Abstract
Bacterial associations with plant roots are thought to contribute to the success of phytoremediation. We tested the effect of addition of a polycyclic aromatic hydrocarbon contaminated soil on the structure of the rhizosphere microbial communities of wheat (Triticum aestivum), lettuce (Lactuca sativa var. Tango), zucchini (Cucurbita pepo spp. pepo var. Black Beauty), and pumpkin (C. pepo spp. pepo var. Howden) 16S rDNA terminal restriction fragment length polymorphism (T-RFLP) profiles of rhizosphere microbial communities from different soil/plant combinations were compared with a pairwise Pearson correlation coefficient. Rhizosphere microbial communities of zucchini and pumpkin grown in the media amended with highest degree of contaminated soil clustered separately, whereas communities of these plants grown in unamended or amended with lower concentrations of contaminated soil, grouped in a second cluster. Lettuce communities grouped similarly to cucurbits communities, whereas wheat communities did not display an obvious clustering. The variability of 16S rDNA T-RFLP profiles among the different plant/soil treatments were mostly due to the difference in relative abundance rather than presence/absence of T-RFLP fragments. Our results suggest that in highly contaminated soils, the rhizosphere microbial community structure is governed more by the degree of contamination rather than the plant host type.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Background
Polycyclic aromatic hydrocarbons (PAHs) are toxic, persistent, and recalcitrant to biodegradation (Ritter et al. 1995; Shuttleworth and Cerniglia 1995). In soils, hydrophobic PAHs strongly bind to soil and root surfaces making accumulation within plants highly unlikely (Simonich and Hites 1995; Wild and Jones 1992). Nevertheless, some plants, namely cucurbits, are known to accumulate significant amount of PAHs, which is likely due to production of low molecular weight organic acids (LMWOA) by plant roots (Parrish et al. 2006).
In addition, there is some evidence that the rhizosphere can facilitate relief of phytotoxicity imposed by the soil chemical conditions. Numerous reports indicate that pollutant degradation in rhizosphere is significantly faster than in surrounding bulk soils (Miya and Firestone 2000, 2001; Nichols et al. 1997). The mechanism of phytotoxicity relief is not yet understood, although rhizosphere microorganisms may play a major role. Some believe plant growth promoting rhizobacteria reduce plant stress in contaminated soils by reducing the concentration of ethylene below stress levels (Glick 2003), thus promoting phytoremediation indirectly. Others believe that rhizosphere bacterial communities relieve phytotoxicity through the stimulatory effect of rhizodeposition (carbon exudation) on microbial metabolic and cometabolic pathways that allow contaminant degradation (Kamath et al. 2004; Miya and Firestone 2001).
The composition of rhizosphere bacterial communities typically differs greatly from those of the surrounding soil, and from one plant species to another, as a result of diverse plant–microbe interactions (Grayston et al. 1998; Marschner et al. 2001; Smalla et al. 2001). Although the root exudate composition is a main factor setting the rhizosphere microbial community structure, other factors such as soil physicochemical properties (Zhou et al. 2002), plant age and root architecture (Kuske et al. 2003), plant nutritional status (Matz and Jürgens 2003; Yang and Crowley 2000), presence of plant parasites (Vestergård et al. 2004), protozoan grazing (Ronn et al. 2002), as well as chemical stress may lead to dramatic changes in rhizosphere community structure (Kerkhof et al. 2000).
In the case that contamination level governs the structure of rhizosphere bacterial communities, one would expect significant community shifts as the level of imposed stress, and associated degree of phytotoxicity, increases. The purpose of our study is, therefore, to examine for a set of flowering plants, one monocot (Poacea), and three dicots (Cucurbitaceae, Asteraceae), how the degree of soil chemical contamination (and associtated phytotoxicity) affected rhizosphere microbial community structure. The used soil was obtained by mixing an uncontaminated and a heavily PAH-contaminated soil in varying proportions. We postulated that plant type was the dominant factor in determining rhizosphere bacterial community structure.
2 Materials and Methods
2.1 Sample Description
We examined rhizosphere microbial communities of zucchini (Cucurbita pepo sp. pepo var. Black Beauty), pumpkin (C. pepo sp. pepo var. Howden), lettuce (Lactuca sativa var. Tango), and wheat (Triticum aestivum). These plants were chosen because cucurbits (zucchini and pumpkin) were shown to translocate persistent organic pollutants (Huelster et al. 1994; Mattina et al. 2002; White and Kottler 2002). This is unusual considering the low solubility of many organic pollutants. Cucurbits are believed to exude LMWOA, which facilitate the movement of hydrophobic contaminants into the aqueous phase where degradation and other forms of removal are possible (White and Kottler 2002; White et al. 2003). Wheat and lettuce plants have not demonstrated this capability. Therefore, these four plants should provide a spectrum of contaminant removal capabilities.
Seeds were pretreated before packaging to minimize fungal growth, and no additional surface sterilization was performed. Seeds were germinated in layers of wet paper towel at room temperature and transferred with tweezers to the pots with 400 g of potting media.
The potting media was created artificially by mixing pristine soil with historically contaminated soil in different proportions. Despite problems inherited with this design, it has a practical advantage of simulating a field situation. In bioremediation practice, mixing highly contaminated soil with pristine soil is one way of reducing the toxic effects and promoting biodegradation. Therefore, it is of interest to know the structure and dynamics of microbial communities of mixed soils and the contribution of plants. Uncontaminated Cheshire fine sandy loam collected at the Connecticut Agricultural Experiment Station Research Farm in Hamden, CT is used to grow a variety of flowering plants and expected to bear high bacterial diversity. Winsted soil (Winsted, CT) derived from a former Manufactured Gas Plant (MGP) site was highly contaminated with a suite of PAHs (total 1.1 ± 0.1 g/kg) and expected to have low bacterial content. It was mixed with an uncontaminated Cheshire soil to 0%, 0.3%, 3%, and 30% (w/w) in the final potting media. The Winsted soil and Cheshire soils were comparable in particle size distribution and total organic carbon content (e.g., 87.2/56 sand, 10/36 slit, 2.8/8 clay, 2.27/1.4 TOC %, respectively). To ensure homogeneity of potting media, soils were laid in thin layers on a sterile aluminum tray, mixed thoroughly with sterile spatulas section by section, and then mixed manually in a large bucket.
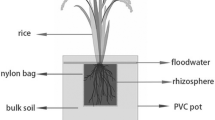
Each plant was grown in pots in quadruplicate in four different soil mixtures at room temperature and 16 h of incandescent light. The soil was kept moist with sterile water. Once a week, 30 ml of 10% sterile Hoagland’s solution (5 mM KNO3, 5 mM Ca (NO3)2, 2 mM MgSO4, 1 mM KH2PO4, 100 μM Fe(III)NaEDTA, 230 μM H3BO3, 45 mM MnCl2, 1.5 μM CuSO4, 1.5 μM ZnSO4, 0.4 μM (NH4)6Mo7O24) was supplied per pot. After 3 months of growth under defined laboratory conditions, the plants were harvested from the potting media. Freshly cut shoots were weighted. Standard t test (α = 0.05) was performed to check for effect of soil treatment on shoot weight. Roots were carefully removed from potting media under sterile conditions; large dry clumps of soil were shaken off the roots. The soil particles closely adhering to the roots were included as part of the rhizosphere, which is defined as the zone of physical and chemical interaction between the root and the surrounding soil. Roots (the whole root) were placed in 40 mL of 0.1 M potassium phosphate buffer (PPB) and sonicated using a sonic dismembrator wand for 30 s at setting 4 (which translates to 8–11 W depending on solution density). Sonicant was allowed to settle for 10 min, and the liquid phase was collected. These suspensions was spun down at 10,000×g for 20 min, the supernatant removed, the pellet re-suspended in 1 mL of 0.1 M PPB, and frozen for storage. These suspensions as well as 1 g of bulk Cheshire and Winsted soil were used for DNA extraction.
2.2 DNA Extraction Procedure
Community genomic DNA was extracted with UltraClean Mega Soil DNA Kit (#12900-100) under gentle lysis treatment as described by the manufacturer (MoBio, Inc., Solana Beach, CA). The resulting DNA extract was eluted in 60 μl of MoBio S5 solution. A 10-μl aliquot was used for evaluation of DNA quality and mass via gel electrophoresis. The remaining portion of the extract was kept at −80°C. DNA concentrations were estimated with PicoGreen (Molecular Probes, Eugene, OR) staining and fluorescence spectrophotometry.
2.3 16S rDNA Amplification
Eubacterial 16S rRNA genes were amplified with forward S-D-Bact-49-a-S-20 (5′-TNANACATGCAAGTCGAICG-3′) and reverse S-D-Bact-1492-a-A-19 (5′-GGYTACCTTGTTACGACTT-3′) primers (Moyer et al. 1994), renamed based on (Alm et al. 1996). For use in terminal restriction fragment length polymorphism (T-RFLP), the forward primer was 5′-labeled with 4,7,2′,4′,5′,-7′-hexachloro-6-carboxyfluorescein (Operon, Inc., Alameda, CA). The Qiagen Taq PCR core kit with Q solution (Qiagen, Valencia, CA) was used for all PCRs. Each 100-μl reaction mixture consisted of 1× PCR buffer, 1× Q solution, 5 mM Mg2+, 0.4 μM of each primer, 200 μl dNTPs, and 2.5 U Taq polymerase. The amplification was carried out in Perkin Elmer thermocyclers (Perkin Elmer Lifesciences Inc, Boston, MA, models 2400 and 9700) with following cycling program (Initial denaturation 2 min at 94°C; followed by 33 cycles of denaturation 30 s at 94°C, annealing 45 s at 60°C, elongation 1.5 min at 72°C, and a final elongation step 7 min at 72°C).
2.4 Gel Electrophoresis
Quality of DNA and PCR products were evaluated via standard gel electrophoresis, stained with SybrGold (Molecular Probes, Eugene, OR, USA), and viewed on a transilluminator chamber (Bio-Rad, Hercules, CA, USA). Gel images were captured with CCD camera and viewed and analyzed with Multi-Analyst software (Bio-Rad, Hercules, CA, USA). GeneRuler™ DNA ladder mix (Fermentas Inc., Hanover, MD #SM033) was used as the size and mass standard.
2.5 Restriction Digest
Amplicons from triplicate reactions were pooled and purified with DNA Clean and Concentrator-5 (Zymo Research, Orange, CA) and eluted in 20 μl of TE (pH 8.0). The final concentration of amplicons was established via fluorescent spectroscopy using Picogreen® DNA Quantification Reagent (Molecular Probes, Eugene, OR). Volumes were adjusted to obtain a final concentration 20 ng/μl. Restriction digests were performed in 20 μl final volume with 2 μl of 10× buffer, 15 U of enzyme, and 200 ng of the amplicons. The reactions were incubated for 3.5 h at 37°C. The following enzymes were used: RsaI, HhaI, and HpaII (all obtained from Invitrogen Inc., Carlsbad, CA).
2.6 T-RFLP Profiling
Restriction digests were purified with DNA Clean and Concentrator-5 (Zymo Research, Orange, CA), eluted in 10 μl of sterile ultrapure water, and concentrated by drying in a Speed-Vac. Each dried sample was re-suspended in a mixture of 0.5 μl of loading buffer, 0.5 μl milli-Q water, and 1 μl of TAMRA2500 standard (ABI, Foster City, CA) before loading on polyacrylamide gel. The 5% Long Ranger gel (5.0 ml 50% Long Ranger gel stock solution, 1.25 g glycerol, 5.0 ml 10× TBE) was cast using a 36-cm well-to-read distance plates and comb with 0.2 mm well and 64 lanes. After sample loading gels were run for 4 h under native conditions (30°C) controlled by an ABI Prism 377 DNA sequencer. The data were collected with ABI 672 software and analyzed with GeneScan 3.1 software (ABI). Fragment sizes of the terminal restriction fragments were called by the Local Southern method.
2.7 Data Analysis
BioNumerics software (Applied-Maths, Belgium) was used for all subsequent numerical analyses. Sample files from GeneScan software were directly imported into Bionumerics, and gel images were recreated. Based on densitometric curve analysis, independently for each gel, Wiener cutoff points and background scale values were used to set an arithmetic averaging filter and the background noise removal, respectively. Due to variability among sample replicates, densitometric curves of replicate treatments were averaged using a Bionumerics script and analyzed further using Pearson correlation coefficient for standardized characters to build pairwise similarity matrix and UPGMA to cluster the patterns. Unfortunately, Bionumerics does not allow bootstrap analysis of curve-based data. Therefore, we used the cophenetic correlation value and error bars to validate clusters. The cophenetic correlation is a parameter of a consistence of a cluster. This method calculates the correlation between the dendrogram-derived similarities and the Pearson product–moments for each cluster. The error bars indicate consistence of a cluster as an average standard deviation of similarity. The standard deviation of a branch is obtained by reconstructing the similarity values from the dendrogram branch and comparing the values with the original similarity values.
3 Results and Discussion
We performed a study whereby soil treatments were created by mixing an agricultural soil with a heavily contaminated MGP soil in different proportions. We realize that in this way, mixtures of different bacterial content were created. However, considering the low bacterial content and diversity in the MGP soil due to the high level of PAH contamination, we can assume that most of the bacterial diversity in the soil mixture originated from the agricultural soils. These treatments, furthermore, mimic practical scenarios of reducing soil contamination by diluting highly contaminated soils of low bacterial diversity with high-quality soils of high bacterial diversity.
Clustering of averaged TRF profile curves (Fig. 2) resulted in three groups, each at 90% similarity (clusters I, II, and III). Cluster I contained rhizosphere communities of three plants grown in potting media amended with 30% of MGP soil (pumpkin, lettuce, and zucchini), as well as of wheat and zucchini grown in media with 3% and 0.3% MGP, respectively.
Cluster II consisted of communities of lettuce, pumpkin, and zucchini grown in media with 3% of MGP soil, as well as communities of zucchini and pumpkin grown in media with 0.3% of MGP soil. The third cluster consists of communities of lettuce, pumpkin, and zucchini grown in media with 0% MGP soil as well as communities of wheat grown in media with 0.3% and 30% of MGP soil. Unfortunately, cluster separation was not supported statistically with cophenetic correlation less than 70% and large error bars at the node of each cluster. Lack of statistical support suggests clusters are not significantly different.
Our data suggest, then, that rhizosphere bacterial communities from different plant species respond to a high level of PAH (30% MGP soil or 30,000 ppm of total PAH) contamination in a similar fashion. With the exception of wheat, this response overshadows plant-specific effects. The clustering pattern is governed by the contaminant rather than plant type.
The bacterial community of the wheat rhizosphere responded differently to the level contamination and suggests a stronger effect of plant type over soil environment.
Microbial communities of the pumpkin, zucchini, and lettuce rhizosphere are similar in both contrasting environments (uncontaminated and highly contaminated). The rhizosphere microbial communities of pumpkin and zucchini (Cucurbitaceae) cluster with lettuce (Asteraceae), but not with communities of wheat (Poaceae). It is possible that this phenomenon could be explained by the physiological uniqueness of wheat, the only monocot in the experiment. Monocots are known to differ from dicots not only in morphology and structure of their vascular system but also at the root biochemical level. For example, grass monocots are known to employ a different strategy of Fe sequestration than dicots and non-grass monocots (Cohen et al. 1997).
Bacterial communities of cucurbit rhizospheres grown in the media with intermediate levels of MGP soil (0.3% and 30%) are more similar than communities of lettuce and wheat suggesting uniqueness of the cucurbits. This is consistent with previous research that showed cucurbits and lettuce can accumulate persistent organic pollutants from the soil environment, possibly due to differences in their rhizosphere exudate chemistry, high above ground biomass and large surface area of leaves providing greater transpiration rates (Huelster et al. 1994). Limited chemical analysis shows that cucurbit root exudates have markedly high protein content and a low sugar level (Richardson and Baker 1982). Moreover, the types of sugars and proteins as well as range of organic acids are different in cucurbits compared to other plants (Richardson and Baker 1982). These differences might, at least partially, explain the ability of cucurbits to take up persistent organic pollutants and also may impact microbial community structure.
Despite the high similarity within the clusters, some trends are obvious. Bacterial communities of cucurbits grown in media with intermediate level of contamination (0.3% and 3% MGP soil, resulting in approximately 300 and 3,000 ppm of total PAH, respectively) group closer together, whereas communities of lettuce and wheat grown in media with 0.3% MGP soil group closer with the same communities grown with 30% MGP soil. Bacterial community of lettuce rhizosphere grown in media with 0% and 3% MGP, but not 0.3% or 30% MGP soil, group together with bacterial communities of cucurbits.
Our data suggest that at intermediate level of contamination, the plant effect dominates the effect of contamination level. Notably, rhizosphere bacterial communities of cucurbits responded to the intermediate contamination level in more similar way compared to other plants. Indeed, the averaged T-RFLP profiles of microbial communities of lettuce and cucurbits grown in presence of these moderate amounts of MGP soil cluster with profiles of the uncontaminated soils with over 90% similarity. It is unclear why the low contamination level has little effect on community structure, but it suggests that the rhizosphere conditions (type and amount of exudates) are little altered by the contamination level. On the other hands, the composition of root exudates is known to depend on many factors, including type, physiological status, and growth stage of the plant (Fan et al. 2001; Gransee and Wittenmayer 2000; Hamlen et al. 1972; Hutsch et al. 2002; Vancura and Hovadik 1965) and stress conditions (Gagnon and Ibrahim 1997; Walker et al. 2003).
T-RFLP profiles show high variability within the replicate treatments (50.0–97.5% of replicate treatment similarity), masking variability between samples. Such a high variability is not unusual, since replicate treatments are plants grown in distinct pots, watered manually. Variability within the replicates was explained by unequal distribution rather than presence/absence of terminal fragments. Moreover, fragment distribution accounts for most of the variability (60–98% similarity) among the different treatments as well (Fig. 1). In fact, no new prominent fragments appeared even in rhizosphere communities grown in media containing 30% of MGP soil. Absence of new prominent peaks in profiles of all contaminated soil treatments suggests that the effect of the MGP soil presence on overall rhizosphere microbial community was minimal. T-RFLP profiles of MGP soil contain significantly less fragments as compared to uncontaminated Cheshire soil (Fig. 3), validating our decision to exclude bacterial diversity of MGP soil from consideration as having low input into bacterial recruitment by plant roots.
While we observed both clustering effects associated with the degree of soil contamination and plant host (lettuce and cucurbits vs. wheat), cluster similarities were explained by variability in evenness of rhizosphere microbial communities rather than their richness (number of TRFs). Since the bulk soil is the main source for the recruitment of the rhizosphere microbial communities (all seeds were sterilized—and all contaminated treatments received Cheshire and MGP soil as bulk soil) but different plant exudates vary in content and amounts, the resulting rhizosphere microbial community is believed to be plant-specific. Earlier microcosm studies on rhizosphere microbial communities of various plants (Marschner et al. 2001; Smalla et al. 2001) also indicated plant specificity. However, detailed fragment analysis indicated that the majority of the DGGE fragments were shared between the plants and at maximum, 25% of the fragments were plant-specific (Marschner et al. 2001). Our findings support that the microbial community of bulk soil differs from that of rhizosphere soil, as neither the MGP nor the Cheshire soil microbial community profiles relate to the defined rhizosphere clusters (Fig. 1).
We did not include a control bulk soil with appropriate combinations of Winsted and Cheshire soils. Although the absence of the controls limits our investigation of the structure of the microbial population of a particular soil mixture, we still had an opportunity to compare the effect of different plants on the bacterial community structure of a particular soil mix. The Cheshire and Winsted (MGP) bulk soil controls are distinct from all the rhizosphere samples and can easily serve as an outgroup (Fig. 2).
Distribution of 16S rDNA terminal fragments in rhizosphere microbial communities (RsaI digest) depending on plant type and fraction of MGP soil (%). Top five most abundant terminal fragments are depicted in separate panel for each plant. In bulk control soils (data not shown), four of these fragments had relative abundances below 50 and fragment 644.62 bp (marked with asterisk) was undetected
To estimate the effect of contamination on plant growth, we measured the plant shoot weight at harvest (Fig. 3). After 3 months of growth, the addition of MGP soil had no negative effect on shoot weights of cucurbits. Shoot weights of lettuce and wheat were, however, negatively affected by addition of 3% and 30% of MGP soil. Addition of 0.3% of MGP soil had no affect on the dry weights of lettuce and wheat (Fig. 4).
While the underlying mechanisms are not definite, it is interesting to note that at the highest level of contamination, cucurbits displayed a clear physiological response: their shoot weight increased significantly compared to the replicates grown in less contaminated matrices (Fig. 4). A less striking but significant effect was also noted for lettuce grown at the highest contamination level: its shoot weight increased significantly above the replicate grown at 3% contamination. The manifestation of this positive physiological response at the highest contamination level suggests that the plants were able to relieve phytotoxicity, which was apparent at the lower contamination levels. While the mechanisms of phytotoxicity relief are unclear and may be manifold, they are likely associated with changes in the amount and type of root exudates, which could in turn affect the relative make up of the rhizosphere microbial community.
In contrast to the other plants, the shoot weights of wheat decreased steadily with degree of soil contamination. This might suggest that wheat is not able to mount some sort of phytotoxicity relief mechanism, and the rhizosphere chemical composition is not altered with level of soil contamination, resulting in a more constant microbial community profile. Despite the plant specificity of rhizosphere microbial communities, there are no data suggesting profound plant specificity in wheat over other plants (Fig. 3).
We conclude that the structure of the microbial communities of pumpkin, zucchini, and lettuce rhizospheres is primarily affected by the degree of MGP soil contamination, although there is a certain sensitivity threshold such that those effects are only manifest at high levels of contamination. However, in case of wheat, the plant effect is more dominant in shaping the rhizosphere microbial community.
References
Alm, E., Oerther, D., Larsen, N., Stahl, D., & Raskin, L. (1996). The oligonucleotide probe database. Applied and Environmental Microbiology, 62, 3557–3559.
Cohen, C., Norvell, W., & Kochian, L. (1997). Induction of the root cell plasma membrane ferric reductase (an exclusive role for Fe and Cu). Plant Physiology, 114, 1061–1069.
Fan, T., Lane, A., Shenker, M., Bartley, J., Crowley, D., & Higashi, R. (2001). Comprehensive chemical profiling of gramineous plant root exudates using high-resolution NMR and MS. Phytochemistry, 57, 209–221.
Gagnon, H., & Ibrahim, R. (1997). Effects of various elicitors on the accumulation and secretion of isoflavonoids in white lupin. Phytochemistry, 44, 1463–1467.
Glick, B. (2003). Phytoremediation: synergistic use of plants and bacteria to clean up the environment. Biotechnology Advances, 21, 383–393.
Gransee, A., & Wittenmayer, L. (2000). Qualitative and quantitative analysis of water-soluble root exudates in relation to plant species and development. Journal of Plant Nutrition and Soil Science, 163, 381–385.
Grayston, S. J., Wang, S., Campbell, C., & Edwards, A. (1998). Selective influence of plant species on microbial diversity in the rhizosphere. Soil Biology and Biochemistry, 30, 369–378.
Hamlen, R., Lukezic, F., & Bloom, J. (1972). Influence of age and stage of development on the neutral carbohydrate components in root exudates from alfalfa plants grown in agnotobiotic environment. Canadian Journal of Plant Science, 52, 633–642.
Huelster, A., Mueller, J. F., & Marschner, H. (1994). Soil-plant transfer of polychlorinated dibenzo-p-dioxins and dibenzofurans to vegetables of the cucumber family (Cucurbitaceae). Environmental Science & Technology, 28, 1110–1115.
Hutsch, B., Augustin, J., & Merbach, W. (2002). Plant rhizodeposition - an important source for carbon turnover in soils. Journal of Plant Nutrition and Soil Science, 165, 397–407.
Kamath, R., Schnoor, J., & Alvarez, P. (2004). Effect of root-derived substrates on the expression of nah-lux genes in Pseudomonas fluorescens HK44: implications for PAH biodegradation in the rhizosphere. Environmental Science & Technology, 38, 1740–1745.
Kerkhof, L., Santoro, M., & Garland, J. (2000). Response of soybean rhizosphere communities to human hygiene water addition as determined by community level physiological profiling (CLPP) and terminal restriction fragment length polymorphism (T-RFLP) analysis. FEMS Microbiology Letters, 184, 95–101.
Kuske, C., Ticknor, L., Busch, J., Gehring, C., & Whitham, T. (2003). The pinyon rhizosphere, plant stress, and herbivory affect the abundance of microbial decomposers in soils. Microbial Ecology, 45, 340–352.
Marschner, P., Yang, C., Lieberei, R., & Crowley, D. (2001). Soil and plant specific effects on bacterial community composition in the rhizosphere. Soil Biology and Biochemistry, 33, 1437–1445.
Mattina, M., White, J., Eitzer, B., & Iannucci-Berger, W. (2002). Cycling of weathered chlordane residues in the environment: compositional and chiral profiles in contiguous soil, vegetation, and air compartments. Environmental Toxicology and Chemistry, 21, 281–288.
Matz, C., & Jürgens, K. (2003). Interaction of nutrient limitation and protozoan grazing determines the phenotypic structure of a bacterial community. Microbial Ecology, 45, 384–398.
Miya, R., & Firestone, M. (2000). Phenanthrene-degrader community dynamics in rhizosphere soil from a common annual grass. Journal of Environmental Quality, 29, 584–592.
Miya, R., & Firestone, M. (2001). Enhanced phenanthrene biodegradation in soil by slender oat root exudates and root debris. Journal of Environmental Quality, 30, 1911–1918.
Moyer, C., Dobbs, F., & Karl, D. (1994). Estimation of diversity and community structure through restriction fragment length polymorphism distribution analysis of bacterial 16S rRNA genes from a microbial mat at an active, hydrothermal vent system, Loihi Seamount Hawaii. Appl Environ Microbiol., 60, 871–879.
Nichols, T., Wolf, D., Rogers, H., & Beyrouty, C. (1997). Rhizosphere microbial populations in contaminated soils. Water, Air, and Soil Pollution, 95, 165–167.
Parrish, Z., White, J., Isleyen, M., & Gent, M. (2006). Accumulation of weathered polycyclic aromatic hydrocarbons (PAHs) by plant and earthworm species. Chemosphere, 64, 609–618.
Richardson, P., & Baker, D. (1982). The chemical composition of cucurbit vascular exudates. Journal of Experimental Botany, 33, 1239–1247.
Ritter, L., Solomon, K., Forget, J., and Stemeroff, M. (1995). A review of selected persistent organic pollutants. The International Programme on Chemical Safety (IPCS) within the framework of the Inter-Organization Programme for the Sound Management of Chemicals (IOMC). 1–145.
Ronn, R., McCaig, A., Griffiths, B., & Prosser, J. (2002). Impact of protozoan grazing on bacterial community structure in soil microcosms. Applied and Environmental Microbiology, 68, 6094–6105.
Shuttleworth, K. L., & Cerniglia, C. E. (1995). Environmental aspects of PAH biodegradation. Applied Biochemistry and Biotechnology, 54, 291–302.
Simonich, S. L., & Hites, R. A. (1995). Organic pollutant accumulation in vegetation. Environmental Science & Technology, 29, 2095–2103.
Smalla, K., Wieland, G., Buchner, A., Zock, A., Parzy, J., Kaiser, S., et al. (2001). Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: plant-dependent enrichment and seasonal shifts revealed. Applied and Environmental Microbiology, 67, 4742–4751.
Vancura, V., & Hovadik, A. (1965). Root exudates of plants. II. Composition of root exudates of some vegetables. Plant and Soil, XXII, 21–32.
Vestergård, M., Bjørnlund, L., & Christensen, S. (2004). Aphid effects on rhizosphere microorganisms and microfauna depend more on barley growth phase than on soil fertilization. Oecologia, 141, 84–93.
Walker, T., Bais, H., Halligan, K., Stermitz, F., & Vivanco, J. (2003). Metabolic profiling of root exudates of Arabidopsis thaliana. Journal of Agricultural and Food Chemistry, 51, 2548–2554.
White, J., & Kottler, B. (2002). Citrate-mediated increase in the uptake of weathered 2,2-bis(p-chlorophenyl)1,1-dichloroethylene residues by plants. Environmental Toxicology and Chemistry, 21, 550–556.
White, J., Mattina, M., Lee, W., Eitzer, B., & Iannucci-Berger, W. (2003). Role of organic acids in enhancing the desorption and uptake of weathered p, p'-DDE by Cucurbita pepo. Environmental Pollution, 124, 71–80.
Wild, S. R., & Jones, K. C. (1992). Organic chemicals entering agricultural soils in sewage sludges: screening for their potential to transfer to crop plants and livestock. The Science of the Total Environment, 119, 85–119.
Yang, C., & Crowley, D. (2000). Rhizosphere microbial community structure in relation to root location and plant iron nutritional status. Appl Environ Microbiol, 66(1), 345–351.
Zhou, J., Xia, B., Treves, D., Wu, L., Marsh, T., O’Neill, R., et al. (2002). Spatial and resource factors influencing high microbial diversity in soil. Applied and Environmental Microbiology, 68, 326–334.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Pritchina, O., Ely, C. & Smets, B.F. Effects of PAH-Contaminated Soil on Rhizosphere Microbial Communities. Water Air Soil Pollut 222, 17–25 (2011). https://doi.org/10.1007/s11270-011-0800-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11270-011-0800-2