Abstract
Porcine circovirus type 2 (PCV2) is the primary causative agent of porcine circovirus-associated diseases in swine and is also described as the modulator of host immunity that exacerbates the clinical outcome of many bacterial and viral infections. To date, it has caused increasingly larger losses in the pig industry worldwide. The genomic DNA of PCV2 is predicted to contain 11 open reading frames (ORFs) and at least seven potential ORFs-encoding proteins larger than 5 kDa. Currently, however, only five virally encoded proteins (Rep, Rep′, Cap, ORF3, and ORF4 protein) have been identified in PCV2 replication. In the present review, we strive to discuss the current understanding of the genomic DNA of PCV2 with the purpose of providing insight into the scientific basis of the pathogenesis of PCV2 and the prevention of its infection.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Porcine circovirus type 2 (PCV2) is classified into the genus Circovirus of the family Circoviridae, which encompasses a group of other animal circoviruses including goose circovirus, canary circovirus, psittacine beak and feather disease virus, chicken anemia virus, and pigeon or columbid circovirus [1–5]. PCV2 was originally identified as the etiological agent of naturally occurring post-weaning multisystemic wasting syndrome (PMWS) in swine [6–9]. This condition has been experimentally reproduced in gnotobiotic, specific pathogen-free (SPF), conventional pigs [10–12] and BALB/c mice [13–15]. Many other conditions like porcine respiratory disease complex, porcine dermatitis and nephropathy syndrome [16, 17], congenital tremors, fetal myocarditis, and reproductive failure are also described to be associated with PCV2 infection [18–21], which are collectively known as porcine circovirus-associated diseases (PCVD/PCVAD) [22].
The PCV2 virus was first identified within high-health herds in western Canada in 1991 [23] and primarily affects 5–18-week-old weanling piglets. Morbidity rates may vary (15–20 %) and occasionally reach 60–80 % in complicated cases [24, 25]. Pigs affected by PMWS show weight loss or unthriftiness, enlarged lymph nodes, dyspnea, tachypnea, anemia, diarrhea, and jaundice. PCV2 infection leads to lymphoid depletion, histiocytic infiltration, and ultimately immunosuppression. PCV2 infection is also found as a coincident co-infection with other pathogens like Haemophilus parasuis, Mycoplasma hyopneumoniae, porcine parvovirus, and porcine reproductive and respiratory syndrome virus [8, 25–28]. PCV2 infection is currently considered to be endemic in most of the swine-producing countries of the world [16].
PCV2 virion is icosahedral, non-enveloped, and 17 nm in diameter [29]. Phylogenetic analysis indicated that PCV2 strains could be divided into five genotypes (PCV2a, PCV2b, PCV2c, PCV2d, and PCV2e) based on performing pairwise sequence comparison analysis of PCV2 isolates [30–34]. In another related study, PCV2 genotype was also defined as two subgroups with eight clusters (1A–1C and 2A–2E) [35].
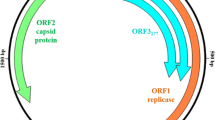
PCV2 genome is a single-stranded, closed circular DNA containing 1,766–1,768 nucleotides (nt). The genome size of PCV2 is reduced to the absolute necessities to perform copying and packaging of the viral genome [36]. The genome encodes proteins by both the encapsidated viral DNA and the complementary DNA of the replicative intermediate (RI) synthesized in the host [37] and is computationally predicted to possess 11 overlapping open reading frames (ORFs) [38]. The 1, 5, 7, and 10 ORFs are located on the viral plus-strand and transcribe clockwise, while the 2, 3, 4, 6, 8, 9, and 11 ORFs are encoded by the complementary strand and transcribe counterclockwise [38]. These ORFs perform overlapping genetic structure that can take full advantage of the limited viral genetic material, which may be the result of natural selection during the process of biological evolution. ORF1 and ORF2 genes are the two major open reading frames (ORFs) and orientated in opposite directions (Fig. 1). This arrangement generates an ambisense genome organization and creates two intergenic regions (IR). The shorter intergenic region is located between the 3′-ends of the ORF1 and ORF2 gene, while the larger one locates between their 5′-ends and contains the origin of viral genome replication. PCV2 replication originates from a putative stem-loop (SL) structure and replicates via rolling-circle replication (RCR) (Fig. 1). Presence of the similarities between genomic organization and replication strategy indicates that PCV2 is closely related to plant geminiviruses and nanoviruses.
Genomic schematic of PCV2 strain pmws PCV (GenBank Accession No. AF027217). Coding sequences of the four ORFs are annotated with the nucleotide coordinates of each gene, and their orientations of translation are indicated with triangle symbols. A stem-loop (SL) structure located in the intergenic region (IR) between ORF1 and ORF2 is illustrated. The Rep is translated from the full-length ORF1, whereas the Rep′, Rep3a, Rep3b and Rep3c are produced via alternative splicing of the Rep transcript (as depicted by dotted lines). NS515, NS672, and NS0 are transcribed from three different promoters inside ORF1 downstream of the Rep promoter
Although there have been extensive studies on PCV2 and associated diseases over the last decade [36], to date, only eleven PCV2-specific RNA transcripts (designated CR, Rep, Rep′, Rep3a, Rep3b, Rep3c, NS515, NS672, NS0, ORF3, and ORF4) have been detected. The viral RNAs of the ORF2, ORF3, and ORF4 genes are transcribed from the cDNA strand, while the several Rep-associated and NS-associated RNAs are transcribed in the opposite orientation [39]. However, only five viral proteins that are serially encoded by ORF1, ORF2, ORF3, and ORF4 have been characterized in detail. More importantly, the underlying mechanisms of PCV2 pathogenesis and immune interactions still remain poorly understood. In this review, we primarily summarize the recent advances in the structure and function of the ORFs of this virus to identify knowledge gaps and future research directions.
Recent advances in ORFs genes
Research on the ORF1 gene
The ORF1 gene, called the rep gene and oriented in the viral plus-strand, is the largest ORF of PCV2 (945 bp, nt 51–995). A functional viral interferon-stimulated response element (ISRE)-like sequence (5′-CTGAAAACGAAAGA-3′, nt 1,737–1,751) has been identified in the rep gene promoter (Prep) of PCV2 and plays an important role in viral transcription start individually [40]. Transcript mapping revealed that the rep gene of PCV2 encodes eight products. They include two major products (designated Rep and Rep′) and six minor products (designated Rep3a, Rep3b, Rep3c, NS515, NS672, and NS0). Members of the Rep-associated RNA cluster all share common 5′ and 3′ nucleotide sequences, and they also share 200 common 3′ nucleotide sequences with the NS-associated RNAs [39]. Although Rep is the primary transcript that gives rise to the other Rep-associated RNAs by alternate splicing, the three NS-associated RNAs are transcribed from three different promoters present inside ORF1, independent from the Rep promoter, which is composed of two single mini-promoters [36, 40]. It has been suggested that the differential splicing of Rep3c and NS0 RNAs and the significant differences in relative expression levels of Rep 3c and NS0 RNAs may contribute to the pathogenesis of PCV2 [41]. However, only Rep and Rep′ transcripts have been proven to be capable of translating into functional proteins, and the simultaneous expression of the full-length Rep protein (314 aa, 35.8 kDa) and the spliced frame-shifted version Rep′ (178 aa) is essential for initiating PCV2 replication [36, 42, 43]. Whether the proteins encoded by several other RNA transcripts are expressed or not and whether the expressed proteins are essential for viral replication await elucidation.
The proteins encoded by ORF1 are of similar size in PCV1 and PCV2, among which the PCV1 and PCV2 Rep proteins have the most highly conserved sequence (approximate 85 % homology within aa) [43]. This is why cross-reaction is present in serology testing. In addition, software projections show that the ORF1-encoded protein in PCV2 has three potential glycosylation sites, at aa 23–25 (NPS), aa 256–258 (NQT), and aa 286–288 (NAT), while the PCV1 Rep protein has only one glycosylation site at aa 20–22 (NPS) [43].
It is perhaps surprising that the Rep and Rep′ proteins encoded by PCV1 or PCV2 all contain three aa motifs, which are conserved in enzymes involved in the initiation of DNA replication in the rolling-circle mechanism [44]. Nevertheless, a deoxynucleoside triphosphate-binding domain has been identified in the Rep protein, but not in the Rep’ protein [45]. The Rep protein is currently considered a significant immunogenic protein of PCV2 that plays an important role in cell-mediated immunity to constrain PCV2 replication and prevent the progression of PCV2 infection toward PMWS [46].
Localization studies revealed that PCV2 Rep protein is localized in the nucleus in infected PK15A cells [47]. With a bacterial two-hybrid approach, Timmusk et al. [48] have identified the intermediate filament protein syncoilin and the transcriptional regulator protein c-Myc as cellular proteins interacting with Rep of PCV2. Subsequently, in 2009, Finsterbusch et al. [49] identified three additional proteins homologous to the zinc finger protein 265 (ZNF265), the thymine DNA glycosylase (TDG), and the angiogenic factor VG5Q interacting with the Rep protein of PCV2 using a yeast two-hybrid assay. Interestingly, while VG5Q and TDG interacted also with the Rep′ protein, ZNF265 bound only the full-length Rep protein. Moreover, all three proteins (ZNF265, TDG and VG5Q) bind to the Rep proteins of both PCV, while c-myc and syncoilin have been described to bind only to Rep of PCV2. Although these interactions between Rep and its cellular partners have been confirmed by GST pull-down assay or co-immunoprecipitation, the biological significance of these viral and cellular protein interactions still remains largely unknown.
Research on the ORF2 gene
On the counterclockwise strand, there is the ORF2 gene, called cap, which encodes the major immunogenic capsid protein (Cap protein) that is 27.8 kDa [44, 50]. The ORF2 gene is composed of 702 bp (nt 1,735–1,034) that code for 234 aa. The protein encoded by ORF2 of PCV2 was identified as a major viral structural protein that independently forms viral capsid-like structures when expressed in insect cells from recombinant baculovirus [50]. Because the ORF2 gene of PCV2 is shorter and has less labor-extensive sequencing than the whole genome, it is always available for epidemiological and phylogenetic studies as a target fragment.
Molecular and epidemiological analyses have shown that ORF2 is highly variable compared with ORF1 and ORF3 and that the polymorphisms in the capsid region of PCV2 are related to the replication cycle of the virus [51–53]. PCV2 interacts with virus attachment receptors including heparin sulfate and chondroitin sulfate B glycosaminoglycans via its capsid protein to complete its cellular entry [51]. Alternatively, the assembly kinetics or structural stability of PCV2 is also affected by the Cap protein [52]. Herein, PCV2 is quite appropriate for carrying certain allelic genes within ORF2 relying on its polymorphisms. Research on the virulence determinant factor(s) of PCV2 has revealed that aa substitutions in the PCV2 capsid protein [A proline at position 110 of the capsid protein changed to an alanine (P110A); an arginine at position 191 of the capsid protein changed to a serine (R191S)] have been demonstrated to enhance the growth ability of PCV2 in vitro and attenuate the virus in vivo after 120 passages in cultured PK-15 cells [53].
Studies have confirmed that the PCV2 Cap protein possesses at least eight different antigenic epitopes. They are three specific antigenic sites (aa 69–83, aa 117–131, and aa 169–183) [54] and five different spatial overlapping antigenic epitopes within aa 47–63, aa 165–200, and the last four aa at the C terminus [55]. Further studies found that the Cap protein contains a highly conserved basic aa sequence (such as arginine) at the former 41 aa of the N terminus resembling that of the major structural protein of chicken anemia virus [9]. This area has a functional nuclear localization signal (NLS) in the Cap protein and is an essential motif affecting PCV2 nuclear distribution. Moreover, site-directed mutagenesis revealed that the aa residues 12–18 and 34–41 of the N terminus play a pivotal role in the nuclear localization of ORF2 [56, 57]. More exciting is that the crystal structure of a monomeric Cap protein subunit and its orientation within a PCV2-like particle has also been elucidated recently. In this model, 60 copies of the Cap protein subunit form an icosahedron with T = 1 symmetry [58, 59]. Consequently, the current research in allusion to PCV2 diagnosis and genetic engineering vaccine is focused on this protein.
The cap proteins of PCV1 and PCV2 differ only marginally in length, but they present a marked degree in amino acid sequence diversion comparing with the Rep proteins of both PCV. Thus, they were candidates for confirming as a molecular marker indicative for the differential pathogenicity of PCV1 and PCV2. PCV2 Cap protein could be detected in the nucleus and cytoplasm, but Cap and Rep protein could be located in different compartments of the nucleus during the early phase of PCV2 infection. Localization of Cap in the nucleoli and Rep in the nucleoplasm was followed by co-location of both proteins in the nucleoplasm [47, 48]. Recently, a total of nine cellular proteins were found to bind the Cap protein of PCV2. They include the complement factor C1qB, the cell adhesion molecule P-selectin, the makorin-1 RING zincfinger protein (MKRN1), the receptor protein for the globular heads of complement component C1q (gC1qR), the nucleosome assembly protein-1 (NAP1), the prostate apoptosis response-4 (Par-4) protein, nucleophosmin-1 (NPM1), the heat-shock protein 40 (Hsp40), and the heat-shock protein 70 (Hsp70) [48, 49, 60]. However, there is no exact description of these interaction partners in the literature, and the functions of the interacting proteins have not yet been wholly elucidated.
Research on the ORF3 gene
The ORF3 gene, called the apoptosis-inducing gene, completely overlaps the ORF1 gene and is oriented in the opposite direction [61]. The ORF3 gene stretches approximately 315 bp (nt 671–357) and encodes 104 aa. Since it was first identified as a nonstructural protein in 2005 in PCV2 productive infections, its structure and function have received attention from scholars worldwide. Studies have revealed that the protein encoded by ORF3 of PCV2 (also called the apoptotic protein, 11.9 kDa) is located predominantly in the nucleus and to a lesser degree in the cytoplasm [61]. The ORF3 protein is not essential for viral replication in cultured cells, but can induce apoptosis in virus-infected cells (such as porcine kidney PK-15 cells, porcine peripheral blood mononuclear cells (PBMC) [62], and lymphocytes) and plays a major role in viral pathogenesis via its apoptotic activity in vitro and in vivo [15, 61]. It is worth noting that this apoptotic activity is correlated with the nuclear localization of ORF3 [63]. Subsequently, evidence was found that the ORF3 protein contributes to enhancing the systemic dissemination of PCV2 infections in mouse models and SPF piglets by inducing the early release of the virus from infected cells [64], and that the ORF3-deficient PCV2 is attenuated in its natural host [65].
A recent report demonstrated that the PCV2 ORF3 protein could specifically and directly interact with the porcine homolog of Pirh2 (pPirh2), a p53-induced ubiquitin-protein E3 ligase involved in the ubiquitination of p53, resulting in decreased levels of Pirh2 and increased cellular levels of p53, thereby leading to apoptosis of the infected cells [63, 66]. More specifically, the aa residues 20–65 of the ORF3 protein are essential in this competitive interaction of ORF3 protein with pPirh2 over p53 [66]. Beyond that, though, the interaction between the ORF3 protein and pPirh2 also leads to altered physiological cellular localization of pPirh2 and a significant reduction in pPirh2 stability [66]. However, whether the ORF3 protein is actually produced during PCV2 infection in pigs and more importantly specific pathogenic role of ORF3 remains under debate. This is indeed reflected by the fact that two bacterial hybrid screens have identified other interacting partners for ORF3. Notably, RGS16, a regulatory protein involved in the signaling by G proteins, is among them [48]. It has been suggested that the PCV2 ORF3 protein co-localized with porcine RGS16 (poRGS16) in LPS-activated porcine PBMC and that poRGS16 may be assisting the translocation of ORF3 protein into the nucleus [67]. Additionally, the ORF3 protein appears to be dispensable for PCV2 replication in pigs [68]. Clearly, more studies are needed to explore the apparent multifunctionality of ORF3 protein in virus replication and pathogenesis.
Research on the ORF4 gene
The ORF4 gene of PCV2 is embedded within ORF3 and oriented in the same direction with approximately 180 bp (nt 565–386) in length [35], while a recent report demonstrated the existence of a novel trans-splicing ORF4 transcript with a size of 355 bp [69]. The ORF4 gene, called the apoptosis-suppressing gene, encodes a new and experimentally confirmed protein that is predicted to be 59-aa long with a molecular mass of 6.5 kDa [70]. In one study, the ORF4 protein was found to have a weak interaction with the Rep protein, although the implication of this possible interaction is currently unknown [48]. Subsequently, mutagenesis studies demonstrated that the ORF4 protein is not essential for PCV2 replication in PK-15 cells or in mice, but plays an important role in suppressing caspase-3 and caspase-8 activity and modulating the host immune system via regulation of CD4+ and CD8+ T lymphocytes during PCV2 infection [70]. In 2014, Gao et al. [71] constructed two PCV2 ORF4 null mutants that had no amino acid mutation in ORF1 and ORF3 compared to the wild-type PCV2, and experimental results indicated that the ORF4 protein may play an important role by restricting ORF3 transcription thereby preventing virus-induced apoptosis.
Meanwhile, findings of peptide dot enzyme-linked immunosorbent assay indicated that the peptide sequence matching aa 19–25 (named 19KSSASPR25) is the common antigenic epitope and core motif for ORF4-specific monoclonal antibodies (mAbs) and that the antigenic epitope recognized by the mAbs to the PCV2 ORF4 protein is a conformational epitope [70]. However, whether the ORF4 protein is truly involved in the pathogenesis of PCV2 is distinctly lack of evidence. After all, currently we do not yet know whether ORF4 protein is actually produced during PCV2 infection in pigs and what are its interacting proteins.
Discussion
Porcine circovirus type 1 (PCV1) was initially described in 1974 as papovavirus- and picornavirus-like particles in a contaminated porcine kidney cell line PK-15 [72]. Fifteen years later, the PCVAD-associated variant strain, PCV2, was isolated from diseased piglets [73]. However, the definitive origin of PCV remains elusive because of the difficulty in experimental verification. Genetically, PCV2 is a relatively stable virus. However, genomic variation in the virus has been noticed all over the world. Among the five distinct PCV2 genotypes identified from pigs worldwide, PCV2a and PCV2b are the most common isolates associated with PCVAD with varying degrees of severity, while PCV2c has only been found in archived samples in Denmark [74]. Two rare genotypes, PCV2d and PCV2e, are newly discovered genotypes in China [75, 76]. Prior to 2003, PCV2a and PCV2b were prevalent in European countries and China, whereas PCV2a was the only recognized genotype in the United States and Canada [59, 74, 77]. Around 2003, a drastic global-scale shift in PCV2 genotypes from PCV2a to PCV2b was evidenced by the availability of more PCV2b sequences in the GenBank [74, 78]. However, the answer as to why there is a sudden genotype prevalence shift since 2003 still remains puzzle. Based on available data, the increasing international trade of pigs and derived products and growing world’s livestock population, coupled with the multiple transmission routes described for PCV2 and its long-lasting viral life strategy, may have played a major role in the global PCV2 genotype replacement (PCV2b over PCV2a) during such period [79–82]. Recently, a distinct signature sequence motif distinguishing PCV2a and PCV2b has been identified in the viral genome [32, 35]. For PCV2a viruses, the motif is 1480–1469 ACC/AAC/AAA/ATC (amino acid sequence TNKI). For the PCV2b viruses, the motif is 1479–1468 TCA/AAC/CCC/CG(T)C [amino acid sequence SNPR(L)]. But there is no significant difference in virulence between the two major genotypes [83]. Therefore, the current available experimental data failed to elaborate the relationship between the amino acid (aa) variations of the PCV2 sequence and the pathogenicity as well as virulence of PCV2 strains.
Nowadays, PCV2 is an emerging swine pathogen causing serious harm and immense economic losses in the global swine industry. Controlling and reducing infection rate of PCV2 in pig farms is not realizable due to unique characteristic of PCV2. However, prevention with vaccines against PCV2 is usually used. So far, series commercial vaccines are available for application in the field, and they have been shown to be effectively protective against PCV2 infection and PCVAD via vaccine trial studies. These vaccines mainly include the inactivated and attenuated PCV1-2a chimeric vaccine Fostera™ PCV (Pfizer Animal Health, Inc.), formerly Suvaxyn® PCV2 One Dose™ (Fort Dodge Animal Health, Inc.), the killed Circovac® vaccine (Merial, Inc.), and three recombinant subunit vaccines [Ingelvac CircoFLEX® vaccine (Boehringer Ingelheim Vetmedica, Inc.), Circumven® vaccine (Intervet/Merck), and Porcilis® PCV (Schering-Plough/Merck)] [77, 84]. However, all these vaccines are developed from the PCV2a virus. Future studies should focus on the development of new vaccines based on the PCV2b genotype that is currently most prevalent worldwide, and even a marker vaccine that can distinguish natural infection from vaccination. Also, additional research is needed to evaluate the effect of vaccine pressure in PCV2b dominance and the potential emergence of variants or vaccine-escape mutants, although these current vaccines play an important role in the control of PCVAD and authentic vaccine failure cases are extremely rare. In addition, many other comprehensive measures including disinfection of pig farm environment, improvement of sanitary level and environmental conditions, effective vaccination for classical swine fever (CSF), porcine reproductive and respiratory syndrome (PRRS), pseudorabies, and control of secondary bacterial infection using antibiotics should be implemented in the pig farms, after all PMWS and other PCVAD are multifactorial diseases.
As the smallest virus known to infect mammals, PCV2 have a very small compact genome and accordingly a highly limited coding capacity. Due to lacking of its own enzyme, the life cycle of PCV2 depends heavily on the host cell machinery [85–87]. Inevitably, during virus infection, PCV2 interacts with the cell, subverts cellular factors or processes to complete the viral replication cycles and modulates the host immune function to cause cytokine imbalance, immunosuppression, and diseases, utilizing its DNA sequences or encoded proteins [77, 86, 88]. During the last years, numerous porcine proteins were identified to interact with PCV2 genomic DNA, Rep/Rep′, Cap and the ORF3 protein, mostly using a yeast or a bacteria-based two-hybrid assay [48, 49, 60, 63]. Almost all interaction partners (summarized in Table 1) have been annotated in the literature as proteins with multiple functions, but there is still no a convincing common ontology. They can be associated with many aspects of the viral molecular biology such as cell tropism and viral replication [51, 89]. However, functions of the described interacting proteins regarding PCV2 infection just stay in the speculation phase, and the domains responsible for the interaction are still unclear.
Genetic characterization and phylogenetic analysis findings have suggested a link between capsid protein variation and PCV2 pathogenicity due to alterations of the determinants involved in tissue tropism or virus–host interactions [90]. A two-aa mutation (sites P110A, R191S) or nt deficiency in the capsid protein of PCV2 have also been found to alter PCV2 virulence [53, 91]. More recently, a pathogenicity study on the observed consistent linear nine-base sequence in the capsid gene of archival PCV2 isolates showed that the mutational events within this 9-bp region can alter the virulence of PCV2 as well [92]. However, other regions outside the capsid protein might be also involved in viral pathogenesis. For example, the chimeric PCV1-2 virus, created by cloning of the pathogenic PCV2 capsid gene into the genomic backbone of nonpathogenic PCV1, can elicit a specific antibody response to the PCV2 Cap protein, but is attenuated in pigs [91, 93]. Recent studies have also revealed that ORF2-, ORF3-, and ORF4-related proteins play important roles in the pathogenesis of PCV2 when ORF3-deficient PCV2 is attenuated in its natural host [65] and ORF4-deficient PCV2 induces a higher viral load as well as more severe microscopic lesions in the spleen at the early stage of infection in mice [70]. In 2011, Ramamoorthy et al. [94] reported that ISRE mutation reduced viral replication in vitro and in vivo and elicited low antibody responses in PCV2 infection. In addition, PCV2 genomic DNA have been confirmed to contain at least five oligodeoxynucleotides (ODNs) containing CpG motifs, and their involvement in the activation of innate immunity of the host via interaction with TLR9 signaling, resulting in the production of IFN and other anti-inflammatory cytokines has also been elucidated [86, 95, 96].
Clearly, all of these research findings indicate that PCV2 pathogenesis is complicated and multigenic and that the viral proteins encoded by ORF2, ORF3, and ORF4 do not solely determine the pathogenicity of PCV2. In addition to the replicase ORF1, capsid protein ORF2, apoptotic protein ORF3, and unidentified anti-apoptotic protein ORF4, a computer search revealed that PCV2 is predicted to contain three additional potential ORF-encoding proteins that are greater than 5 kDa [97]. Further research should be done to clarify whether these potential ORFs are expressed or not and whether other proteins encoded by PCV2 control its pathogenicity.
Apoptosis, a genetically programmed cell death process that eliminates aberrant cells created by DNA damage or infected by viral pathogens, plays essential roles in some developmental pathways and disease processes [98]. However, viruses have evolved versatile strategies that directly or indirectly elicit or inhibit apoptosis during their replication cycles via activating endogenous anti-apoptotic processes and genes or expressing their own anti-apoptotic genes to complete its proliferation [99, 100]. Viruses may benefit from stimulating apoptosis to either hasten the spread of progeny virus particles to neighboring cells while evading the host immune system or induce the breakdown of infected cells in the host, thereby facilitating virus growth [101]. PCV2, which is histopathologically associated with lymphoid depletion and histiocyte infiltration, causes apoptosis in mouse and pig models. Although studies have shown that ORF3 of PCV2 is apoptotic and ORF4 of PCV2 is probably anti-apoptotic, the precise pathogenesis of PCV2-associated diseases and its involvement in apoptosis in particular has yet to be determined. It may be speculated that there are complex functional relationships between ORF3 and ORF4 and that other ORFs may be also triggered alone or simultaneously by common upstream signals in regulating apoptosis. Additionally, recent reports showed that activation of the NF-κB, JNK/p38 MAPK, and PI3 K/Akt pathways influences PCV2-induced apoptosis as well [102–104]. Whether those ORF proteins are involved in these signaling pathways requires further investigation.
Autophagy is a catabolic cellular process conserved in all eukaryotes that involves the degradation and turnover of protein aggregates and damaged organelles in the cytoplasm by lysosomal enzymes. It plays an important role in many disease processes in viral replication and pathogenesis [105–107]. Recent studies have revealed that autophagy is emerging as a process of the host defense mechanism against bacterial and viral infections by influencing the innate and adaptive immune responses [107–109]. Actually, this type of autophagy that is specific to removing bacteria and viruses has been called xenophagy [107]. However, viruses have developed a variety of strategies to subvert the autophagic pathway. The replication processes of some viruses, such as Sindbis virus and tobacco mosaic virus [110, 111], can be successfully repressed by autophagy, while others like human cytomegalovirus, human immunodeficiency virus type 1, and herpes simplex virus type 1 [112–114] are able to inhibit autophagy in favor of their own replication. Some viruses, such as dengue virus and hepatitis B virus [115–117], can make use of autophagosomes for their own replication. Experimental evidence has been discovered that PCV2 could induce autophagy via its capsid protein and employ the autophagic machinery to enhance its replication in host cells [118] and that it induces autophagy by activating the AMPK/ERK/TSC2/mTOR signaling pathway [119]. However, it is worthy to mention that signaling upstream of AMPK after PCV2-induced autophagy remains obscure despite the Cap protein has been confirmed as the signaling molecule that activates AMPK. Considering the fact that great complexity exists in the signaling cross-talk of the autophagic processes, further examinations should be performed to clarify this issue in more detail.
Taken together, it would be very interesting to study how the autophagic and/or apoptotic processes are utilized by PCV2 to induce its active infection (Fig. 2). Meanwhile, investigating how host cells interact with PCV2 virions, taking attachment, internalization and uncoating of PCV2 in target cells for example, will be also meaningful. In addition, the details of interactions between viral and cellular proteins are very cursory at this time, we have yet to identify, characterize, and functionally analyze the other proteins (possibly encoded by ORF5 to ORF11, Rep3a, Rep3b, Rep3c, NS515, NS672, and NS0) produced by PCV2. Development of this work will be of benefit to deepening our understanding of the roles played by these proteins and their different traits (overall aa sequence, antigenic epitopes, hydrophobic and hydrophilic domains, glycosylation sites, localization, and cellular interactome) in relation to PCV2 pathogenesis, clinical signs, and lesions. Also, understanding the important host–virus interactions and revealing their potential roles in virus replication and pathogenesis will be of paramount importance. All of these obtained results will extend our understanding of the pathogenic mechanism and reproduction of PCV2 and thereby shed light upon the vaccines that are used to effectively prevent and control virus-induced diseases.
A hypothetical model depicting some of the mechanisms involved in PCV2 pathogenesis. For one thing, PCV2 expresses Cap protein, which subsequently activates AMPK via an unidentified signaling molecule and then activates ERK1/2 and TSC2 and represses mTOR signaling, thereby inducing autophagy in host cells. For another, PCV2 expresses ORF3, ORF4 and even other ORFs proteins that are most likely to partake in regulating PCV2-induced apoptosis: Despite of heated debates all over the interactions between ORF3 and ORF4, it is a fact that ORF3 protein initiates apoptosis by interacting with pPirh2 and up-regulating p53 expression, while ORF4 protein suppresses apoptosis by inhibiting activation of both the initiator caspase-8 and the effector caspase-3. Finally, autophagy, together with apoptosis, impacts the pathogenesis of PCV2 in an unknown way
References
K.V. Phenix, J.H. Weston, I. Ypelaar, A. Lavazza, J.A. Smyth, D. Todd, G.E. Wilcox, S.R. Raidal, J. Gen. Virol. 82, 2805–2809 (2001)
B.W. Ritchie, F.D. Niagro, P.D. Lukert, W.L. Steffens III, K.S. Latimer, Virology 171, 83–88 (1989)
D. Todd, F.D. Niagro, B.W. Ritchie, W. Curran, G.M. Allan, P.D. Lukert, K.S. Latimer, W.L. Steffens III, M.S. McNulty, Arch. Virol. 117, 129–135 (1991)
D. Todd, J.H. Weston, D. Soike, J.A. Smyth, Virology 286, 354–362 (2001)
A. Kato, M. Fujino, T. Nakamura, A. Ishihama, Y. Otaki, Virology 209(2), 480–488 (1995)
G.M. Allan, F. McNeilly, S. Kennedy, B. Daft, E.G. Clarke, J.A. Ellis, D.M. Haines, B.M. Meehan, B.M. Adair, J. Vet. Diagn. Invest. 10(1), 3–10 (1998)
J. Ellis, L. Hassard, E. Clark, J. Harding, G. Allan, P. Willson, J. Strokappe, K. Martin, F. McNeilly, B. Meehan, D. Todd, D. Haines, Can. Vet. J. 39(1), 44–51 (1998)
G.M. Allan, F. McNeilly, B.M. Meehan, S. Kennedy, D.P. Mackie, J.A. Ellis, E.G. Clarke, E. Saubi, N. Espuna, P. Riera, A. Botner, C.E. Charreyre, Vet. Microbiol. 66, 115–123 (1999)
B.M. Meehan, F. McNeilly, D. Todd, S. Kennedy, V.A. Jewhurst, J.A. Ellis, L.E. Hassard, E.G. Clark, D.M. Haines, G.M. Allan, J. Gen. Virol. 79, 2171–2179 (1998)
R. Magar, R. Larochelle, S. Thibault, L. Lamontagne, J. Comp. Pathol. 123, 258–269 (2000)
Y. Okuda, M. Ono, S. Yazawa, I. Shibata, J. Vet. Diagn. Invest. 15, 107–114 (2003)
G.M. Allan, F. McNeilly, J. Ellis, S. Krakowka, A. Botner, K. McCullough, H. Nauwynck, S. Kennedy, B. Meehan, C.E. Charreyre, Vet. Microbiol. 98, 165–168 (2004)
M. Kiupel, G.W. Stevenson, J. Choi, K.S. Latimer, C.L. Kanitz, S.K. Mittal, Vet. Pathol. 38, 74–82 (2001)
M. Kiupel, G.W. Stevenson, E.J. Galbreath, A. North, H. HogenEsch, S.K. Mittal, BMC Vet. Res. 1, 7 (2005)
J. Liu, I. Chen, Q. Du, H. Chua, J. Kwang, J. Virol. 80, 5065–5073 (2006)
G.M. Allan, J.A. Ellis, J. Vet. Diagn. Invest. 12, 3–14 (2000)
J. Segales, C. Rosell, M. Domingo, Vet. Microbiol. 98, 137–149 (2004)
C. Chae, Vet. J. 168(1), 41–49 (2004)
C. Chae, Vet. J. 169, 326–336 (2005)
T. Opriessnig, X.J. Meng, P.G. Halbur, J. Vet. Diagn. Invest. 19(6), 591–615 (2007)
S. Ramamoorthy, X.J. Meng, Anim. Health Res. Rev. 10(1), 1–20 (2009)
J. Gillespie, T. Opriessnig, X.J. Meng, K. Pelzer, V. Buechner-Maxwell, J. Vet. Intern. Med. 23(6), 1151–1163 (2009)
G.M. Allan, B.M. Meehan, D. Todd, S. Kennedy, F. McNeilly, J.A. Ellis, E.G. Clark, J. Harding, E. Espuna, A. Botner, C.E. Charreyre, Vet. Rec. 142(17), 467–468 (1998)
J. Segales, M. Domingo, Vet. Q. 24(3), 109–124 (2002)
L. Darwich, J. Segales, E. Mateu, Arch. Virol. 149(5), 857–874 (2004)
J. Nielsen, I.E. Vincent, A. Botner, A.S. Ladekaer-Mikkelsen, G. Allan, A. Summerfield, K.C. McCullough, Vet. Immunol. Immunopathol. 92(3–4), 97–111 (2003)
L. Darwich, J. Segales, M. Domingo, E. Mateu, Clin. Diagn. Lab. Immunol. 9(2), 236–242 (2002)
J. Segales, M. Domingo, F. Chianini, N. Majo, J. Dominguez, L. Darwich, E. Mateu, Vet. Microbiol. 98(2), 151–158 (2004)
J. Liu, I. Chen, H. Chua, Q. Du, J. Kwang, Virology 347, 422–433 (2006)
C. de Boisseson, V. Beven, L. Bigarre, R. Thiery, N. Rose, E. Eveno, F. Madec, A. Jestin, J. Gen. Virol. 85, 293–304 (2004)
C.M. Ma, C.C. Hon, T.Y. Lam, V.Y. Li, C.K. Wong, T. de Oliveira, F.C. Leung, J. Gen. Virol. 88, 1733–1737 (2007)
A.K. Cheung, L.M. Lager, O.I. Kohutyuk, A.L. Vincent, S.C. Henry, R.B. Baker, R.R.R. Rowland, A.G. Dunham, Arch. Virol. 152, 1035–1044 (2007)
J. Segale’s, A. Olvera, L. Grau-Roma, C. Charreyre, H. Nauwynck, L. Larsen, K. Dupont, K. McCullough, J. Ellis, S. Krakowka, A. Mankertz, M. Fredholm, C. Fossum, S. Timmusk, N. Stockhofe-Zurwieden, V. Beattie, D. Armstrong, B. Grassland, P. Baekbo, G. Allan, Vet. Rec. 162(26), 867–868 (2008)
F. Wang, X. Guo, X.N. Ge, Z.T. Wang, Y.H. Chen, Z.L. Cha, H.C. Yang, Virus Res. 145, 151–156 (2009)
A. Olvera, M. Cortey, J. Segales, Virology 357, 175–185 (2007)
T. Finsterbusch, A. Mankertz, Virus Res. 143(2), 177–183 (2009)
A.K. Cheung, Virology 313, 452–459 (2003)
A.L. Hamel, L.L. Lin, G.P.S. Nayar, J. Virol. 72, 5262–5267 (1998)
A.K. Cheung, Virology 305, 168–180 (2003)
J.Y. Gu, Y. Zhang, X. Lian, H.L. Sun, J.M. Wang, W.T. Liu, G. Meng, P. Li, D. Zhu, Y.X. Jin, R.B. Cao, Virol. J. 9, 152–161 (2012)
A.K. Cheung, Virology 310, 41–49 (2003)
A. Mankertz, J. Mankertz, K. Wolf, H.J. Buhk, J. Gen. Virol. 79, 381–383 (1998)
A. Mankertz, B. Mueller, T. Steinfeldt, C. Schmitt, T. Finsterbusch, J. Virol. 77(18), 9885–9893 (2003)
E.V. Koonin, T.V. Ilyina, Biosystems 30, 241–268 (1993)
A. Mankertz, B. Hillenbrand, Virology 279, 429–438 (2001)
M. Fort, M. Sibila, M. Nofrarías, E. Pérez-Martín, A. Olvera, E. Mateu, J. Segalés, Vet. Immunol. Immunopathol. 137, 226–234 (2010)
D.F. Gilpin, K. McCullough, B.M. Meehan, F. McNeilly, I. McNair, L.S. Stevenson, J.C. Foster, J.A. Ellis, S. Krakowka, B.M. Adair, G.M. Allan, Vet. Immunol. Immunopathol. 94, 149–161 (2003)
S. Timmusk, C. Fossum, M. Berg, J. Gen. Virol. 87, 3215–3223 (2006)
T. Finsterbusch, T. Steinfeldt, K. Doberstein, C. Rödner, A. Mankertz, Virology 386, 122–131 (2009)
P. Nawagitgul, I. Morozov, S.R. Bolin, P.A. Harms, S.D. Sorden, J. Gen. Virol. 81, 2281–2287 (2000)
G. Misinzo, P.L. Delputte, P. Meerts, D.J. Lefebvre, H.J. Nauwynck, J. Virol. 80(7), 3487–3494 (2006)
M.A. O’Dea, A.P. Hughes, L.J. Davies, J. Muhling, R. Buddle, G.E. Wilcox, J. Virol. Methods 147(1), 61–66 (2008)
M. Fenaux, T. Opriessnig, P.G. Halbur, F. Elvinger, X.J. Meng, J. Virol. 78, 13440–13446 (2004)
D. Mahé, P. Blanchard, C. Truong, C. Arnauld, P.L. Cann, R. Cariolet, F. Madec, E. Albina, A. Jestin, J. Gen. Virol. 81, 1815–1824 (2000)
P. Lekcharoensuk, I. Morozov, P.S. Paul, N. Thangthumniyom, W. Wajjawalku, X.J. Meng, J. Virol. 78(15), 8135–8145 (2004)
Q. Liu, S.K. Tikoo, L.A. Babiuk, Virology 285(1), 91–988 (2001)
Q. Liu, P. Willson, S. Attoh-poku, L.A. Babiuk, Protein Expr. Purif. 21(1), 115–120 (2001)
R. Khayat, N. Brunn, J.A. Speir, J.M. Hardham, R.G. Ankenbauer, A. Schneemann, J.E. Johnson, J. Virol. 85, 7856–7862 (2011)
B.R. Trible, R.R.R. Rowland, Virus Res. 164, 68–77 (2012)
J. Liu, J. Bai, L.L. Zhang, Z.H. Jiang, X.W. Wang, Y.F. Li, P. Jiang, Virology 447, 52–62 (2013)
J. Liu, I. Chen, J. Kwang, J. Virol. 79, 8262–8274 (2005)
W.L. Lin, M.S. Chien, P.C. Wu, C.L. Lai, C.J. Huang, Open. Virol. J. 5, 148–153 (2011)
J. Liu, Y. Zhu, I. Chen, J. Lau, F. He, A. Lau, Z.L. Wang, A.K. Karuppannan, J. Kwang, J. Virol. 81, 9560–9567 (2007)
A.K. Karuppannan, J. Kwang, Virology 410, 248–256 (2011)
A.K. Karuppannan, M.H. Jong, S.H. Lee, Y. Zhu, M. Selvaraj, J. Lau, Q. Jia, J. Kwang, Virology 383, 338–347 (2009)
A.K. Karuppannan, S. Liu, Q. Jia, M. Selvaraj, J. Kwang, Virology 398, 1–11 (2010)
S. Timmusk, E. Merlot, T. Lovgren, L. Jarvekulg, M. Berg, C. Fossum, J. Gen. Virol. 90, 2425–2436 (2009)
N.M. Juan, T. LeRoith, T. Opriessnig, X.J. Meng, Virus Res. 147, 60–66 (2010)
Z.Z. Gao, Q.F. Dong, Y.G. Jiang, T. Opriessnig, J.X. Wang, Y.P. Quan, Z.Q. Yang, Virus Genes 47(2), 268–275 (2013)
J.L. He, J.J. Cao, N. Zhou, Y.L. Jin, J.S. Wu, J.Y. Zhou, J. Virol. 87(3), 1420–1429 (2013)
Z.Z. Gao, Q.F. Dong, Y.H. Jiang, T. Opriessnig, J.X. Wang, Y.P. Quan, Z.Q. Yang, Virus Res. 183, 56–62 (2014)
I. Tischer, W. Mields, D. Wolff, M. Vagt, W. Griem, Arch. Virol. 91(3–4), 271–276 (1986)
G. Allan, S. Krakowka, J. Ellis, C. Charreyre, Virus Res. 164, 4–9 (2012)
K. Dupont, E.O. Nielsen, P. Bækbo, L.E. Larsen, Vet. Microbiol. 128, 56–64 (2008)
F. Wang, X. Guo, X. Ge, Z. Wang, Y. Chen, Z. Cha, H. Yang, Virus Res. 145(1), 151–156 (2009)
L. Guo, Y. Lu, Y. Wei, L. Huang, C. Liu, J. Virol. 7, 273 (2010)
X.J. Meng, Annu. Rev. Anim. Biosci. 1, 43–64 (2013)
M. Cortey, E. Pileri, M. Sibila, J. Pujols, M. Balasch, J. Plana, J. Segales, Vet J. 187, 363–368 (2011)
C. Firth, M.A. Charleston, S. Duffy, B. Shapiro, E.C. Holmes, J. Virol. 83, 12813–12821 (2009)
A.R. Patterson, T. Opriessnig, Anim. Health Res. Rev. 11, 217–234 (2010)
L.J. Pe´rez, H.D. de Arce, M. Cortey, P. Dominguez, M.I. Percedo, C.L. Perera, J. Tarradas, M.T. Frias, J. Segale´s, L. Ganges, J.I. Nunez, Vet. Microbiol. 151, 245–254 (2011)
N. Rose, T. Opriessnig, B. Grasland, A. Jestin, Virus Res. 164, 78–89 (2012)
T. Opriessnig, S. Ramamoorthy, D.M. Madson, A.R. Patterson, N. Pal, S. Carman, X.J. Meng, P.G. Halbur, J. Gen. Virol. 89, 2482–2491 (2008)
N.M. Beach, X.J. Meng, Virus Res. 164, 33–42 (2012)
I. Tischer, D. Peters, R. Rasch, S. Pociuli, Arch. Virol. 96, 39–57 (1987)
A. Mankertz, Virus Res. 164, 54–60 (2012)
A.K. Cheung, Virus Res. 164, 46–53 (2012)
R. Ramamoorthy, X.J. Meng, Anim. Health Res. Rev. 10, 1–20 (2009)
H.J. Nauwynck, R. Sanchez, P. Meerts, D.J. Lefebvre, D. Saha, L. Huang, G. Misinzo, Virus Res. 164, 43–45 (2012)
R. Larochelle, R. Magar, S. D′Allaire, Virus Res. 90, 101–112 (2002)
S.B. Shang, Y.L. Jin, X.T. Jiang, J.Y. Zhou, X. Zhang, G. Xing, J.L. He, Y. Yan, Mol. Immunol. 46, 327–334 (2009)
S. Krakowka, G. Allan, J. Ellis, A. Hamberg, C. Charreyre, E. Kaufmann, C. Brooks, B. Meehan, Virus Res. 164, 90–99 (2012)
M. Fenaux, T. Opriessnig, P.G. Halbur, X.J. Meng, J. Virol. 77, 11232–11243 (2003)
S. Ramamoorthy, T. Opriessnig, N. Pal, F.F. Huang, X.J. Meng, Vet. Microbiol. 147(1–2), 49–58 (2011)
F.C. Hasslung, M. Berg, G.M. Allan, B.M. Meehan, F. McNeilly, C. Fossum, J. Gen. Virol. 84, 2937–2945 (2003)
F.H. Wikström, B.M. Meehan, M. Berg, S. Timmusk, J. Elving et al., J. Virol. 81, 4919–4927 (2007)
B.M. Meehan, J.L. Creelan, M.S. McNulty, D. Todd, J. Gen. Virol. 78, 221–227 (1997)
K. Labbe, M. Saleh, Cell Death Differ. 15, 1339–1349 (2008)
A. Busca, M. Saxena, M. Kryworuchko, A. Kumar, Curr. Genomics 10, 306–317 (2009)
L. Galluzzi, O. Kepp, E. Morselli, I. Vitale, L. Senovilla, M. Pinti, L. Zitvogel, G. Kroemer, J. Intern. Med. 267, 526–542 (2010)
L. Galluzzi, C. Brenner, E. Morselli, Z. Touat, G. Kroemer, PLoS Pathog. 4(5), e1000018 (2008)
L. Wei, J. Kwang, J. Wang, L. Shi, B. Yang, Y.Q. Li, J. Liu, Virology 378, 177–184 (2008)
L. Wei, Z. Zhu, J. Wang, J. Liu, J. Virol. 83, 6039–6047 (2009)
L. Wei, S.S. Zhu, J. Wang, J. Liu, J. Virol. 86(24), 13589–13597 (2012)
D.J. Klionsky, J. Cell Sci. 118, 7–18 (2005)
A. Esclatine, M. Chaumorcel, P. Codogno, Curr. Top. Microbiol. Immunol. 335, 33–70 (2009)
D. Sir, J.J. Ou, Autophagy in viral replication and pathogenesis. Mol. Cells 29, 1–7 (2010)
V. Deretic, B. Levine, Cell Host Microbe 5(6), 527–549 (2009)
J.M. Yuk, T. Yoshimori, E.K. Jo, Exp. Mol. Med. 44, 99–108 (2012)
X.H. Liang, L.K. Kleeman, H.H. Jiang, G. Gordon, J.E. Goldman, G. Berry, B. Herman, B. Levine, J. Virol. 72(11), 8586–8596 (1998)
Y. Liu, M. Schiff, K. Czymmek, Z. Talloczy, B. Levine, S.P. Dinesh-Kumar, Cell 121(4), 567–577 (2005)
A. Orvedahl, D. Alexander, Z. Tallóczy, Q. Sun, Y. Wei, W. Zhang, D. Burns, D. Leib, B. Levine, Cell Host Microbe 1(1), 23–35 (2007)
M. Chaumorcel, S. Souquere, G. Pierron, P. Codogno, A. Esclatine, Autophagy 4, 46–53 (2008)
G.B. Kyei, C. Dinkins, A.S. Davis, E. Roberts, S.B. Singh, C. Dong, L. Wu, E. Kominami, T. Ueno, A. Yamamoto, M. Federico, A. Panganiban, I. Vergne, V. Deretic, J. Cell Biol. 186(2), 255–268 (2009)
Y.R. Lee, H.Y. Lei, M.T. Liu, J.R. Wang, S.H. Chen, Y.F. Jiang-Shieh, Y.S. Lin, T.M. Yeh, C.C. Liu, H.S. Liu, Virology 374(2), 240–248 (2008)
D. Sir, Y. Tian, W. Chen, D. Ann, T. Yen, J.H. Ou, Proc. Natl. Acad. Sci. USA. 107(9), 4383–4388 (2010)
J. Li, Y. Liu, Z. Wang, K. Liu, Y. Wang, J. Liu, H. Ding, Z. Yuan, J. Virol. 85(13), 6319–6333 (2011)
B.L. Zhu, F. Xu, J. Li, J.B. Shuai, X.L. Li, W.H. Fang, Virus Res. 163, 476–485 (2012)
B.L. Zhu, Y.S. Zhou, F. Xu, J.B. Shuai, X.L. Li, W.H. Fang, J. Virol. 86(22), 12003–12012 (2012)
Acknowledgments
This review was supported in part by a grant from the National Natural Science Foundation of China (Project No. 30972186).
Conflict of interest
The authors declare that they do not have any conflict of interests with respect to the research, authorship, and/or publication of this article.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lv, Qz., Guo, Kk. & Zhang, Ym. Current understanding of genomic DNA of porcine circovirus type 2. Virus Genes 49, 1–10 (2014). https://doi.org/10.1007/s11262-014-1099-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-014-1099-z