Abstract
Outbreaks of pseudorabies (PRs) have occurred in Yunnan, China, which caused significant economic loss. To determine the prevalence and origin of PR in Yunnan, especially among vaccinated pigs, overall 791 samples of blood, tissue, semen, and sera were analyzed by serological methods, PCR, and sequence analysis of gD gene. Detection with viral gI antibody or PCR showed that the yearly positive rates of PR virus (PRV) in Yunnan from 2010 to 2014 were 48.15, 21.26, 2.17, 5.22, and 0.35%, respectively, with an average of 15.43%. In general, the incidence declined through the period of 2010–2014 probably due to the application of PRV eradication strategies. A phylogenetic tree was constructed based on the complete sequence of gD gene, with all strains clustered into two independent clades, i.e., Asian and European–American clades. The virus isolates from Henan, Tianjin, Heilongjiang, Sichuan, Shandong, Fujian, Xinjiang, Hubei, Guangdong, and Yunnan fell into Asian group, which harbored South Korea isolate. Four Yunnan virus isolates together with South Korean Namyangju fell into in the European–American clade. It showed that PR was pandemic as there was not a clear clue about the geographical origin of the PRV isolates in China since 2010.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pseudorabies (PR) disease was first found from an American cattle population in 1813 and listed as one of the top three epidemic diseases in pig industry. PR is caused by Pseudorabies virus (PRV), which belongs to suid herpesvirus type 1 (SuHV 1), a member of Alphaherpesvirinae subfamily within the Herpesviridae family. Pig is the natural host, reservoir, and source of PRV infection. PRV infection results in some obvious clinical manifestations such as fever, local intense pruritus, and encephalomyelitis in farm animals including cattle, sheep, pig, dog, some avian species, and cat, with exception of higher primates [1,2,3,4,5]. Hunting dogs and wild boars [6, 7] are also important potential carriers, which aggravated the PRV control. The PRV infections were found around the world and widely prevailed in China in the past decades, causing enormous economic loss in pig industry [1, 8,9,10,11]. The symptoms differed greatly from swine ages, such as 100% mortality rate in piglets, stillbirths, mummification and/or abortions in pregnant sows, and low growth rates in asymptomatic chronic infections in adult pigs [4,5,6, 8, 10, 12,13,14]. Although the infections has been controlled by an attenuated vaccines since 1970s, new clinical features had developed in China, in terms of transmission diversity, infected area and rate, severe clinical signs, and coinfection. From a sudden break-out of pseudorabies, Gu et al. [15] collected 38 tissue samples from pigs with clinical signs of pseudorabies on 13 farms from 4 provinces in southern China in 2012–2013. After PCR detection, 29 samples (76%) showed wild-type PRV infection, indicating that a novel, highly virulent PRV strain with antigenic variance had spread widely in southern China. Therefore, new PR pathogenicity has been continuously revealed since late 2011 in China [5, 8, 10, 14,15,16,17,18,19).
There are a number of reports about the genetic information and the origin of the PRV genome [2, 10, 20,21,22,23,24]. The PRV genome is a double-stranded linear DNA molecule with 14.36 kb in length and contains 67–72 open reading frames (ORFs), which show significant variations when compared with previous isolates in most viral coat proteins including glycoproteins E, B, C, D, and I, which are critical factors that mediate viral attachment (gC and gD), viral spread (gE), and induction of host immunity (gG and gI). The glycoproteins play a major role for viral interaction and are of importance for pathogenesis [3, 4, 25]. Phylogenetic analysis based on the PRV genes was documented to reveal the origin and relationships between the PRV isolates [6, 8, 10, 12, 14]. In this study, we investigated the prevalence of PRV in Yunnan of China during the time period of 2010–2014 and performed a phylogenetic analysis of our isolates and some reference strains based on the PRV gD gene sequences attempting to investigate the genetic and variant features of PRV in Yunnan province.
Materials and methods
Sample collection and serological detection
Seven hundred ninety-one samples were collected including blood, semen, sera, and tissues from sows and boars in pig farms from 15 prefectures in Yunnan province and two adjoining provinces, China, during 2010–2014 Serological detection with Pseudorabies Virus gI Antibody (PRV gI Ab) Test Kit were performed to verify if the suspected sera samples were positive for PR virus according to the manufacturer’s instructions (IDEXX Laboratories, Inc., Beijing Yuanxiang Biological Tech. Co. Ltd.).
Isolation of viral DNA and PCR amplification
The blood, tissue, and semen samples were subjected to DNA extraction according to the instructions of DNA extraction kit (Sangon Biological Engineering Co. Ltd.). The complete gD gene fragment of PRV was amplified by PCR from the extracted DNA using a pair of specific primers (Sangon Biological Engineering Co. Ltd.), which were designed according to the reference sequences in GenBank database (Accession Numbers AJ271966, BK001744 and JF797218). The primer sequences were 5′-CCCCAGGTTCCCATACACTC-3′ (forward, genomic position 118840–118859 nt) and 5′-TCATCATCGACGCCGGTACT-3′ (reverse, genomic position 120083–120102 nt), with the amplification resulting in a 1263 bp long product. PCR was performed in a 25-μL volume containing 2.5 U DNA polymerase, 2.5 μL buffer II, 10 μL GC enhancer, 10 pmol of each primer, 2 mM dNTP for each, 8.0 μL ddH2O, and 1 μL extracted DNA containing 20–50 ng/μL. The PCR conditions consisted of an initial cycle at 94 °C for 5 min, 35 amplification cycles (94 °C for 1 min, 60 °C for 1 min, and 72 °C for 1 min) and a final extension step at 72 °C for 7 min.
Cloning of gD gene and sequencing
After PCR, the products were subjected to 1.5% agarose gel electrophoresis for detecting the presence of gD gene fragment. Then, the PCR products were electrophoresed on 2.5% agarose gel for purification according to the instructions (Bio Teke Corparaion, Beijing, China). With random orientation, the PCR products were ligated into pMD18-T vectors (Sangon Biological Engineering Co. Ltd., Shanghai, China) at the EcoRV site, followed by transformation into Escherichia coli DH5a cells (Takara Biotch Co. Ltd.). The plasmids were extracted, with the inserts sequenced by Sangon Biological Engineering Co. Ltd.
Phylogenetic analysis of gD sequence
The resulting raw sequences were aligned and checked using DNAStar 6.0 (DNAStar Inc., Madison, WI). The complete gD gene (1203–1209 bp) were identified by comparison with the reference sequences of Suid herpesvirus 1 from GenBank database (Accession Numbers BK001744, JF797218 and AJ271966). Then, a total of 29 gD gene sequences from GenBank were used in the analysis (Table 1). Variant sites and haplotypes were determined using MEGA software, Version 4.0 [26] and DnaSP software, Version 3.00 [27]. The nucleotide homologies were analyzed using DNAman (version 6.0, Lynnon Co.). Phylogenetic tree was constructed using MEGA software under the Kimura 2-parameter substitution model by neighbor-joining method with 1000 bootstrap replicates.
Results
Positive rates of PRV from 2010 to 2014 in Yunnan
The prevalence of PRV in Yunnan province, China, is listed in Table 2. Ninety-nine positive samples were identified from 791 clinical samples during the period of 2010–2014, with an average rate of 15.43%. Declining tendency of PRV positive rates was observed through the 5 years, with the lowest rate in 2014 (0.35%), except for a slight increase in 2013 (5.22%).
PCR amplification, purification, and identification of PRV gD gene
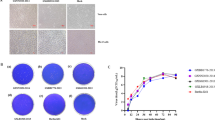
After PCR amplification, the products were electrophoresed on 1.5% agarose gel, and the fragment of gD gene of PRV was about ~1300 bp, which was in accordance with the expected 1263 bp in length (Fig. 1a). The gD gene fragments of PRV were electrophoresed on 2.5% agarose gel for purification according to the manuals displaying ~1300 bp in size (Fig. 1b). The purified PCR products were ligated into pMD18-T vectors at the EcoRV site for transformation into E. coli DH5a cells. After analyzing the sequences of the pMD18-T vector and the amplicons from gD gene, HincII was found able to cut both pMD18-T vector and the amplicons from gD gene at single site, respectively, and therefore selected for digestion, which resulted in two digestion bands. The gel images of the products after digestion are shown in Fig. 1c, of which the fragments in length were as expected. Depending on the ligation orientation of the gD amplicons into pMD18-T vector, two possibilities could be seen after digestion of HincII: 3408 bp band and 547 bp band for samples 1–5, 3231 bp band and 724 bp band for sample 6, which are indicated in Fig. 1c. The plasmids were extracted, and the inserts were sequenced by Sangon Biological Engineering Co, Ltd.
PCR amplification, purification, and identification of PRV gD gene. a The PCR products of gD gene of PRV. Lane 1 Negative control, Lane 2–10 the PCR products of gD gene from nine representative suspected PRV samples. M DNA Marker DL2000. b The electrophoresis image of products of gD gene on 2% gel after purification from a. Lane 1–6 the six clone samples; M DNA Marker DL2000. c Identification of recombinant plasmids digested by HincII. According to the sequences of pMD18T vector and the amplicons of gD gene, the cleavage sequence of HincII and the ligation orientation, the length of fragments from digestion is expected to be 3408 and 547 bp (lanes 1–5), or 3231 and 724 bp (lane 6). M: DNA Marker DL1000
Sequencing, variants, and phylogeny of gD gene of PRV
Six positive clones were sequenced, aligned, and compared with the complete gD gene counterparts of PRV available in GenBank database. The sequences of gD gene were identified in range of 1203–1209 bp in length, encoding 400–403 aa residues, and the GC content was 74.69%. Sixty-seven variant sites were found in the gD gene sequences, including 53 predominant transitions, 6 C/G (at the position 379, 468, 627, 828, 918, and 1023 nt) and 8 C/A (744, 808, 814, 820, 826, 1019, 1030, and 1129 nt) transversions, leading to 45 amino acid variations. The sequence variation of gD genes incurred as well in the gaps covering 829–834 nt which contained (GAACCC) insertions, coding (PR)6–7 repeats residue (Figs. 2, 3).
Nucleotide variants of PRV gD gene. The names of the six gD genes cloned in this study are highlighted in green squares. Sixty-seven variant sites were found in the gD gene sequences, including 53 predominant transitions, 6 C/G (at the position 379, 468, 627, 828, 918, and 1023 nt), and 8 C/A (744, 808, 814, 820, 826, 1019, 1030, and 1129 nt) transversions. The sequence variance of gD genes incurred as well in the gaps covering 829–834 nt which contained (GAACCC) insertions (shown in the red square). Two independent clades, i.e., Asian clade and European–American clade can be seen based on the nucleotide acid sequence comparison. The nucleotide sequences shown here are not consecutive, with only sequence variants shown. The positions of the consensus nucleotide acids are shown above the sequence (Color figure online)
Amino acid variants of PRV gD gene product. The names of the six gD genes cloned in this study are highlighted in green squares. The sixty-seven variant sites found in the gD gene sequences led to 45 amino acid variations (PR)6–7 repeat insertion (shown in the blue square). Two independent clades, i.e., Asian clade and European–American clade can be seen based on the amino acid sequence comparison. The nucleotide sequences shown here are not consecutive, with only sequence variants shown. The positions of the consensus amino acids are shown above the sequence, in column (Color figure online)
The phylogenetic tree was constructed for the 29 complete gD genes together with some available sequences in GenBank using neighbor-joining (N-J) method. Clustering analysis revealed two independent clades, i.e., Asian clade and European–American clade (Fig. 4). The virus isolates from Henan, Tianjin, Heilongjiang, Sichuan, Shandong, Fujian, Xinjiang, Hubei, Guangdong, and Yunnan fell into Asian group, varying from 1203 to 1209 bp in length except for Ea strain (AF086702, 1215 bp), which were relatively closer to previously isolated strains, suggesting that there was a continuous evolutionary process. ZB10142374, SD1404, qihe547, HN1201, TJ, and HLJ8 strains from China shared 100% nucleotide identity with each other, as an Asian classical PRV in swine epidemiology, showing that PR was pandemic, and there was no clear sequence variation from the geographical zones between PRV virus isolates from China since late 2011. The typical isolates of European PRV, such as Kaplan, Batha, and Becker, together with the South Korean Namyangju, were clustered into the same group characterized by 1203 bp in length, including 4 Yunnan virus isolates which formed a separated subclade (Fig. 4). Herein, the virus isolates shared one haplotype comprising of DUL34Pass (JQ809330) and Kaplan (KJ717942, JF797218, and AJ271966). The nucleotide sequences of gD gene were 97.8–100.0% homology between the PRV isolates. It was inferred that PRV isolates in Yunnan province after 2010 were extensively spread with the pig transmission and trade, and escaped from the protection from Bartha-K61 vaccine.
Discussion
Pseudorabies (PR) is caused by PRV infection and of economic importance in pig industry because this viral disease can lead to 100% mortality rate in piglets, stillbirths, mummification, and/or abortions in pregnant sows, as well as lower growth without obvious symptom in adult pigs as a natural reservoir and host. PR has been effectively controlled since 1970s using Bartha-K61 vaccine in China [1, 5, 9, 12, 14]. However, PR outbreak still occurs in China since late 2011, indicating that the vaccination cannot fully protect the pigs from PRV infection. Later, many cases with suspected animals, inoculated with PRV Bartha-K61, were collected to determine the presence and variants of PRV [5, 8, 10, 14, 15, 28]. In this study, we collected 791 suspected samples from pig farms mainly in Yunnan, China, to determine the positive rate by universal PRV gI Ab test kit and PCR technique. The average positive rate of PRV was 15.43% and showed a decline during the period of 2010–2014 (Table 2), except a slight increase in 2013 (5.22%). Some measures have been available for the control and eradication of the PRV, e.g., the application of inactivated vaccine before mating or artificial insemination and 4 weeks before delivery again for sows; intraocular-nasal vaccination for the piglets at the birth day; and vaccination by injection for the growing-finishing pig at the age of 70-day and breeding boars every 6 month.
Based on the gD gene analysis, the six PRV strains from Yunnan of China were separated into two independent branches, with only two strains into Asian clade and the other four into European–American clade which consists of a separated subclade. It inferred that the PR epidemiology in Yunnan was more complicated than other regions in China [5, 8, 10, 14, 28]. The recent isolates from Yunnan intermingled and kept a continuous evolutionary history [10, 17, 18, 28], sharing one haplotype in Asian clade, as well as having an independent subclade in European–American clade. The nucleotide acid similarity of the PRV isolates was 97.6–100.0%. PR could be transmitted by commercial animals including swine and boar. The outbreak of PR in Yunnan since late 2010 might originate from PRV variants, animal introduction, or trade activities [15, 28]. It is of importance that PRV investigation is compulsory to monitor the presence and mutations of new PRV variants and epidemiology due to commercial transmission. More work on the prevalence and pathogenicity of PRV worldwide is needed for the control and eradication of PR.
References
W. Hong, H. Chen, L. Fang, R. Zhou, Q. He et al., Clone and nucleotide sequence analysis of gD gene of pseudorabies virus Ea strain. Acta Vet. Zootech. Sin. 32(3), 235–243 (2001)
B.G. Klupp, C.J. Hengartner, T.C. Mettenleiter, L.W. Enquist, Complete, annotated sequence of the pseudorabies virus genome, J. Virol. 78(1), 424–440 (2004). Erratum in: J. Virol. 78(4), 2166
T.C. Mettenleiter, Aujeszky’s disease (pseudorabies) virus: the virus and molecular pathogenesis—state of the art, June 1999. Vet. Res. 31(1), 99–115 (2000)
L.E. Pomeranz, A.E. Reynolds, C.J. Hengartner, Molecular biology of pseudorabies virus: impact on neurovirology and veterinary medicine. Microbiol. Mol. Biol. Rev. 69(3), 462–500 (2005)
Q.Y. Yang, Z. Sun, F.F. Tan, L.H. Guo, Y.Z. Wang et al., Pathogenicity of a currently circulating Chinese variant pseudorabies virus in pigs. World J. Virol. 5(1), 23–30 (2016). doi:10.5501/wjv.v5.i1.23
A. Moreno, E. Sozzi, G. Grilli, L.R. Gibelli, D. Gelmetti et al., Detection and molecular analysis of pseudorabies virus strains isolated from dogs and a wild boar in Italy. Vet. Microbiol. 177(3–4), 359–365 (2015). doi:10.1016/j.vetmic.2015.04.001
S. Verpoest, A.B. Cay, N. de Regge, Molecular characterization of Belgian pseudorabies virus isolates from domestic swine and wild boar. Vet. Microbiol. 172, 72–77 (2014)
T.Q. An, J.M. Peng, Z.J. Tian, H.Y. Zhao, N. Li et al., Pseudorabies virus variant in Bartha-K61-vaccinated pigs, China, 2012. Emerg. Infect. Dis. 19(11), 1749–1755 (2013). doi:10.3201/eid1911.130177
W. Fan, J. Hu, J. Wu, X. Yuan, Z. Zhang et al., Isolation and identification of LA strain of pseudorabies virus. Chin. J. Vet. Sci. 23(6), 538–540 (2003). doi:10.16303/j.cnki.1005-4545.2003.06.008
Y. Luo, N. Li, X. Cong, C.H. Wang, M. Du et al., Pathogenicity and genomic characterization of a pseudorabies virus variant isolated from Bartha-K61-vaccinated swine population in China. Vet. Microbiol. 174(1–2), 107–115 (2014). doi:10.1016/j.vetmic.2014.09.003
X. Zeng, Y. Wu, J. Chen, J. Yao, B. Wu, Isolation and identification of porcine pseudorabies virus FZ strain and sequence analysis of the gD gene of the strain. J. Fujian Agric. For. Univ (Nat. Sci. Ed.) 40(5), 501–510 (2011). (in Chinese)
L. Chen, T. Zhao, L. Zhang, Y. Li, Clone and sequencing of gD and gE gene of PRV XIN-W isolate of pseudorabies virus, Prog. Vet. Med. 26(1), 66–70, 88 (2005) (in Chinese)
W.A. Mulder, J.M. Pol, E. Gruys, L. Jacobs, M.C. De Jong et al., Pseudorabies virus infections in pigs. Role of viral proteins in virulence, pathogenesis and transmission. Vet. Res. 28, 1–17 (1997)
Y. Wang, S. Qiao, X. Li, W. Xie, J. Guo et al., Molecular epidemiology of outbreak-associated pseudorabies virus (PRV) strains in central China. Virus Genes 50(3), 401–409 (2015). doi:10.1007/s11262-015-1190-0
Z. Gu, C. Hou, H. Sun, W. Yang, J. Dong et al., Emergence of highly virulent pseudorabies virus in southern China. Can. J. Vet. Res. 79, 221–228 (2015)
J.M. Peng, T.Q. An, H.Y. Zhao, Y.M. Liu, J.Z. Chen et al., Identification and antigenic variation of new epidemiology of pseudorabies virus from swine. Chin. J. Prev. Vet. Med. 35(1), 1–4 (2013). (in Chinese)
C. Ye, J.C. Guo, J.C. Gao, T.Y. Wang, K. Zhao et al., Genomic analyses reveal that partial sequence of an earlier pseudorabies virus in China is originated from a Bartha-vaccine-like strain. Virology 491, 56–63 (2016). doi:10.1016/j.virol.2016.01.016
C. Ye, Q.Z. Zhang, Z.J. Tian, H. Zheng, K. Zhao et al., Genomic characterization of emergent pseudorabies virus in China reveals marked sequence divergence: evidence for the existence of two major genotypes. Virology 483, 32–43 (2015). doi:10.1016/j.virol.2015.04.013
X. Zhang, J. Liu, C. Wei, A. Dai, X. Li et al., Cloning and sequences analysis of gB and gD gene of porcine pseudorabies virus Fujian strain. China Anim. Husb. Vet. Med. 41(5), 62–65 (2014). (in Chinese)
J.F. Kreuze, A. Perez, M. Untiveros, D. Quispe, S. Fuentes et al., Complete viral genome sequence and discovery of novel viruses by deep sequencing of small RNAs: a generic method for diagnosis, discovery and sequencing of viruses. Virology 388, 1–7 (2009)
P. Oláh, D. Tombácz, N. Póka, Z. Csabai, I. Prazsák et al., Characterization of pseudorabies virus transcriptome by Illumina sequencing. BMC Microbiol. 15, 130 (2015). doi:10.1186/s12866-015-0470-0
E. Sozzi, A. Moreno, D. Lelli, S. Cinotti, G.L. Alborali et al., Genomic characterization of pseudorabies virus strains isolated in Italy. Transbound. Emerg. Dis. 61(4), 334–340 (2014). doi:10.1111/tbed.12038
M.L. Szpara, Y.R. Tafuri, L. Parsons, S.R. Shamim, K.J. Verstrepen et al., A wide extent of inter-strain diversity in virulent and vaccine strains of Alphaherpesviruses. PLoS Pathog. 7(10), e1002282 (2011). doi:10.1371/journal.ppat.1002282
S. Xiang, Z. Zhou, X. Hu, Y. Li, C. Zhang et al., Complete genome sequence of a variant pseudorabies virus strain isolated in central China. Genome Announc. 4(2), 1–2 (2016). doi:10.1128/genomeA.00149-16
T.C. Mettenleiter, Immunobiology of pseudorabies (Aujeszky’s disease). Vet. Immunol. Immunopathol. 54, 221–229 (1996)
K. Tamura, J. Dudley, M. Nei, S. Kumar, MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599 (2007)
F. Rozas, R. Rozas, DnaSP version 3.0: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15, 174–175 (1999)
X. Yu, Z. Zhou, D. Hu, Q. Zhang, T. Han et al., Pathogenic pseudorabies virus, China, 2012. Emerg. Infect. Dis. 20(1), 102–104 (2014). doi:10.3201/eid2001.130531
Acknowledgements
The work was jointly supported by Applied Basic Research Key Project of Yunnan (Grant No. 2016FA018) and Key Laboratory of Veterinary Public Health of Higher Education of Yunnan Province.
Author contributions
G.F.Y. and X.H.S. conceived and designed the study. W.B.B. and X.Q.Z. collected the samples. C.L.S. and L.G. carried out the experiments. C.L.S., G.F.Y. and X.H.S. drafted the manuscript. All authors read and approved the final manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors have declared no competing interests.
Human and animal consent
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Edited by Zhen F. Fu.
Rights and permissions
About this article
Cite this article
Song, C., Gao, L., Bai, W. et al. Molecular epidemiology of pseudorabies virus in Yunnan and the sequence analysis of its gD gene. Virus Genes 53, 392–399 (2017). https://doi.org/10.1007/s11262-017-1429-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-017-1429-z