Abstract
The longitudinal dispersion coefficient (Kx) is fundamental to modeling of pollutant and sediment transport in natural rivers, but a general expression for Kx, with applicability in low or high flow conditions, remains a challenge. The objective of this paper is to develop a Pareto-Optimal-Multigene Genetic Programming (POMGGP) equation for Kx by analyzing 503 data sets of channel geometry and flow conditions in natural streams worldwide. In order to acquire reliable data subsets for training and testing, Subset Selection of Maximum Dissimilarity Method (SSMD), rather than the classical trial and error method, was used by a random manipulation of these data sets. A new hybrid framework was developed that integrates SSMD with Multigene Genetic Programming (MGP) and Pareto-front optimization to produce a set of selected dimensionless equations of Kx and find the best equation with wide applicability. The POMGGP-based final equation was evaluated and compared with 8 published equations, using statistical indices, graphical visualization of 95% confidence ellipse, Taylor diagram, discrepancy ratio (DR) distribution, and scatter plots. Besides being simple and applicable to a broad range of conditions, the proposed equation predicted Kx more accurately than did the other equations and can therefore be used for the prediction of longitudinal dispersion coefficient in natural river flows.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Nowadays, it is not uncommon to find dumping of biological, chemical and physical contaminants in rivers across the world, despite full knowledge that they pose a potential danger to public health. Contaminants propagate in vertical, longitudinal and transverse directions (Tayfour and Singh 2005), however, at a considerable distance downstream from the source and after mixing, just the longitudinal dispersion is dominant (Riahi-Madvar et al. 2009). Therefore, the longitudinal dispersion coefficient is fundamental in one dimensional (1-D) modeling of river water quality. Several water quality models, such as QUAL2E (Parveen and Singh 2016), QUAL2K (Hadgu et al. 2014), SIAQUA (Fan et al. 2015) and WASP (Moses et al. 2016), use the 1-D advection-dispersion equation expressed as (Eq. 1) (Noori et al. 2016):
where C is cross-sectionally averaged concentration, u is the longitudinal velocity, Kx is the longitudinal dispersion coefficient, x is the longitudinal coordinate parallel to the mean flow direction, and t is the time in unsteady modeling (Noori et al. 2016).
If a set of concentration and velocity measurements in a river are available, then it is simple to calculate the longitudinal dispersion coefficient. However, these measurements are time consuming and costly (Najafzadeh and Tafarojnoruz 2016). An alternative is to develop an equation incorporating the factors that affect the longitudinal dispersion coefficient. Seo and Cheong (1998) showed that the longitudinal dispersion coefficient can be expressed as a function of several factors as:
Which H is the flow depth, U* is the bed shear velocity, U is flow velocity, Sn is the sinuosity of the river, B is the channel width and D is the longitudinal dispersion coefficient (K). Several investigations have been carried out from the middle of the twentieth century to find an equation for the longitudinal dispersion coefficient. First, analytical and empirical approaches were developed (Elder 1959; Fischer et al. 1979; Kashefipour and Falconer 2002; Li et al. 2013). Empirical approaches were based on regression analysis using limited database.
Recently, Artificial Intelligence (AI) approaches have been used. For example, Artificial Neural Network (ANN) (Alizadeh et al. 2017), Adaptive Neuro-Fuzzy Inference System (ANFIS) (Riahi-Madvar et al. 2009), Support vector Machine (SVM) (Noori et al. 2009), and genetic programming (Rajeev and Dutta 2009) were used and found satisfactory in determining the longitudinal dispersion coefficient. However, most of these studies utilized small datasets (less than or equal to 150 data sets) of natural rivers. Although their results were promising, they may not be generalized to all rivers or for the extended data bases. Also, selection of subsets for training and testing of the AI models must be carefully done as it impacts the results. The training subset must encompass the high/low flow conditions to decipher the best pattern of the data. Finally, the optimization of equations is important to improve the robustness of results.
Considering these three points, (i.e. subset selection, extended data base, and optimal solution) the objective of this study was to develop a predictive equation for the longitudinal dispersion coefficient, using the integrated multigene genetic programming. To that end, the Subset Selection of Maximum Dissimilarity Method combined with the Multigene Genetic Programming based on Pareto Optimal solution (POMGGP) was utilized for developing predictive equations of longitudinal dispersion coefficient in natural rivers with 503 worldwide data sets. This optimization method is capable of selecting the best generation among all possible/existing generations. The reminder of the paper is organized as follows. In section 2 the methodology of model development and data sets are presented. Section 3 discusses the results of prediction and compares them with several existing methods. Finally, section 4 includes the conclusion of the study.
2 Methodology and Data Analysis
2.1 Longitudinal Dispersion Data
Among the geometrical and hydrodynamic parameters, flow depth (H), bed shear velocity (U*), flow velocity (U), and channel width (B) have the most effect on the longitudinal dispersion coefficient (K) (Noori et al. 2016). In this study, non-dimensional parameters of B/H, U*/U, and Kx/U*H were used to predict the longitudinal dispersion coefficient. A field dataset comprising 503 samples representing different river flow and pollutant conditions was collected from the literature (Deng et al. 2001; Kashefipour and Falconer 2002; Carr and Rehmann 2007; Riahi-Madvar et al. 2009; Ahmad 2013). The Kx values in these data sets are calculated based on the measured concentration profiles of tracer (C-t curves). Statistical characteristics, such as minimum (Min), maximum (Max), average (Mean) and standard deviation (SD) of the total, training, and testing datasets, were selected by the SSMD, as given in Table 1. The SSMD procedure and its results are discussed in section 2.2.
2.2 Data Pre-Processing by SSMD
The Subset Selection of Maximum Dissimilarity (SSMD) method was used to select training and testing subsets by random data manipulation. Due to the variability of dataset, this categorization and data-assimilation method was crucial for MGGP for accurate prediction (May et al. 2008). It may be noted that too few data create noise on one hand and large database creates complex equations with over-fitting challenges on the other hand (Yapo et al. 1998). The SSMD, developed by Kennard and Stone (1969), avoided these problems. The SSMD algorithm generates a subset from the master set in such a way that the subset data includes the highest dissimilarities. The selected subset does not concentrate on a specified area or the data from the edge of dataset.
The Kennard and Stone (1969) algorithm chose a subset of N-dimensional points that were distributed uniformly in the experimental space. If parameter X is the dataset as X = (x1, x2, …, xp) and a set of m = 1, 2, …, N points are defined as candidates of the subset for training.
The design points were selected sequentially. If the squared distance between the ith and jth points is defined as \( {D}_{i,j}^2 \) and k points have already been chosen (k < p), then the minimal distance from candidate point of N to k points can be defined as
The point of N is not in the training subset yet where the k points already are in the training subset. The (k + 1)th point in the training subset was selected from the remaining (N-k) points by:
where the N point belongs to the remaining dataset that is farthest from an existing point. In this study, 70% of the dataset was selected as the training subset, and the remaining points of dataset made the test subset. Therefore, the steps of SSMD are as follows (Wang and Huai 2016):
-
1-
Normalize the points of dataset.
-
2-
Choose the first number from the X dataset with the highest x1N and place it in the M subset as s1.
-
3-
Choose the second point of dataset with the largest distance to s1 and put it in the M subset as s2.
-
4-
After k steps, the dataset of X includes (p-k) data, where the M subset contains k data. The distances between M subset components and the remainder of X dataset components are considered. The minimum value of distance is designated to acquire (N-k) dpmin. The maximum dpmin is selected as mk = 1
-
5-
Repeat step 4 until 70% of dataset points are put in M or k = MN.
-
6-
De-normalize the selected data in the M and X subsets.
By the SSMD method, 70% and 30% of the data were selected for training and testing of the derived models, respectively, as shown in Table 1. Figure 1 shows the distribution of data selected subsets and shows that the training subset covers the boundaries of the data set and the testing subset is located inside the training subset. Therefore, the SSMD was able to discover the information on the boundaries of data space. Variations in the training subset were more extensive than those in the testing subset and will lead to more general prediction models. The most significant feature of the SSMD is that it encompasses outlier data in the training set.
2.3 Genetic Programming (GP)
Genetic Programming (GP), proposed by Koza (1992), automatically solves expressional optimization problems using Darwinian Theory of Evolution by natural selection. GP reproduces random population of species to create a new population to find the best fit solution (DanandehMehr and Kahya 2017). The final GP model may include the program’s external inputs, functions, and constants. To choose the best function set from all functions, a precise guess is required. The GP produces a series of formulas with distinct complexities, but the formulas that are moderately difficult are preferred. A simple equation may not be precise, but a complicated one may be over-trained and will result in over-fitting.
2.3.1 Multigene Genetic Programming (MGGP)
Recently, MGGP has been developed by modifying the classical GP. It linearly integrates small complexity GP expressions (Searson 2015). Every computer program or individual in MGGP is a weighted linear manipulation of genes (i.e. tree) plus a bias (noise) term. The linear constants for every MGGP individual are calculated by the usual least squares technique. MGGP has been shown to simulate more accurately the nonlinear behavior than does the classical linear regression method (DanandehMehr and Kahya 2017). Throughout the MGGP development, genes are acquired and removed by a two-point top level crossover that allows replacing genes between individuals (DanandehMehr and Kahya 2017).
2.3.2 Pareto Optimal-Multigene Genetic Programming (POMGGP)
Due to its simplicity in producing different levels of Pareto frontiers, the Pareto optimal is used in hybrid with MGGP. The Pareto solution set operates, based on a balance between multiple optimization goals, so that the results are more in line with the actual state of the problem, which can be used as a new way in multi-objective problems. The feasible solutions of a multi-objective problem are determined by the disassembly sequences that satisfy the disassembly priority relation.
A collection of the entire Pareto optimal solutions is entitled as the final Pareto optimal solutions set, and a set of values of the target function that are related to the disassembly sequence is called the Pareto optimal frontier (Zhang et al. 2017).
Figure 2 exhibits the implementation process and a flowchart of the SSMD-POMGGP modeling. The SSMD-POMGGP combines three robust techniques of input subset selection, optimal solution, and multi-expression findings in an integrated framework as presented in this figure.
2.4 Performance criteria
The proposed framework was assessed, using statistical evaluation criteria such as the root mean square error (RMSE), mean absolute error (MAE), Nash-Sutcliffe Efficiency (NSE), coefficient of determination (R2), index of agreement (d), persistence index (PI), confidence index (CI), and relative absolute error (RAE). Also, several pre- and post-processing visualization approaches were used to assess model predictions. Model performance was assessed using estimated and measured Kx values to calculate the Standard Deviation (SD), Centered Root Mean Square Difference (RMSD), and correlation coefficient (R2), as summarized by the Taylor diagram (Taylor 2001).
Taylor (2001) presented a single diagram to abbreviate several accuracy indices (RMSD and R2) to compare models. This is mentioned as the Taylor diagram and works as a complete technique of evaluating the performance of different estimators. It graphically illustrates a series of points on a polar diagram. The azimuth angle shows the correlation coefficient between the estimated and measured values. The radial location from the beginning characterizes the proportion of the normalized standard deviation (SD) of the estimated values from their equivalent measured values.
3 Results and Discussion
3.1 Pareto Optimal MGGEP Model Results
The authors explored the use of subset selection (SS) technique for pre-processing of 503 field data of natural rivers. By combining the SSMD outputs with MGGP, predictive equations were derived. In the MGGP algorithm, different parameters which affect the general applicability of derived equations should be justified. These parameters were adjusted in a trial-and-error manner based on the published values (Gandomi and Alavi 2012a, 2012b). From Pareto analysis the MGGP models for the prediction of Kx had the lowest prediction error. The number of derived models was determined by the population size. The complexity of optimization was controlled by the number of populations. The maximum acceptable number of genes in a multi-gene program and the maximum tree depth directly determined the size of the search field and the number of expressions discovered inside the search field. These parameters were set as a compromise between the running time and the complexity of developed expressions. The parameters used to find the best MGGP models are presented in Table 2.
Figure 3 shows the changes of log values of the best and average model fits in different generations during training. By increasing the generation size, the model fitness value decreases and the model tends to converge. The optimal fitness value was established at the 55th generation (fitness = 2868.494). It is worth noting that in each generation the RMSE was calculated and when the change in RMSE in two successive generations was less than 10%, then the generation stopped and the final fitness value was selected as the optimal fitness.
Although two variables of B/H and U*/U and three genes were used in the present paper, MGGP extract too complicated expressions to increase the estimation accuracy that would be unsuitable for practical applications. The complexity of derived multigene solutions is discussed by (Gandomi and Alavi 2012a, 2012b; DanandehMehr and Kahya 2017; DanandehMehr and Nourani 2017; Wang et al. 2017). In order to meet this challenge, Pareto front analysis of model population was used. The Pareto front plots the best model population versus its estimation performance and the complexity of model. Figure 3 shows the population of the developed expressions as a function of their complexity, which is stated by the number of nodes, on the horizontal axis, as well as their predictive performance (1-R2 parameter) on the vertical axis. The smaller values in Pareto front plot of each population are desirable. The produced equations that perform somewhat accurate with less complexity than the best equation in the population can be clarified in this plot.
When the Pareto-front location in the solution space is recognized, one can simply choose between the accuracy and complexity of solutions and pick up a thrifty model. In this study the Pareto-optimal predictive equation was chosen in such a way that the Pareto-optimal solution had statistical significant R2 value and enveloped the input variables of B/H and U*/U in the best solution. The critical R value for the training stage with 351 degrees of freedom was 0.148 (R2 > 0.022, α = 0.01 level of significance), As illustrated in Fig. 3, the solution with the smallest complexity that positioned below the horizontal line of 0.56 and had included two input variables can be picked as the Pareto optimal solution. As the Pareto optimal solution was derived based on the model performance in the training step, its accuracy should be verified at the testing step and then used as the Pareto-optimal equation of Kx. As shown in Fig. 3, the Pareto front in the population is depicted by blue circle and the best solution in the population is colored in green circle as the Pareto-optimal equation. Finally, the equation with the expressional complexity 116 was chosen as the POMGGP equation for the estimation of Kx in natural rivers.
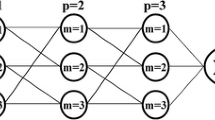
The final multigene model of Kx, derived by the Pareto solution, is shown in Fig. 4. The final Pareto solution had 3 individual genes. Although the structure of the final best model for Kx included some nonlinear components (e.g. divide, power, square), but in the form of its coefficients, it is a linear multigene model with specified weights of its genes. The tree structure of individual genes that comprise the model is shown in Fig. 4 and the mathematical form of each gene includes its weighting coefficient, as presented in Table 3, in which numerical precision reduced for display purposes and x1 = (B/H)0.5, x2 = U*/U. The structural properties relating to the GP tree representation were as genes equal to 3, Nodes were equal to 38, model complexity was 121 with a depth of 4. The last two columns in Table 3 show gene weights and statistical significance of each of the three selected genes of the best Pareto model. The weight of gene 3 was greater than the other genes and bias term. Also, the grade of importance of each gene was assessed by means of p values. As is seen, the involvement of genes, regardless of bias term with p value = 0.1152, to explain changes in Kx was strong, as their p values were small and seemed equal to 0. The statistical importance of the bias term, because of its highest p value, was lower than the genes.
The simplified mathematical expression of the Kx equation was derived according to the bias term and genes coefficients. The predictive PO-MGGEP model in the form of dimensionless parameters can be written as:
In which Kx is dispersion coefficient (m2/s); H is flow depth (m); U*is bed shear velocity (m/s); U is flow velocity (m/s); and B is channel width (m); Kx/HU* is Dimensionless dispersion coefficient, B/H is width to depth ratio and U*/U is ratio of shear velocity to mean velocity.
In Fig. 5, the scatter plot of Kx/HU* and POMGGP predictions along with their 95% confidence ellipse (Johnson and Wichern 2007) are presented. These confidence ellipses show the predictability of observations by the developed equation in their acceptable statistical bounds and covering zones. As is seen, the POMGGP model was capable to estimate the observed points with the acceptable amount of accuracy. There are 16, 6 and 22 points outside of 95% confidence ellipse in train, test subsets and all data respectively.
3.2 Comparison of POMGGP Model and Existing Equations
The longitudinal dispersion coefficient was calculated for the testing phase using eight formulas from the literature. Comparison between the results of the Pareto optimal equation and those of the available empirical equations based on a set of statistical indices over testing data set is illustrated in Table 4. The statistical criteria were computed for all previously published equations, which are presented in Table 5. According to these criteria, the Pareto model performed much better than the previous formulae. The coefficient of determination for the Pareto model was 0.41 in the testing phase, while it was near zero for all other formulae. As an example, the Liu (1977) formula showed a value about 0.1. Also, the NSE values were negative for all formulae except for POMGGP model. Negative NSE values mean the application of model or formula was worse than no knowledge model in which the prediction was simply the average of observed values. Based on RMSE and MAE values which show the error in modeling, the POMGGP model performed better than all other formulae. Just for MAE, the Alizadeh et al. (2017) equation reached a better value than POMGGP model, although the difference was negligible and its R2 was lower. For the RAE criterion, the values oscillated between 0.81 and 5.5. Again POMGGP model and the Alizadeh et al. (2017) formula showed the best performance and the Wang et al. (2017) formula was in the next rank. Based on the RAE values, the Fischer et al. (1979), Liu (1977) and Seo and Cheong (1998) formulae performed as estimation of the mean values. The performance of other formulae was worse than forecasting the mean observed values. According to the PI and d values, the POMGGP model performance is far better than other formulae. Finally, the CI values as a product of NSE and d showed that the POMGGP model was superior to other formulae. Only POMGGP model reached a positive value, while the others gained negative values which showed worse forecasting than the mean observation.
Results showed that nearly all previous equations lost their applicability in the independent data set that had not been used in their calibration, while the POMGGP equation had an acceptable outcome in terms of statistical measures. The given assessment metrics also indicate that all the previously empirical equations were not capable of accurate prediction of Kx and were very inaccurate than was POMGGP equation. The Taylor diagram (Fig. 6) was used to visually compare different performance indices and which plots a series of points on a polar plot for the eight equations and POMGGP. The Taylor diagram demonstrated the normalized Standard Deviation (SD) between estimated and measured Kx values along the circular distances with normalized origins and R2 values used as the azimuth angles. The measured Kx values had a particular demonstration on the Taylor diagram and the implication is that whenever the closer model accuracy indices to the measurements, the superior the model prediction. Figure 6 displays the Taylor diagram based on training, testing and all data sets, which indicates that POMGGP had a major enhancement in the Kx estimation and the efficiencies of previous equations can be graded as: POMGGP, Liu (1977), Alizadehet al. (2017), Sattar and Gharabaghi (2015) and etc., cover all the data sets. The superiority of the POMGGP method over the previous equations is clear from the Taylor diagram and its performance measures.
Finally, for comparison of errors of all formulae, the discrepancy ratio (DR) was utilized. The standardized normal DR is capable of showing the error distribution. The formulation of DR is as follows:
In the case of DR = 0.0, there is a correct estimation while in the case of DR > 0.0 there is an overestimation; or underestimation when DR < 0. The standardized normal DR was calculated and plotted for all formulas (Fig. 7). From Fig. 7, all formulas had wider error distribution than the POMGGP. It should be noted that the figure was plotted between −4 and 5 on horizontal axis, while the error distribution in some formulae reached over 1000. Thus, the POMGGP model outperformed the next ranking Alizadeh et al. (2017) and Liu (1977) models.
4 Conclusions
Estimation of the longitudinal dispersion coefficient (Kx) is important for modeling pollutant and suspended sediment transport in streams and there are significant contributions in the literature on this topic. The emphasis in this paper is on the automatic equation finding of the Kx by POMGGP by a large database than the previous studies. It is notable that by using the SSMD resulted in fairly well distribution of data in train and test stages. The data cannot depict the entire of process if they are too few because uncertainties will result in wrong equation outcomes. Also too much data increase the burden on model optimization and produce a too complex equation but in this study it is eliminated by preprocessing of database with SSMD algorithm. The benefit of applying a robust subset selection in the training of predictive models is twofold and it is possible to use SSMD in training all predictive models.
One advantage of POMGGP is that with using only a few hormonal parameters as input vector, it will result in superior outcomes without the necessity for further data processing or expression findings. Unlike previous equations in Table 4, whose are single expression equations and their exponents are not integers, the new equation shows a linear combination of nonlinear expressions with a concise form, in which all exponents are integers or 3/2. In point of error classification mean absolute error (MAE) of eq. 9 varies with Kx/HU* drastically. For Kx/HU* values ranges from 1 to 100 the MAE of equation are 385.6 and 146.6 in train and test, for Kx/HU* values of 100–1000 the MAE values are 552 and 389.1 in train and test and for Kx/HU* values of 1000–6400 the MAE values are 2850.9 and 1544.4 in train and test respectively. The NSE for the present model is about 0.39 and as provided in Table 5, eq. 9 gives a relatively high value of NSE and proves the superiority of it than all the existing equations in terms of accuracy and NSE significantly. The new derived equation outperformed the previous equations because it use the gene expression based models by using Pareto optimality, employing a large number of datasets to train the model, dividing the datasets to train and test automatically by SSMD to overcome the over fitting problem in training. Despite the improvements in new equation, the performance and accuracy of the equation is not as much reliable as expected. Consequently, more studies are required in order to improve the efficiency of the models against concentration profiles.
The Kx in open channel flow is influenced by the velocity gradients in the width direction and requires information about the velocity distribution over the width of channel for turbulent flow and mixing coefficient in the transverse direction of flow. These two matters are not considered in the present study and require further studies over velocity profile and turbulence characters and should put more effort on kinematic viscosity and velocity measurements in future studies to refine the equations. Future studies are required to focus on evaluating other types of POMGGP optimization and other training algorithms. Also further tracer studies are required to verify the applicability of Kx equation for concentration profile modeling. The tracer studies over Kx are more accurate than the rigid values of Kx because they account for the conditions for the specific reach of the river being investigated, including the geometry, flow, and weather.
References
Alizadeh MJ, Shabani A, Kavianpour MR (2017) Predicting longitudinal dispersion coefficient using ANN with metaheuristic training algorithms. Int J Environ Sci Technol 14:2399
Ahmad Z (2013) Prediction of longitudinal dispersion coefficient using laborary and field data: relationship comparisons. Hydrol Res 44(2)
Carr ML, Rehmann CR (2007) Measuring the dispersion coefficent with acoustic doppler current profilers. J HydraulEng-Asce 133(8):977–982
DanandehMehr AD, Kahya E (2017) A Pareto-optimal moving average multigene genetic programming model for daily streamflow prediction. J Hydrol 549:603–615
DanandehMehr AD, Nourani V (2017) A Pareto-optimal moving average-multigene genetic programming model for rainfall-runoff modelling. Environ Model Softw 92:239–251
Deng Z-Q, Singh VP, Bengtsson L (2001) Longitudinal dispersion coefficient in straight rivers. J Hydraul Eng 127:919–927
Elder JW (1959) The dispersion of a marked fluid in turbulent shear flow. J Fluid Mech 5(04):544–560
Fan FM, Fleischmann AS, Collischonn W, Ames DP, Rigo D (2015) Large-scale analytical water quality model coupled with GIS for simulation of point sourced pollutant discharges. Environ Model Softw 64:58–71
Fischer BH, (1975) Discussion of ‘‘simple method for predicting dispersion in streams,’’ by R.S. McQuivey and T.N. Keefer. J Environ Eng Div 101:453
Fischer HB, List EJ, Koh RCY, Imberger J, Brooks NH (1979) Mixing in Inland and Coastal Waters. Academic, New York
Gandomi AH, Alavi AH (2012a) A new multi-gene genetic programming approach to nonlinear system modeling. Part I: materials and structural engineering problems. Neural Comput & Applic 21(1):171–187
Gandomi AH, Alavi AH (2012b) A new multi-gene genetic programming approach to non-linear system modeling. Part II: geotechnical and earthquake engineering problems. Neural Comput & Applic 21(1):189–201
Hadgu LT, Nyadawa MO, Mwangi1 JK, Kibetu PM, Mehari BB (2014) Application of Water Quality Model QUAL2K to Model the Dispersion of Pollutants in River Ndarugu, Kenya. Computational Water, Energy, and Environmental Engineering 3:162–169
Johnson RA, Wichern DW (2007) Multivariate analysis. Encyclopedia of Statistical Sciences, 8. [Chapter 4 (result 4.7 on page 163)
Kashefipour MS, Falconer RA (2002) Longitudinal dispersion coefficients in natural channels. Water Res 36(6):1596–1608
Kennard RW, Stone LA (1969) Computer aided design of experiments. Technometrics 11(1):137–148
Koza JR (1992) Genetic programming: on the programming of computers by means of natural selection (Vol. 1). MIT press
Li X, Liu H, Yin M (2013) Differential evolution for prediction of longitudinal dispersion coefficients in natural streams. Water ResourManag 27:5245–5260
Liu H (1977) Predicting dispersion coefficient of streams. J Environ Eng Div 103:59–69
May RJ, Maier HR, Dandy GC, Fernando TG (2008) Non-linear variable selection for artificial neural networks using partial mutual information. Environ Model Softw 23(10):1312–1326
Moses SA, Janaki L, Joseph S, Joseph J (2016) Water quality prediction capabilities of WASP model for a tropical lake system. Lake and Reservoirs 20(4):285–299
Najafzadeh M, Tafarojnoruz A (2016) Evaluation of neuro-fuzzy GMDH-based particle swarm optimization to predict longitudinal dispersion coefficient in rivers. Environ Earth Sci 75(2):1–12
Noori R, Deng Z, Kiaghadi A, Kachoosangi FT (2016) How reliable are ANN, ANFIS, and SVM techniques forpredicting longitudinal dispersion coefficient in natural rivers? J Hydraul Eng 142:04015039
Noori R, Karbassi A, Farokhnia A, Dehghani M (2009) Predicting the longitudinal dispersion coefficient using support vector machine and adaptive neuro-fuzzy inference system techniques. Environ Eng Sci 26(10):1503–1510
Parveen N, Singh SK (2016) Application of Qual2e Model for River Water Quality Modelling. International Journal of Advance Research and Innovation 4(2):429–432
Rajeev RS, Dutta S (2009) Prediction of longitudinal dispersion coefficients in natural rivers using genetic algorithm. Hydrol Res 40(6):544–552
Riahi-Madvar H, Ayyoubzadeh SA, Khadangi E, Ebadzadeh MM (2009) An expert system for predicting longitudinal dispersion coefficient in natural streams by using ANFIS. Expert Syst Appl 36(4):8589–8596
Sattar AM, Gharabaghi B (2015) Gene expression models for prediction of longitudinal dispersion coefficient in streams. J Hydrol 524:587–596
Searson DP (2015) GPTIPS 2: an open-source software platform for symbolic data mining. In: Handbook of genetic programming applications (pp. 551–573). Springer International Publishing
Seo IW, Cheong TS (1998) Predicting longitudinal dispersion coefficient in natural streams. J Hydraul Eng 124:25
Tayfour G, Singh VP (2005) Predicting longitudinal dispersion coefficient in natural streams by artificial neural network. J Hydraul Eng 131(11):991–1000
Taylor KE (2001) Summarizing multiple aspects of model performance in a single diagram. J Geophys Res-Atmos 106(D7):7183–7192
Wang YF, Huai WX, Wang WJ (2017) Physically sound formula for longitudinal dispersion coefficients of natural rivers. J Hydrol 544:511–523
Wang Y, Huai W (2016) Estimating the longitudinal dispersion coefficient in straight natural rivers. J Hydraul Eng 142(11):04016048
Yapo PO, Gupta HV, Sorooshian S (1998) Multi-objective global optimization for hydrologic models. J Hydrol 204(1–4):83–97
Zhang T, Georgiopoulos M, Anagnostopoulos GC (2017) Pareto-optimal model selection via SPRINT-race. IEEE Transactions on Cybernetics
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Riahi-Madvar, H., Dehghani, M., Seifi, A. et al. Pareto Optimal Multigene Genetic Programming for Prediction of Longitudinal Dispersion Coefficient. Water Resour Manage 33, 905–921 (2019). https://doi.org/10.1007/s11269-018-2139-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11269-018-2139-6