivity of 4.5 mS cm−1 (around 2.4 g L−1NaCl) followed by 6 h of regular urban wastewater electric conductivity (around 0.5 g L−1 NaCl); (b) cycle of 6 h maximum electric conductivity of 8.5 mS cm−1 (around 4.8 g L−1NaCl) followed by 6 h of regular urban wastewater electric conductivity (around 0.5 g L−1 NaCl). A schematic of the daily cycle of operation is given in Fig. 1.
The bioreactors were operated controlling the hydraulic retention time to 6 h and the total solids to 2500 mg L−1. The biofouling on the membrane was controlled by periodical cleaning of the membrane to avoid transmembrane pressures higher than 0.5 bar. The total solids in the bioreactor were purged in order to maintain them at 2500 mg L−1 concentration. In this sense, the calculated solids retention time for the bioreactors was of 22.32 days.
2.2 Sampling of Biomass, DNA Extraction, and Next-Generation Sequencing
When the bioreactors were operating under steady-state conditions, biomass samples were collected from the membrane tank. Two sampling dates were considered for each bioreactor, separated in time by 2 weeks of operation under the same conditions. The collection of samples was done from the mixed liquor in the membrane tank and from the membrane biofilm following Gonzalez-Martinez et al. (2014). To collect the mixed liquor biomass, 100 mL was taken from the completely mixed membrane tank. For the membrane biofilm, a part of the membrane was sonicated at room temperature for 2 min at 40 kHz in a bath sonicator (Ultrasonic bath, Selecta, Barcelona, Spain), then placed in an orbital shaker at 155 rpm for 1 h. In parallel, suspensions of mixed liquor and membrane biofilm were centrifuged at 3500 rpm (821.73 g) during 10 min at room temperature, then discarding the supernatant and subjecting the biomass to DNA extraction process.
The DNA extraction was done using the FastDNA SPIN Kit for Soil (MP Biomedicals, Solon, OH, USA) and the FastPrep apparatus, following the instructions given by the manufacturer. DNA was extracted from each of the biomass samples in five replicates, then the five replicates were merged into the same DNA pool, as previously done (Gonzalez-Martinez et al. 2016). The extracted DNA pools were kept at − 20 °C and sent to RTLGenomics laboratory (Lubbock, TX, USA) in order to proceed with the next-generation sequencing procedure using the Illumina MiSeq equipment and the Illumina MiSeq Reagents Kit v3. The primer pair 28F (5′-GAGTTTGATCNTGGCTCAG-3′)-519R (5′-GTNTTACNGCGGCKGCTG-3′) (Rodriguez-Sanchez et al. 2018a, b) was used for the amplification of the hypervariable regions V1-V2-V3 of the 16S rRNA gene of the domain Bacteria. The PCR conditions for the amplification of such region were preheating at 94 °C during 120 s; then 32 cycles of 94 °C for 30 s, 40 °C for 40 s, and 72 °C for 60 s; a final elongation at 72 °C for 300 s.
2.3 Ecological Data from the Biological Samples
The raw data provided by the Illumina MiSeq sequencing was treated to yield valuable ecological data of the biological samples. Mainly mothur v1.34.4 (Schloss et al. 2009) was used for the treatment of such data. In the first place, the paired-end reads generated by the Illumina MiSeq equipment were aligned against each other using Needleman conditions to create contigs, avoiding the generation of ambiguous bases in the overlap region due to differences in overlapping nucleotides Phred score values (Unno 2015). Then, the contigs were screened to eliminate those with > 0 ambiguous bases and > 8 homopolymers. The remaining contigs were aligned against the SiLVA SEED v132 database using the k-nearest neighbor algorithm with k-mer search methodology using a k-mer size of 8 bp and under Needleman conditions. The contigs that failed to start and end at the positions of the primers of choice were discarded as mis-target errors. The remanent contigs were then preclustered to eliminate sequencing noise, following a preclustering threshold of 1 bp difference for each 100 bp in the sequence (Huse et al. 2010). The contigs were then checked for their chimeric nature using VSEARCH algorithm (Rognes et al. 2016) implemented in mothur software. Non-chimeric sequences were then classified against the SiLVA nr v132 database using the k-nearest neighbor algorithm with k-mer search methodology using a k-mer size of 8 bp under a taxonomic cutoff of 80% in order to remove sequences that could not affiliate to the domain Bacteria. After removal of cross-domain contamination, the high-quality contigs remaining were used to construct OTUs.
Construction of OTUs was done using a cutoff of 97% similarity, using the abundance-based greedy algorithm implemented in VSEARCH (Westcott and Schloss 2015; Schloss 2016), taking into consideration the Matthew’s correlation coefficient as metric of clusterization quality. A taxonomic consensus for all sequences within an OTU was calculated, thus assigning a taxonomy to each OTU based on SiLVA nr v132. The taxonomic consensus of each OTU was then used for the creation of the taxonomic consensus of the biological sample. Taxonomic consensus of sequences within each OTU and of OTUs within a sample was calculated using a cutoff of 80%. The taxonomic consensus of OTUs within all samples was considered as relevant ecological data.
2.4 Analysis of Similarity of Bacterial Community Structures
The analysis of similarity of the bacterial community structures was done in three different ways: PERMANOVA analyses (Weiss et al. 2017), singular value decomposition of centered log-ratio-transformed OTU distribution (Bian et al. 2017), and Dirichlet Multinomial Mixture model analysis (Holmes et al. 2012). The PERMANOVA analyses were computed based on Bray-Curtis distance over the taxonomic consensus of OTU distribution and under 9999 bootstrap replications using PASTv3.The singular value decomposition analyses used a transformation of the taxonomic consensus of OTU distribution was done by zero-value correction through a Bayesian multiplicative replacement method using zCompositions package implemented in R software followed by a centered log-ratio calculation using robComposition package implemented in R software. The singular value decomposition computation was done over such transformed data. The calculation of Dirichlet multinomial mixture models was done by partitioning the taxonomic consensus of OTUs distribution into metacommunities with minimum partition of 2 and maximum partition of 8, assessing the most appropriate partitioning as the minimum value reported by Laplace approximation calculations for each partitioning model.
2.5 Diversity Indices and Detection of Sensitive OTUs and the Correlations Between Them
The α-diversity indices of Shannon-Wiener (H) and Simpson (1D) were calculated for each of the biological samples taking into account the consensus taxonomy OTU distribution. The sensitivity of the OTUs among the different samples was observed by the means of expected effect size and SIMPER analysis. The expected effect size used the OTU distribution was done over the zero-corrected and centered log-ration transformed data obtained from the posterior distribution of 128 Monte Carlo simulations drawn from a Dirichlet distribution, computed through the ALDEx2 package implemented in R (Bian et al. 2017). The SIMPER analyses were done over the OTU distribution using PASTv3 software. The sensitivity of OTUs in both cases was assessed for the variables of maximum influent salinity (4.5 vs 8.5 mS cm−1) and biomass type (membrane biofilm vs mixed liquor).
2.6 Culture and Isolation of Mineral-Formation-Mediating Bacteria
The biomass samples from the membrane biofilm were used as inoculum for the culture of bacterial strains with capacity to mediate the formation of carbonate minerals and phosphate minerals, following previous research in MBBR-MBR systems (Gonzalez-Martinez et al. 2017). This biomass was obtained by sonication of a part of the membrane at room temperature for 2 min at 40 kHz in a bath sonicator (Ultrasonic bath, Selecta, Barcelona, Spain).
The biomass was used as seed for growth in two different media specifically designed for the formation of carbonate minerals (MC) and phosphate minerals (ME). The composition of the two media was as follows: 18 g L−1 agar, 10 g L−1 yeast extract, 5 g L−1 protease peptone, and 1 g L−1 glucose, 4 g L−1 calcium acetate for the MC medium and 8 g L−1 magnesium acetate for ME medium. The media was autoclaved at 112 °C for 20 min avoiding the precipitation of carbonate minerals in the MC medium due to intense heating. The pH of the media was set to 7.0 by addition of 0.1 M KOH. Additional ME liquid medium was done using the following composition: 10 g L−1 yeast extract, 5 g L−1 protease peptone, and 1 g L−1 glucose, 4 g L−1 calcium acetate for the MC medium and 8 g L−1 magnesium acetate. The difference between the ME solid and ME liquid media was the presence or absence of agar, respectively.
Samples of membrane biofilm were serially diluted up to 10−6, then 1 mL of concentrations of 10−4, 10−5, and 10−6 were used for inoculation of the media by surface-spreading and streaking. Each dilution for each medium had five replicates in total. The liquid ME medium was inoculated with 10−4 dilution in a 1% v/v. These were incubated at 25 °C in the dark and examined every 24 h to check for the apparition of minerals using optical microscopy. Uninoculated media and media inoculated with autoclaved membrane biofilm were used as control.
Colonies detected for relevant biomineralization-mediation capacity were isolated into the respective medium MC or ME in order to obtain a pure culture for further taxonomic identification of their 16S rRNA gene by using Sanger sequencing and the primer pair fD1-rD1, conducted at the University of León, Spain.
2.7 Characterization of Isolated Strains
The 16S rRNA gene of the isolated strains with relevant capacity to mediate mineral formation were used for their taxonomic classification. The strains were classified by BLAST search of highly similar sequences (MegaBLAST) against the nt database of the NCBI. The taxonomic affiliation of the strains was assigned by the first BLAST match with high quality (max score > 2500, e-value ~0, query cover ≥ 98%, identity ≥ 99%, curated RNA (NR) collection) of a sequence with known species level that was detected in the next-generation sequencing. Strains that did not pass these criteria were discarded as contamination. The remaining sequences were aligned against themselves using CLUSTALW algorithm (Larkin et al. 2007), and the alignment was used to construct a phylogenetic tree under the neighbor-joining method using the Jukes-Cantor model with a bootstrap test of 1000 replications, developed in MEGA7 (Kumar et al. 2016).
2.8 Extraction of Minerals and Analysis Using Optical Microscopy, Scanning Electron Microscopy, and X-Ray Diffractometry
Optical microscopy was used for the evaluation of mineral formation in the solid media. After a period of 14 days of incubation, the minerals formed in the solid MC medium were extracted by cutting the medium to pieces, submerged in water and boiled to dissolve the agar. The minerals obtained were then washed with distilled water for impurity removal, then let to dry at 37 °C. Accordingly, the minerals from the liquid ME medium were obtained by sedimentation, then washed with distilled water to remove impurities and let to dry at 37 °C.
The obtained minerals were observed by the means of scanning electron microscopy. The samples were carbon-coated and seen by using a high-resolution field emission scanning electron microscope Carl Zeiss, Supra 40 V (Carl Zeiss, Oberlocken, Germany). Samples of interest were also analyzed for their chemical composition using energy-dispersive X-ray (EDX) microanalysis (Aztec 350, Oxford Instruments, Abingdon, UK) and using the AMSCD mineral database.
3 Results and Discussion
3.1 Operation of the MBBR-MBR Systems
Data for the performance of the MBBR-MBR systems is shown in Table 1. From there, it was inferred that the removal performances were substantially different between the bioreactors in terms of organic matter and nitrogen (99.47 vs 91.60% for BOD5, 81.72 vs 60.72% for COD, 56.09 vs 27.11% for NH4+, 54.29 vs 26.77% for TN for the 4.5 and the 8.5 mS cm−1 scenarios, respectively) (Rodriguez-Sanchez et al. 2018b). Based on these results, the MBBR-MBR system working under the 4.5 mS cm−1 loading performed better (about 10–30%) in terms of BOD5 removal and COD removal. In this way, the higher salinity concentrations mildly affected the removal of organic matter in the bioreactors. Also, the 4.5 mS cm−1 scenario presented much higher NH4+ oxidation and TN removal values (around 200%) than the 8.5 mS cm−1 scenario.
Importantly, none of the systems could successfully form attached biomass to the carriers, as confirmed by low biofilm density values (< 50 mg L−1), which demonstrated the low contribution in terms for the total solids as these were controlled at 2500 mg L−1. Therefore, the contribution of the attached biofilm to the MBBR-MBR systems was less than 2% (Rodriguez-Sanchez et al. 2018b). It has been shown that introduction of saline influent in a steady-state MBR system treating regular urban wastewater exerts a high pressure over the attached biofilms of carriers, lowing important amounts in the process (Di Trapani et al. 2014). Moreover, difficulties for the formation of attached biofilm to carriers in MBBR-MBR systems treating constant salinity and variable salinity wastewater has been reported before, and the explanation of the inadequacy of the wastewater microbiota to develop biofilm in the presence of high salt concentrations was given as the reason for the absence of carrier biomass (Rodriguez-Sanchez et al. 2017a, b, 2018a). For the purposes of MBBR-MBR engineering, the low attached biofilm concentrations found under saline wastewater conditions had two valuable future implications. First, the MBBR-MBR systems under salinity conditions operated as MBR systems in practice. Second, the presence of carriers did not support higher total solids with reduced suspended solids, which in turn did not protect the membrane against operation at high solids concentrations, which has been claimed as one of the advantages of the MBBR-MBR systems over the MBR systems with respect to prevention of membrane fouling.
According to the results obtained, the implications of the treatment of saline wastewater using MBBR-MBR technologies are of concern from the biofouling point of view. More research should be conducted over the influence that the improvement of attached biofilm over carriers has on the biofouling of MBBR-MBR systems operating under saline effluents.
3.2 Analyses of Similarity of Biological Samples
The analyses of similarity of biological samples based on their consensus taxonomy of OTUs was done by quantitative and qualitative methodologies. For quantitative methodologies, the PERMANOVA analysis discriminating the samples in terms of maximum influent salinity (4.5 and 8.5 mS cm−1) and biomass type (membrane biofilm and mixed liquor) showed statistically significant differences (p < 0.05) for the different salinity conditions (Table S1). On the other hand, the biomass type did not have any statistically significant difference (p > 0.05). The pairs of samples sharing maximum influent salinity conditions and biomass type did not show any statistically significant difference (p > 0.05) as well, suggesting that the whole taxonomic consensus of OTUs between both scenarios and biomass were not that different overall. The other quantitative analyses, the Dirichlet multinomial mixing modelization, suggested that the best partitioning model accounted for 2 partitions (minimum Laplace approximation value), and the model grouped the samples according to maximum influent salinity (Fig. 2), corroborating the results obtained by the PERMANOVA analyses. Moreover, the results of the qualitative analysis of similarity was in accordance with these of the quantitative analyses of similarity, suggesting that the taxonomic consensus of OTUs was more affected by the maximum influent salinity than by the biomass type (Fig. 3).
The analyses of similarity showed that there was evidence of the strong effect of the influent maximum salinity over the consensus taxonomy of OTUs of both mixed liquor and membrane biofilm in the hybrid MBBR-MBR systems studied. In this sense, the attached biomass and the suspended biomass were similar in terms of consensus taxonomy of OTUs, suggesting that under variable influent salinity, the specialization of biomass did not appear in the attached biomass with respect to the mixed liquor, on the contrary, as reported for hybrid MBBR-MBR systems treating regular-salinity urban wastewater (Leyva-Diaz et al. 2015). Thus, the most important factor for the development of microbial communities in the hybrid MBBR-MBR subjected to influent variable salinities was found to be the maximum influent salinity concentration.
3.3 Ecology of the Biological Samples
The ecology of the 20 dominant consensus taxonomy OTUs in the biological samples studied is shown in Fig. 4. The relative abundance of these consensus taxonomy OTUs as well as their contribution to dissimilarity among groups of samples as determined per SIMPER analyses is presented.
Interestingly, the dominant genus found in all samples was related to Rhodanobacter. This genus had a mean relative abundance for the 4.5 mS cm−1 scenario than for the 8.5 mS cm−1 scenario, as calculated by Dirichlet multinomial mixing modelization—29.42% mean (26.26–32.96% as 95% confidence interval) for 8.5 mS cm−1, 46.04% mean (41.58–50.97% as 95% confidence interval) for 4.5 mS cm−1 (Table S2). Rhodanobacter genus has been reported as a dominant phylotypes in MBR and hybrid MBBR-MBR systems treating saline wastewater under constant and variable conditions (Rodriguez-Sanchez et al. 2017b; Rodriguez-Sanchez et al. 2018a). It has been reported that Rhodanobacter was positively correlated with heterotrophic and nitrification kinetics of these bioreactors, suggesting possible roles in organic matter removal, nitrification, and denitrification. Moreover, Rhodanobacter seemed to compete with autotrophic nitrifier Nitrobacter, which aims to consider its possible role as nitrite oxidizer in membrane technologies treating saline wastewater (Rodriguez-Sanchez et al. 2017b; Rodriguez-Sanchez et al. 2018a).
Another Rhodanobacteraceae family member, affiliated to genus Chujaibacter, was found to be important for the 4.5 mS cm−1 scenario (9.19%, (8.17–10.33%)), but less important for the 8.5 mS cm−1 conditions (2.49%, (2.02–3.08%)). The only strain isolated belonging to the genus Chujaibacter showed strict aerobic metabolism and no growth in 1% NaCl concentration, no capacity for nitrate reduction but positive assimilation of N-acetylglucosamine (Kim et al. 2015). This is therefore the first report of Chujaibacter in wastewater treatment systems, and it could be inferred that this strain should be different from the previously described Chujaibacter soli. This strain could thrive in the hybrid MBBR-MBR by aerobic degradation of organic matter with special attention to N-acetylglucosamine, which is readily available in the bioreactor since variable salinity conditions exerts high pressure over the microbial communities and thus increase mortality of cells (Rodriguez-Sanchez et al. 2017b). More research should focus on the elucidation of the ecological role of Chujaibacter in hybrid MBBR-MBR systems in particular and wastewater treatment bioprocesses in general.
On the other hand, Thermomonas genus accounted for a higher representation in the 8.5 mS cm−1 scenario (6.30%, (5.4–7.35%)) compared to the 4.5 mS cm−1 scenario (0.64% (0.55–0.83%)). It has been found that the Thermomonas genus thrives in MBR and hybrid MBBR-MBR systems treating saline wastewater, and their metabolic capability for oxidation of both organic matter, including N-acetylglucosamine, and ammonium has been reported (Wang et al. 2014; Ali et al. 2016). Indeed, kinetic analyses showed that Thermomonas relative abundance was linked to faster autotrophic kinetics but slower heterotrophic kinetics, and therefore, its ecological role in the MBR and hybrid MBBR-MBR systems treating saline wastewater has been suggested as heterotrophic nitrifier outcompeting autotrophic nitrifier for ammonium substrate due to lack of adaptation of ammonium oxidizing bacteria to saline wastewater environment (Rodriguez-Sanchez et al. 2017b).
Among all the dominant consensus taxonomy OTUs, the one that accounted for the highest dissimilarity contribution was the Rhodanobacter genus, followed by Chujaibacter genus (Fig. 4). The contributions to dissimilarity between biological samples for Rhodanobacter and Chujaibacter were very high (27.82–33.51% and 8.00–10.65%, respectively), except for the comparison between mixed liquor and fixed film biomass at the 8.5 mS cm−1 scenario, were their dissimilarity contributions were lower (15.24 and 1.15%, respectively). For the comparison between mixed liquor and fixed film biomass at the 8.5 mS cm−1 scenario, the top dissimilarity contributor was Ferruginibacter (10.76%). On the contrary, for the other groups of samples, Rhodanobacter and Chujaibacter together summed up to 44.16% of dissimilarity. In this sense, the SIMPER results showed that the proliferation of Rhodanobacter and Chujaibacter were major driving factors for the differences among the biological samples. This could be caused by the competitive nature of these genera in the hybrid MBBR-MBR systems treating variable influent salinity wastewater, thus reducing the diversity in the systems due to outcompetition of these groups with respect to other phylotypes. The diversity and evenness in the biological samples was confirmed by the Shannon-Wiener and Simpson indices, which have been claimed as the most robust indices to compare diversity of microbial communities (Haegeman et al. 2013). In this sense, the reduction of diversity and evenness in the biological samples could be linked to the relative abundance of dominant Rhodanobacter and Chujaibacter, since all samples at 4.5 mS cm−1 had lower evenness (as per lower values in Simpson index) and lower diversity (as per lower Shannon-Wiener index values) (Table S3). Thus, the importance of Rhodanobacter and Chujaibacter, and of Rhodanobacteraceae family in general, in variable salinity wastewater treatment processes should be further analyzed.
Quantitative differential analyses over the consensus taxonomy OTU distribution showed 4 consensus taxonomies with significant different relative abundance between mixed liquor and membrane biofilm (expected effect size < − 1 or expected effect size > 1) (Fig. S1). Only one of these phylotypes was among the dominant 20 but did not have major importance in terms of relative abundance for the bioreactors compared to other phylotypes (0.63%, (0.48–0.81%) for 4.5 mS cm−1, 1.00%, (0.73–1.35%) for 8.5 mS cm−1). On the other hand, 119 phylotypes showed significant relative abundance between the different maximum influent salinities. Among them, the dominant Rhodanobacter, Chujaibacter, and Thermomonas genera were present. As previously shown, maximum influent salinity levels selected for different dominance of Rhodanobacter, Chujaibacter, and Thermomonas in terms of relative abundance.
These dominant genera showed statistically significant correlation as shown by calculation of Erb’s ρ correlation coefficient (Fig. S2). Rhodanobacter genera had statistically significant positive correlation (ρ > 0.65) with Chujaibacter and a dominant unclassified Rhodanobacteraceae family member. On the other hand, Chujaibacter had statistically significant negative correlation (ρ < − 0.65) with Thermomonas genus. In this sense, the competition among the dominant phylotypes in both salinity scenarios showed a competition between Thermomonas and Chujaibacter. Since Thermomonas has been linked to autotrophic kinetics and had been suggested as an important nitrifier in hybrid MBBR-MBR systems treating saline wastewater (Rodriguez-Sanchez et al. 2017b), it is possible that Chujaibacter is able to develop this type of metabolism in the hybrid MBBR-MBR systems treating variable-salinity influent wastewater.
These results were also correlated with those from the SIMPER analysis developed over the 95% confidence interval relative abundance values of the Dirichlet multinomial mixing partitions showed that the most significant contributions to dissimilarity belonged to Rhodanobacter (15.81%), Chujaibacter (6.29%), and Thermomonas (5.20%) (Table S3). Moreover, the relative abundances of these OTUs were significantly different between the 8.5 and the 4.5 mS cm−1 salinity scenarios (26.9 vs 43.6%, 2.55 vs 9.25%, 6.37 vs 0.67%, respectively). In this way, significantly different higher presence of Rhodanobacter and Chujaibacter could be related to higher organic matter and ammonium oxidation potential in the MBBR-MBR systems. More research on the metabolic capabilities with respect to contributions to organic matter removal and ammonium oxidation of Rhodanobacter and Chujaibacter should be performed, as well as over the capacity of Chujaibacter to oxidize ammonium.
3.4 Formation of Minerals Mediated by Bacterial Activity from Membrane Biofilm
It was found that the growth of bacterial colonies in the MC and ME media was accompanied by the appearance of mineral-like structures (Fig. 5). The morphology of the minerals formed in the media were different for the two salinity scenarios. In the MC medium, the minerals formed by membrane biofilm collected under 4.5 mS cm−1 were of smaller size and sphere-like (Fig. 5a, c) in comparison to those obtained in MC medium inoculated with membrane biofilm collected under 8.5 mS cm−1 operation, which had a rhomboid-like shape (Fig. 5e, g). With respect to the ME medium, the minerals formed in plates inoculated with 4.5 mS cm−1 membrane biofilm presented a marked tetrahedral structure (Fig. 5b, d) when compared to those in the plates with 8.5 mS cm−1 membrane biofilm, which were smaller and more compact (Fig. 5f, h). The plates seeded with autoclaved media did not show formation of any mineral, as occurred with the plates not inoculated. In this sense, the culture of membrane biofilm from both salinity scenarios showed that formation of minerals in the MC and ME media was mediated by microbial activity and that different minerals were formed for the biomass from different maximum influent salinity conditions.
The colonies that showed a significant number of biominerals around them were isolated in respective MC or ME media in order to determine the taxonomy of such mineral-formation-mediating strains. Construction of a phylogenetic tree and taxonomic affiliation of their almost-complete 16S rRNA gene showed that, among the 20 successfully cultured strains, 5 different species were found (Fig. 6). For the membrane biomass of salinity scenario 4.5 mS cm−1, only the species Bacillus stratosphericus was identified three times from the MC medium and once from. On the other hand, the membrane biofilm for the 8.5 mS cm−1 scenario yielded five different strains with capability to mediate mineral formation. In the MC, medium 6 strains were identified and all were affiliated to Bacillus toyonensis species. Among those identified in the ME medium, 1 was affiliated to Bacillus stratosphericus, 1 to Microbacterium esteraromaticum, 1 to Janibacter melonis, and 2 to Comamonas testosteroni.
Bacillus stratosphericus was firstly isolated from air samples collected at great altitude and was reported to have very high resistance to NaCl (up to 17.4%) (Shivaji et al. 2006). Bacillus toyonensis has been reported to have resistance to high salinities (up to 5% NaCl) (Jiménez et al. 2013a) and has been used widely in animal nutrition since 1975 as it is the basis of the probiotics food additive Toyocerin® (Jiménez et al. 2013b). Neither Bacillus stratosphericus nor Bacillus toyonensis have previously been reported to mediate mineral formation. Nevertheless, many Bacillus strains have been found to help calcite and phosphate mineral formation in wastewater systems and high-salinity environments, such as Bacillus marisflavi, Bacillus pumilus, Bacillus megaterium, Bacillus subtilis, Bacillus thuringensis, or Bacillus flexum (Gonzalez-Martinez et al. 2017; Uad et al. 2014; Silva-Castro et al. 2015).
Microbacterium esteraromaticum was firstly described in 1993 (Yokota et al. 1993) as Aureobacterium esteraromaticum and unified in 1998 to the genus Microbacterium (Takeuchi and Hatano 1998).The strain Microbacterium esteraromaticum DSM 8609 has been reported for ammonium oxidation under low concentrations and low temperatures (Zhang et al. 2016), which could be explained as a heterotrophic nitrification-aerobic denitrification metabolism as observed in other strains of Microbacterium esteraromaticum (Vinothkumar et al. 2016). A strain of the Microbacterium genus has been reported for mediation of mineral formation of calcite nature (Xu et al. 2017). Nevertheless, no reports to this date have informed about mineralization of phosphate minerals mediated by Microbacterium genus.
Janibacter melonis was firstly isolated from an abnormally spoiled melon in Korea, showing strict aerobic growth (Yoon et al. 2004). Further than infecting melons, Janibacter melonis has been reported a cause of bacteremia in humans causing low fever, skin swelling, pain, and erythema (Elsayed and Zhang 2005) and has been noticed that some strains of Janibacter melonis have higher virulence than Janibacter terrae, known to cause infections in humans (Chander et al. 2018). No Janibacter strains have been reported for mineral precipitation.
Comamonas testosteroni has been reported as NaCl tolerant (up to 3% concentration) and facultative anaerobic bacterium (Zhu et al. 2014). It has been found that Comamonas testosteroni could be of importance for the development of membrane biofilm since it has a strong capacity for biofilm formation (Li et al. 2009). Comamonas isolated from membrane biofilm of hybrid MBBR-MBR system treating urban wastewater have previously been reported to mediate the precipitation of carbonates (Gonzalez-Martinez et al. 2017).
The relative abundances of the consensus taxonomy OTUs affiliated to the same genus that the stains isolated from the mineralization cultures (except for Janibacter, which was not found but attributed to the only unclassified Intrasporangiaceae member present among the taxonomic consensus) showed that their presence in the bacterial community structure in the hybrid MBBR-MBR systems treating variable-salinity influent wastewater was not dominant. In terms of biofouling risk, as assessed by Gonzalez-Martinez et al. (2015), the most dangerous phylotype at 4.5 mS cm−1 scenario was Janibacter melonis, and for the 8.5 mS cm−1, it was Microbacterium. Nevertheless, summed presence of the total consensus taxonomy OTUs related to isolated strains with mediation in mineral formation process was of 0.29–0.41% for the 4.5 mS cm−1 and 0.31–0.76% for the 8.5 mS cm−1 scenarios (Fig. 7).
3.5 Nature of Minerals Extracted from the MC and ME Media Formed by Mediation of Bacterial Activity
Differences were observed between the minerals extracted from the MC and ME media for the two maximum salinity scenarios (Fig. 8). Moreover, the results of the X-ray microanalyses for regions of interest on the mineral surfaces are shown in Fig. 9. For the minerals extracted from the MC media, their external morphology was similar for the scenarios of 4.5 and 8.5 mS cm−1, showing rhombohedral and sphere-like structures with either a smooth surface or rough surface with presence of bacterial footprints (Fig. 8a, b, e, g). Their X-ray microanalysis spectra demonstrated similar elemental formation with main Ca element composition regardless of the roughness of the surface, the presence of the bacterial footprints, or the morphology of the mineral tested (Fig. 9a, b). The similarity in morphology and composition for minerals in MC medium culturing membrane biofilm from 4.5 and 8.5 mS cm−1 scenarios could potentially be caused by the presence of only one strain with capacity for mediation of mineral formation in such medium, related to genus Bacillus. Therefore, it is possible that the mediation of Bacillus stratosphericus from the 4.5 mS cm−1 membrane biofilm and Bacillus toyonensis from the 8.5 mS cm−1 membrane biofilm resulted in the formation of similar minerals. In this sense, it is possible that these two Bacillus strains have the same behavior in terms of mineral formation mediation.
On the other hand, the morphologies of the minerals extracted from the ME medium were different between the two salinity scenarios (Fig. 8c, d, f, h). For the 4.5 mS cm−1 scenario, the mineral observed had a tetrahedral morphology. On the other hand, the morphology of minerals for the 8.5 mS cm−1 scenario showed compact minerals with polished surfaces marked with bacterial footprints. The differences in morphology were also correlated to differences in composition as determined by X-ray microanalysis. For the 4.5 mS cm−1 membrane biofilm, the minerals generated in the ME medium has a composition made mainly of Mg and P elements. On the other hand, the elemental composition of the minerals extracted from the ME medium inoculated with membrane biofilm from the 8.5 mS cm−1 scenario consisted on Ca and P. Differences in the composition and morphology of the minerals formed could depend on the taxonomy of isolated strains with capacity to mediate mineral formation from ME media: Bacillus stratosphericus only for the 4.5 mS cm−1 membrane biofilm, and Bacillus stratosphericus, Microbacterium esteraromaticum, Janibacter melonis, and Comamonas testosterone for the 8.5 mS cm−1 membrane biofilm. In this sense, presence of the three non-Bacillus strains could lead to formation of a majority of minerals in the ME medium with different characteristics than those for the Bacillus stratosphericus strains.
4 Conclusions
Two hybrid MBBR-MBR systems were operated at 23.6 L h−1 m−2 flux rate under 6 h hydraulic retention time and 2500 mg L−1 total solids for the treatment of tidal-like variable-salinity influent with maximums of 4.5 and 8.5 mS cm−1, respectively. Maximum influent salinity showed to affect organic matter removal and ammonium oxidation between the MBBR-MBR, favoring the lowest influent salinity conditions (99.47 vs 91.60% for BOD5, 81.72 vs 60.72% for COD, 56.09 vs 27.11% for NH4+, 54.29 vs 26.77% for TN). The effect of the maximum influent salinity over the bacterial communities and their capability for mediating carbonate and phosphate mineral precipitation was analyzed. The culture of mineral-precipitation-mediating bacteria from membrane biomass under both conditions showed that Bacillus stratosphericus was the only mineralization-inducing strain under 4.5 mS cm−1, with 8.5 mS cm−1 membrane biofilm showing a higher diversity with Bacillus stratosphericus, Bacillus toyonensis, Comamonas testosteroni, Microbacterium esteroaromaticum, and Janibacter meloni being found as important for biological-induced mineralization. Carbonate-based minerals were identical in morphology and composition among the two salinity scenarios, while the phosphate-based minerals did differ in structure and chemical composition. The whole bacterial community structure for membrane biofilm was very similar than that of the mixed liquor at each salinity scenario, with maximum influent salinity driving the diversity of the system by diminishing the relative abundance of dominant Rhodanobacter (26.26–32.96% vs 41.58–50.97%), Chujaibacter (8.17–10.33% vs 2.02–3.08%) and Thermomonas (5.4–7.35% vs 0.55–0.83%) genera with increasing salinity.
References
Ali, M., Chai, L. Y., Min, X. B., Tang, C. J., Afrin, S., Liao, Q., et al. (2016). Performance and characteristics of a nitritation air-lift reactor under long-term HRT shortening. International Biodeterioration and Biodegradation, 111, 45–53. https://doi.org/10.1016/j.ibiod.2016.04.003.
Bassin, J. P., Kleerebezem, R., Muyzer, G., Rosado, A. S., Van Loosdrecht, M. C. M., & Dezotti, M. (2012). Effect of different salt adaptation strategies on the microbial diversity, activity, and settling of nitrifying sludge in sequencing batch reactors. Applied Microbiology and Biotechnology, 93(3), 1281–1294. https://doi.org/10.1007/s00253-011-3428-7.
Bian, G., Gloor, G. B., Gong, A., Jia, C., Zhang, W., Hu, J., et al. (2017). The gut microbiota of healthy aged Chinese is similar to that of the healthy young. mSphere, 2(5), e00327–e00317. https://doi.org/10.1128/mSphere.00327-17.
Castillo-Carvajal, L. C., Sanz-Martin, J. L., & Barragan-Huerta, B. E. (2014). Biodegradation of organic pollutants in saline wastewater by halophilic microorganisms: a review. Environmental Science and Pollution Research, 21(16), 9578–9588. https://doi.org/10.1007/s11356-014-3036-z.
Chander, A. M., Kochhar, R., Dhawan, D. K., Bhadada, S. K., & Mayilraj, S. (2018). Genome sequence and comparative genomic analysis of a clinically important strain CD11-4 of Janibacter melonis isolated from celiac disease patient. Gut Pathogens, 10(1), 1–8. https://doi.org/10.1186/s13099-018-0229-x.
Cortes-Lorenzo, C., Rodriguez-Diaz, M., Lopez-Lopez, C., Sanchez-Peinado, M., Rodelas, B., & Gonzalez-Lopez, J. (2012). Effect of salinity on enzymatic activities in a submerged fixed bed biofilm reactor for municipal sewage treatment. Bioresource Technology, 121, 312–319. https://doi.org/10.1016/j.biortech.2012.06.083.
Cortés-Lorenzo, C., González-Martínez, A., Smidt, H., González-López, J., & Rodelas, B. (2016). Influence of salinity on fungal communities in a submerged fixed bed bioreactor for wastewater treatment. Chemical Engineering Journal, 285, 562–572. https://doi.org/10.1016/j.cej.2015.10.009.
Di Trapani, D., Di Bella, G., Mannina, G., Torregrossa, M., & Viviani, G. (2014). Comparison between moving bed-membrane bioreactor (MB-MBR) and membrane bioreactor (MBR) systems: influence of wastewater salinity variation. Bioresource Technology, 162, 60–69. https://doi.org/10.1016/j.biortech.2014.03.126.
Elsayed, S., & Zhang, K. (2005). Bacteremia caused by Janibacter melonis. Journal of Clinical Microbiology, 43(7), 3537–3539. https://doi.org/10.1128/JCM.43.7.3537-3539.2005.
Gao, D. W., Wen, Z. D., Li, B., & Liang, H. (2014). Microbial community structure characteristics associated membrane fouling in A/O-MBR system. Bioresource Technology, 154, 87–93. https://doi.org/10.1016/j.biortech.2013.11.051.
Gonzalez-Martinez, A., Leyva-Díaz, J. C., Rodriguez-Sanchez, A., Muñoz-Palazon, B., Rivadeneyra, A., Poyatos, J. M., et al. (2015). Isolation and metagenomic characterization of bacteria associated with calcium carbonate and struvite precipitation in a pure moving bed biofilm reactor-membrane bioreactor. Biofouling, 31(4), 333–348. https://doi.org/10.1080/08927014.2015.1040006.
Gonzalez-Martinez, A., Rodriguez-Sanchez, A., Lotti, T., Garcia-Ruiz, M. J., Osorio, F., Gonzalez-Lopez, J., & van Loosdrecth, M. C. M. (2016). Comparison of bacterial communities of conventional and A-stage activated sludge systems. Scientific Reports, 6, 18786. https://doi.org/10.1038/srep18786.
Gonzalez-Martinez, A., Rodriguez-Sanchez, A., Rivadeneyra, M. A., Rivadeneyra, A., Martin-Ramos, D., Vahala, R., & Gonzalez-Lopez, J. (2017). 16S rRNA gene-based characterization of bacteria potentially associated with phosphate and carbonate precipitation from a granular autotrophic nitrogen removal bioreactor. Applied Microbiology and Biotechnology, 101(2), 817–829. https://doi.org/10.1007/s00253-016-7914-9.
Haegeman, B., Hamelin, J., Moriarty, J., Neal, P., Dushoff, J., & Weitz, J. S. (2013). Robust estimation of microbial diversity in theory and in practice. ISME Journal, 7(6), 1092–1101. https://doi.org/10.1038/ismej.2013.10.
Holmes, I., Harris, K., & Quince, C. (2012). Dirichlet multinomial mixtures: generative models for microbial metagenomics. PLoS One, 7(2). https://doi.org/10.1371/journal.pone.0030126.
Huse, S. M., Welch, D. M., Morrison, H. G., & Sogin, M. L. (2010). Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environmental Microbiology, 12, 1889–1898. https://doi.org/10.1111/j.1462-2920.2010.02193.x.
Jiménez, G., Blanch, A. R., Tamames, J., & Rosselló-mora, R. (2013a). Complete genome sequence of Bacillus toyonensis BCT-7112 T , the active ingredient of the feed additive preparation toyocerin. Genome Announcements ASM, 1(6), e01080–e01013. https://doi.org/10.1128/genomeA.01080-13.Copyright.
Jiménez, G., Urdiain, M., Cifuentes, A., López-López, A., Blanch, A. R., Tamames, J., et al. (2013b). Description of Bacillus toyonensis sp. nov., a novel species of the Bacillus cereus group, and pairwise genome comparisons of the species of the group by means of ANI calculations. Systematic and Applied Microbiology, 36(6), 383–391. https://doi.org/10.1016/j.syapm.2013.04.008.
Kim, S.-J., Ahn, J.-H., Weon, H.-Y., Hong, S.-B., Seok, S.-J., Kim, J.-S., & Kwon, S.-W. (2015). Chujaibacter soli gen. nov., sp. nov., isolated from soil. Journal of Microbiology, 53(9), 592–597. https://doi.org/10.1007/s12275-015-5136-y.
Kumar, S., Stecher, G., & Tamura, K. (2016). MEGA7 : molecular evolutionary genetics analysis version 7 . 0 for bigger datasets. Molecular Ecology and Evolution, 33(7), 1870–1874. https://doi.org/10.1093/molbev/msw054.
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., Mcgettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics, 23(21), 2947–2948. https://doi.org/10.1093/bioinformatics/btm404.
Leyva-Diaz, J. C., González-Martinez, A., Gonzalez-Lopez, J., Muñio, M. M., & Poyatos, J. M. (2015). Kinetic modeling and microbiological study of two-step nitrification in a membrane bioreactor and hybrid moving bed biofilm reactor—membrane bioreactor for wastewater treatment. Chemical Engineering Journal, 259, 692–702. https://doi.org/10.1016/j.cej.2014.07.136.
Li, M. Y., Zhang, J., Lu, P., Xu, J. L., & Li, S. P. (2009). Evaluation of biological characteristics of bacteria contributing to biofilm formation. Pedosphere, 19(5), 554–561. https://doi.org/10.1016/S1002-0160(09)60149-1.
Rivadeneyra, A., Gonzalez-Martinez, A., Gonzalez-Lopez, J., Martin-Ramos, D., Martinez-Toledo, M. V., & Rivadeneyra, M. A. (2014). Precipitation of phosphate minerals by microorganisms isolated from a fixed-biofilm reactor used for the treatment of domestic wastewater. International Journal of Environmental Research and Public Health, 11(4), 3689–3704. https://doi.org/10.3390/ijerph110403689.
Rivadeneyra, A., Gonzalez-Martinez, A., Portela, G. R., Martin-Ramos, D. J., Gonzalez-Lopez, J., & Rivadeneyra, M. A. (2017). Biomineralisation of carbonate and sulphate by the halophilic bacterium Halomonas maura at different manganese concentrations. Extremophiles, 21(6), 1049–1056. https://doi.org/10.1007/s00792-017-0965-8.
Rodriguez-Sanchez, A., Leyva-Diaz, J. C., Gonzalez-Martinez, A., & Poyatos, J. M. (2017a). Performance and kinetics of membrane and hybrid moving bed biofilm-membrane bioreactors treating salinity wastewater. AICHE Journal, 63(8), 3329–3342. https://doi.org/10.1002/aic.15694.
Rodriguez-Sanchez, A., Leyva-Diaz, J. C., Gonzalez-Martinez, A., & Poyatos, J. M. (2017b). Linkage of microbial kinetics and bacterial community structure of MBR and hybrid MBBR–MBR systems to treat salinity-amended urban wastewater. Biotechnology Progress, 33(6). https://doi.org/10.1002/btpr.2513.
Rodriguez-Sanchez, A., Leyva-Diaz, J. C., Gonzalez-Lopez, J., & Poyatos, J. M. (2018a). Membrane bioreactor and hybrid moving bed biofilm reactor-membrane bioreactor for the treatment of variable salinity wastewater: influence of biomass concentration and hydraulic retention time. Chemical Engineering Journal, 336. https://doi.org/10.1016/j.cej.2017.10.118.
Rodriguez-Sanchez, A., Leyva-Diaz, J. C., Poyatos, J. M., & Gonzalez-Lopez, J. (2018b). Influent salinity conditions affect the bacterial communities of biofouling in hybrid MBBR-MBR systems. Journal of Water Process Engineering. https://doi.org/10.1016/j.jwpe.2018.07.001.
Rognes, T., Flouri, T., Nichols, B., Quince, C., & Mahé, F. (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ, 4, e2584. https://doi.org/10.7717/peerj.2584.
Schloss, P. D. (2016). Application of a database-independent approach to assess the quality of. mSystems, 1(2), 2–5. https://doi.org/10.1128/mSystems.00027-16.Copyright.
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Applied and Environmental Microbiology, 75(23), 7537–7541. https://doi.org/10.1128/AEM.01541-09.
Shivaji, S., Chaturvedi, P., Suresh, K., Reddy, G. S. N., Dutt, C. B. S., Wainwright, M., et al. (2006). Bacillus aerius sp. nov., Bacillus aerophilus sp. nov., Bacillus stratosphericus sp. nov. and Bacillus altitudinis sp. nov., isolated from cryogenic tubes used for collecting air samples from high altitudes. International Journal of Systematic and Evolutionary Microbiology, 56(7), 1465–1473. https://doi.org/10.1099/ijs.0.64029-0.
Silva-Castro, G. A., Uad, I., Gonzalez-Martinez, A., Rivadeneyra, A., Gonzalez-Lopez, J., & Rivadeneyra, M. A. (2015). Bioprecipitation of calcium carbonate crystals by bacteria isolated from saline environments grown in culture media amended with seawater and real brine. BioMed Research International, 2015. https://doi.org/10.1155/2015/816102.
Takeuchi, M., & Hatano, K. (1998). Union of the genera Microbacterium Orla- Jensen and Aureobacterium Collins et a/. in a redefined genus Microbacterium. International Journal of Systematic Bacteriology, 48, 739–747.
Uad, I., Gonzalez-Lopez, J., Silva-Castro, A. G., Vílchez, J. I., Gonzalez-Martinez, A., Martin-Ramos, D., et al. (2014). Precipitation of carbonates crystals by bacteria isolated from a submerged fixed-film bioreactor used for the treatment of urban wastewater. International Journal of Environmental Research, 8(2), 435–446.
Unno, T. (2015). Bioinformatic suggestions on MiSeq-based microbial community analysis. 25(6), 765–770.
Vinothkumar, R., Bharti, V. S., Vennila, A., Kumar, H. S., & Pandey, P. K. (2016). Isolation and characterization of heterotrophic nitrifying—denitrifying bacteria from shrimp farming ponds. Ecology, Environment and Conservation, 22(1), 275–279.
Wang, L., Zheng, S., Wang, D., Wang, L., & Wang, G. (2014). Thermomonas carbonis sp. nov., isolated from the soil of a coal mine. International Journal of Systematic and Evolutionary Microbiology, 64(Pt 11), 3631–3635. https://doi.org/10.1099/ijs.0.063800-0.
Weiss, S., Xu, Z. Z., Peddada, S., Amir, A., Bittinger, K., Gonzalez, A., et al. (2017). Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome, 5(1), 1–18. https://doi.org/10.1186/s40168-017-0237-y.
Westcott, S. L., & Schloss, P. D. (2015). De novo clustering methods outperform reference-based methods for assigning 16S rRNA gene sequences to operational taxonomic units. PeerJ, 3, e1487. https://doi.org/10.7717/peerj.1487.
Xu, G., Li, D., Jiao, B., Li, D., Yin, Y., Lun, L., et al. (2017). Biomineralization of a calcifying ureolytic bacterium Microbacterium sp. GM-1. Electronic Journal of Biotechnology, 25, 21–27. https://doi.org/10.1016/j.ejbt.2016.10.008.
Yokota, a., Takeuchi, M., Sakane, T., & Weiss, N. (1993). Proposal of six new species in the genus Aureobacterium and transfer of Flavobacterium esteraromaticum Omelianski to the genus Aureobacterium as Aureobacterium esteraromaticum comb. nov. International Journal of Systematic Bacteriology, 43(3), 555–564. https://doi.org/10.1099/00207713-43-3-555.
Yoon, J. H., Lee, H. B., Yeo, S. H., & Choi, J. E. (2004). Janibacter melonis sp. nov., isolated from abnormally spoiled oriental melon in Korea. International Journal of Systematic and Evolutionary Microbiology, 54(6), 1975–1980. https://doi.org/10.1099/ijs.0.63167-0.
Zhang, D., Huang, X., Li, W., Qin, W., & Wang, P. (2016). Characteristics of heterotrophic nitrifying bacterium strain SFA13 isolated from the Songhua River. Annals of Microbiology, 66(1), 271–278. https://doi.org/10.1007/s13213-015-1105-2.
Zhu, D., Xie, C., Huang, Y., Sun, J., & Zhang, W. (2014). Description of Comamonas serinivorans sp. nov., isolated from wheat straw compost. International Journal of Systematic and Evolutionary Microbiology, 64(2014), 4141–4146. https://doi.org/10.1099/ijs.0.06668.
Acknowledgments
We would like to acknowledge the support given by the Institute of Water Research, the Department of Civil Engineering and the Faculty of Pharmacy of the University of Granada.
Funding
Funding was given by the Ministerio de Economia, Industria y Competitividad of the Government of Spain by the project with reference CTM2013-48154-P and the grant with reference BES-2014-067852.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that there are no conflicts of interest.
Electronic supplementary material
ESM 1
(DOCX 129 kb)
Rights and permissions
About this article
Cite this article
Rodriguez-Sanchez, A., Muñoz-Palazon, B., Hurtado-Martinez, M. et al. Maximum Influent Salinity Affects the Diversity of Mineral-Precipitation-Mediating Bacterial Communities in Membrane Biofilm of Hybrid Moving Bed Biofilm Reactor-Membrane Bioreactor. Water Air Soil Pollut 229, 342 (2018). https://doi.org/10.1007/s11270-018-4020-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11270-018-4020-x