Abstract
Hepatitis C virus (HCV) is a worldwide problem which does not have an effective vaccine and more than 170 million people worldwide are chronically infected by HCV. T cell responses are associated with spontaneous clearance of HCV infection. We report here the development of recombinant Lambda bacteriophage nanoparticles encoding HCV Core antigen. The aim of this study was to investigate the antigen-specific immune responses triggered in mice by different prime–boost combinations of DNA and Lambda phage nanoparticles encoding the HCV Core. The homologous prime/boost with recombinant Lambda nanoparticles induced higher levels of cellular and humoral immune response than the DNA vaccines. However, a heterologous prime/boost of HCV Core protein, using DNA vaccine priming followed by Lambda boost, induced highest level of lymphocyte proliferation, CD8 lymphocytes with cytotoxic function, and shifting the immune response toward a T helper (Th1) pattern and in overall improved immunity. Our study provides a new, safe, and effective vaccine for the prime–boost regimen which augments robust immunity and highlights novel promising strategies in HCV vaccine development.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hepatitis C virus (HCV) is one of the main causative agents of liver disease worldwide and a major problem of public health. About 180 million people worldwide are chronically infected with HCV, with 350,000 deaths each year from hepatitis C-related liver failure and cancer [1]. Moreover, the World Health Organization estimates that three to four million people are newly infected every year [2]. No vaccine is available so far, and the current combination treatment of interferon-alpha (IFNα) and ribavirin is not effective against all HCV genotypes and often not well tolerated by the patients [3].
Design of an effective vaccine to protect against HCV infection has been defined as a difficult challenge due to the heterogeneous nature of the genome and substantial replication rate of the virus [4].
Lack of antigen-specific cellular immunity in chronic HCV patients may be the main reason for persistent infections. Therefore, development of HCV therapeutic vaccines, which can activate the specific immunity in chronic HCV infection and eradicate the virus, may be an effective strategy against HCV [5, 6].
Among different HCV genes, Core antigen exhibits the most conserved viral antigen among the different viral genotypes/subtypes. The antigen has been employed extensively for induction of cellular immunity in animal models, as well as the human models in various vaccine development studies [7–9]. The Core antigen also contains some well-known T cell and B-cell epitopes [9]. Therefore, designing a HCV vaccine based on the Core gene could be a suitable antigenic candidate for vaccine development against HCV infection.
Several strategies to HCV vaccine development have been studied and include recombinant proteins [10], synthetic peptides [11], DNA [12], and prime–boost strategies [13].
Recent studies have shown that DNA vaccines are effective for priming immune responses. A prime–boost strategy with DNA vaccines and attenuated viral vectors expressing similar antigens has been successful in stimulating a protective cell-mediated immune response against viral infection [13]. The prime–boost vaccination regime comprising naked DNA priming which followed by a boost with recombinant modified vaccinia virus Ankara vaccine (MVA) has been successful in simian immunodeficiency virus/human immunodeficiency virus (SHIV)-macaque model of AIDS [14].
Bacteriophages offer an attractive alternative as a natural tool for delivery of therapeutic genes. As a gene delivery system, bacteriophage offers certain advantages compared to viral vectors that can produce endogenous recombination, oncogenic effects, and immunological reactions leading to potentially serious complications [15, 16]. Particularly, lambda bacteriophages having various appealing characteristics as gene/vaccine delivery vehicles, possess a high degree of stability, high production capacity [17], genetic tractability, and inherent biological safety for eukaryotic cells [18].
In recent years, we have shown that recombinant lambda phage nanoparticles containing expression cassette of human papillomavirus E7 gene (HPV-16 E7) under control of a CMV promoter, induce specific cell immune response and antitumor immunity in a mouse model [19]. In other studies, it has been demonstrated that lambda phage vaccine-delivered hepatitis B surface antigen (HBsAg) expression cassette was able to raise antibody levels significantly higher than with standard plasmid-based DNA vaccination in rabbit model [20].
Being particulate antigens, bacteriophage is taken up by antigen-presenting cells (APCs), cleared from the circulation and targeted to the spleen and liver Kuppfer cells [21]. After macropinocytosis and internalization, phage must gain access to the cytoplasm, uncoat and deliver its DNA payload to the nuclei. Lambda phage showed one of the most compact delivery platforms currently available among the various phage systems and synthetic peptide/DNA complexes by passing through nuclear core complex [22]. Thus, phage nanoparticles represent an extremely effective technology for targeting therapeutic vaccines to immune cells.
To achieve a more significant comparison of immune responses against HCV, we have compared the immunogenicity of Core when delivered as naked DNA, as the recombinant Lambda nanoparticles, or in combinations, and the optimal prime–boost regimen was analyzed in C57BL/6 mice. A comparison of various regimes demonstrated that priming with DNA encoding Core, followed by boosting with recombinant Lambda nanoparticles encoding Core results in optimal Core-specific immune responses. Findings from this study may facilitate improvements in HCV vaccine strategies.
Materials and methods
Plasmid construction and amplification
HCV Core gene was amplified using sera of HCV infected patients (genotype 1). The PCR products were cloned into the cloning vector which was then confirmed by sequencing.
The Core gene was digested with EcoRI-XhoI sites and cloned into the digested expression vector pcDNA3 under human cytomegalovirus immediate-early promoter using T4 ligase (Promega, Madison, WI). pcDNA3-HCV Core and pcDNA3 plasmids were prepared using Endo-free Plasmid Purification Kit (Qiagen, Hilden, Germany) according to manufacturer’s instructions.
Construction of Lambda vaccines
Lambda-ZAP®-CMV vector (Stratagene, USA) was used for construction of recombinant Lambda bacteriophages. The vector has potential characteristics for expression in eukaryotic cells. Eukaryotic expression of inserts is driven by the cytomegalovirus (CMV) immediate-early (IE) promoter with the SV40 transcription terminator and polyadenylation signal. Lambda-ZAP- HCV Core vector was prepared by digesting the pcDNA3-HCV Core with EcoRI and XhoI, and cloning the Core gene into the EcoRI and XhoI sites of Lambda-ZAP®-CMV vector. The Packaging extracts (Gigapack® III Gold packaging extract, Stratagene, USA) are used to package recombinant lambda phage nanoparticles with high efficiency as described previously [18]. Wild Lambda-ZAP phage was used as a negative control for further experiments. Bacteriophages were amplified on E. coli strain XL1-Blue MRF´. The phages were purified and concentrated using standard microbiological techniques, then the viral titer was determined by plaque-forming units per milliliter (PFU/ml) as described before [19].
The bacteriophage pellet was resuspended in 1–2 ml of SM buffer (SM buffer included 5.8 g NaCl, 2.0 g MgSO4·7H2O, 50.0 ml 1 M Tris–HCl (pH 7.5), 5.0 ml 2 % (w/v) gelatin up to one liter distilled water) at 4 °C overnight, pelleted once more, and then resuspended in SM solution prior to storage or further manipulation.
In vitro expression studies
Chinese hamster ovary (CHO) cells were cultured in RPMI 1640 (Roswell Park Memorial Institute), (Gibco BRL, Paisley, UK) supplemented with 10 % fetal bovine serum (FBS), and 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine (Sigma, Germany) at 70 % confluence. The subconfluent CHO eukaryotic cells were transfected with the pcDNA3-HCV Core plasmid using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
For lambda nanoparticles assay, Lambda-ZAP- HCV Core nanoparticles at a multiplicity of infection (MOI) of 5 × 105 in a final volume of 100 μl in culture medium were added to CHO cell line. Transduction of target cells was enhanced using centrifugal enhancement [1,200×g for 1 h at 37 °C] after addition of phage particles. The expression of HCV Core protein in transfected and transduced cells was evaluated by SDS-Western blotting. Transduced cells by wild lambda phages and transfected cell by pcDNA3 were used as negative control.
After 48 h, cell monolayers were washed 3 times with phosphate buffered saline (PBS) and scraped into 1 ml of PBS. The cells were lysed in 100 µl of lysis buffer containing 0.01 M Tris–HCl (pH 7.4), 0.15 M NaCl, 1 mM EDTA, 0.1 % SDS, 1 % Triton X-100, 1 % sodium deoxycholate, and 1 mM PMSF. The proteins in the lysate were quantitated and diluted in sodium dodecyl sulfate. Then, the samples were loaded onto a 10 % polyacrylamide/SDS gel (PAGE) and the proteins were isolated by electrophoresis and transferred onto a PVDF membrane. After fixation with 95 % alcohol, washing with PBST, and blocking with PBST containing 5 % dried skim milk, the membranes were incubated with 1:500 dilution of monoclonal mouse anti-hepatitis C Core antigen antibody (Abcam Cambridge, UK) at the room temperature for 2 h. Thereafter, the membranes were extensively washed with PBS-Tween and incubated with goat anti-mouse secondary antibody conjugated to alkaline phosphatase (Sigma, St. Louis, Mo., USA) in secondary antibody solution at room temperature for 1 h. Color was developed by incubating the membrane in alkaline phosphate buffer containing tetramethyl benzidine substrate solution.
Immunization of mice
All experiments were carried out on female 6–8-week-old C57BL/6 mice that were purchased from the Institute Pasteur of Iran. Mice were housed for 1 week before the experiment, given free access to food and water and maintained in a light/dark cycle. All experiments were carried out in accordance with the Animal care and use protocol of Golestan University of Medical Sciences of Iran.
Based on the following program, seven groups each containing ten mice were injected sub-cutaneously (s.c.) three times at 2-weeks intervals.
Group l (DNA–DNA) was immunized with three doses of 100 µg pcDNA3-Core, Group 2 (Lambda–Lambda), three doses of 2 × 1012 pfu recombinant Lambda-HCV Core phage nanoparticles (2 × 1012 phages equal to 100 µg DNA per mouse), Group 3 [DNA–Lambda], one dose of 100 µg pcDNA3-Core, boosted with two doses of 2 × 1012 Lambda-HCV Core phage nanoparticles, and Group 4 [Lambda–DNA], one dose of 2 × 1012 recombinant Lambda-HCV Core phage nanoparticles, boosted with two injection of 100 µg pcDNA3-Core. Group 5 received three doses of 2 × 1012 wild Lambda-ZAP phage and was used as phage control. Groups 6 and 7 were injected with three doses of the pcDNA3 and PBS, respectively, as negative controls as has been exhibited in Table 1.
Five mice per group were sacrificed 1 week following the third immunization and the spleens were removed aseptically, and then cell proliferation, cytolytic activity, and cytokine secretion were assayed. Serum samples were collected from other five mice per group 1 week after last immunization.
Preparation of splenocytes
Mice were sacrificed and spleens removed using aseptic technique. Spleens were removed, and the resulting single-cell suspension was pelleted, and the red blood cells were lysed using a lysis buffer (0.15 M NH4Cl; 1 mM KHCO3; 0.1 mM Na2EDTA; pH 7.2). Cells were then washed and counted. Splenocytes were resuspended in RPMI 1640 supplemented with 10 % FCS, 1 % l-glutamine, 1 % HEPES, 0.1 % 2-mercaptoethanol, and 0.1 % penicillin/streptomycin (all from Gibco).
Determination of Ag-Specific Antibody Responses
The humoral immune response was assessed by testing sera from vaccinated mice for antibodies against recombinant HCV Core (Abcam, Cambridge, MA). To check the level of anti-HCV-Core antibodies, the mice were bled from the tail and 5sera/group were collected 1 week after last immunization. To assay anti-HCV-Core antibody activity, ELISA method was employed.
Briefly, 96-well microtiter plates were coated overnight at 4◦C with Core protein in 100 µl of 50 mM sodium carbonate buffer (pH 9.6) per well. The coated wells were blocked for 1 h at 37 °C with 300 µl of 1 % BSA in PBS. Twofold serial diluted serum samples were allowed to react with coated plates at 37 °C for 2 h, and then incubated with 1:5,000–10,000 dilution of goat anti-Mouse IgG conjugated to horse radish peroxidase (Sigma–Aldrich) at 37 °C for 2 h. Plates were incubated with (3,5,3′,5′)-tetramethylbenzidine (TMB) substrate (Sigma-Aldrich), and the colorimetric reaction was stopped by adding 50 µl of 2 M H2SO4. The absorbance of each well was measured at 450 nm with microtiter plate reader (Bio-Rad). Specific IgG antibody isotypes were measured as described above using mouse IgG1 (Sigma) and IgG2c (eBioscince) specific HRP-labeled conjugates.
Lymphocyte proliferation assay
One week after the third immunization, the splenocytes at a concentration of 2 × 105 cells/well were cultured in 96-well flat-bottom culture plates (NalgeNunc International, Denmark) in the presence of 1 μg/ml Core antigen (Abcam, Cambridge, MA), 5 μg/ml PHA (Sigma Chemicals) or media at 37 °C for 48 h in a humidified 5 % CO2 atmosphere. The preparations were cultured in RPMI 1640 supplemented with 10 % FCS. After 48 h of incubation, 10 μg/ml of MTT [3-(4,5-dimethylthiazol-2-yl)-2,5 diphenyltetrazolium bromide]; (Sigma chemicals) was added to each well and incubated for 4 h at 37 °C in 5 % CO2. Following incubation, the supernatant from each well was removed, and formazan crystals were solubilized by adding 100 μl dimethyl sulfoxide into each well.
The absorbance of each well was then determined at a wavelength of 540 nm, and the results expressed as a stimulation index (SI). SI was calculated as follows: SI = OD of stimulated culture/OD of unstimulated culture. All tests were performed in triplicate for each mouse.
In vitro cytotoxic activity
The cytolytic activity of the splenocytes was determined by lactate dehydrogenase (LDH) release assay. One week after the last immunization; single-cell suspension of splenocytes was prepared and used as effector cells. A precise number of 4 × 104 EL4 cells (EL4 was established in tissue culture from a lymphoma Induced in a C57BL/6 mouse by 9, 10-dimethyl-1,2-benzanthracene) in a volume of 100 μl (as a target cells) were incubated with effector cells (100 μl) at different effector/target ratios. For preparation of the target cells, EL4 cells were stimulated with 1 µg/ml HCV Core antigen (Abcam). Released LDH due to cell lysis was measured by LDH release assaying kit (Takara) according to the manufacturer’s instruction. For low and high control wells (spontaneous releasing and maximum releasing, respectively) instead of effector cell suspension, 100 μl of assay medium or 2 % Triton X-100 in assay medium was added. The percentage of specific cytolysis was determined by the following formula:
Specific cytolysis (%) = [optical density (OD) of experimental LDH release − OD of spontaneous LDH release of effector cells − OD of spontaneous LDH release from target cells]/(maximum LDH release of target cells − OD of spontaneous LDH release of target cells) × 100 %. All determinations were performed in triplicate.
Cytokine assay
One week after the final administration, spleens of individual mice were removed and homogenized in RPMI 1640 medium supplemented with 10 % FCS. Red blood cells were osmotically lysed using ammonium chloride buffer (NH4Cl 0.16 M, Tris 0.17 M). Cells were washed twice with RPMI 1640 medium supplemented with 10 % FCS and counted, and the viability was determined by trypan blue exclusion (0.4 % w/v). A total of 1 × 106 spleen cells were added to each well of a 24-well plate. Three wells were considered for each mouse. The cells were restimulated in vitro with 1 µg/ml HCV Core Antigen (Abcam, Cambridge, MA). Plates were incubated at 37 °C in 5 % CO2, and 48 h after stimulation, the supernatants were removed and kept at −70 °C for evaluation of the secreted IFN-γ and IL-4 levels. The concentration of IFN-γ and IL-4 in the supernatants was estimated using a commercial ELISA kit (R & D systems).
Statistical analysis
The results are depicted as the mean ± SD of triplicate determination. Statistical analysis was performed using the ANOVA. A value of p < 0.05 was considered to be significant statistically. All statistical analysis was accomplished with SPSS 18 software.
Results
Confirmation of the HCV Core protein expression
Cloning of Core gene into pcDNA3 vector was verified by restriction analysis and sequencing (Data not shown).
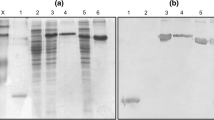
To evaluate the expression of HCV Core gene in the CHO cells, Western blot analysis using monoclonal mouse anti-Hepatitis C Core antibody was used. The transfected CHO lysate with pcDNA3-HCV Core plasmid and infected CHO cell with Lambda-ZAP-HCV Core nanoparticles showed a single band at about 21 kDa for Core in the Western blot. Transfected CHO Cell lysate with pcDNA3 and wild Lambda phage were used as negative control (Fig. 1).
Western blot analysis of HCV core antigen expression by DNA-transfected and lambda-transduced CHO cells. Cell extracts or supernatants were prepared as described in “Materials and methods” section; proteins were separated by SDS polyacrylamide gel electrophoresis, blotted on nitrocellulose membranes, and incubated with the specific anti-Core monoclonal antibody. The arrow indicates the Core protein of approximately 21 kDa of molecule weight. Lane 1 Wild Lambda as lambda negative control, Lane 2 pcDNA3.1 as plasmid negative control, Lane 3 Pre-stained protein markers, lane 3: pcDNA3-Core and Lane 4 Lambda-Core with a molecular weight of 21 kDa
Anti-core antibody response
To compare anti-HCV Core antibodies in the sera induced by DNA and/or recombinant Lambda nanoparticles, seven different immunization groups were tested as follows: priming with DNA and boosting with either DNA (DNA–DNA) or recombinant Lambda nanoparticles (DNA–Lambda); priming with recombinant Lambda nanoparticles and boosting with either recombinant Lambda nanoparticles (Lambda–Lambda) or DNA (Lambda–DNA); wild Lambda as wild bacteriophage, pcDNA3 as negative plasmid and PBS groups. To evaluate different prime–boost strategies, ten mice in each group were immunized thrice at 2-week intervals with 100 µg of DNA or 2 × 1012 pfu of recombinant Lambda nanoparticles encoding HCV Core gene. Figure 2 shows the OD values obtained by direct ELISA performed with diluted sera (1:100) from which the background values obtained with Core coated plates were subtracted.
To determine the anti-HCV core antibodies in the sera of the vaccinated mice (n = 5), a direct ELISA was performed. The absorbance of each well was measured at 450 nm with microtiter plate reader. The highest OD values of Core-specific antibodies were found in the sera of mice with Lambda–Lambda regime. DNA priming/Lambda boosting does not enhance Core-specific antibody than that in the mice immunized with Lambda-DNA and DNA–DNA regimes. ***Indicates statistically significant difference between the DNA–Lambda group with that of Lambda-DNA, DNA- Lambda, and DNA–DNA groups. The graph also shows the statistical significant differences between all treatments and control groups (P < 0.05)
The highest values of Core-specific antibodies were found in the sera of mice with Lambda–Lambda group (P < 0.001) (Fig. 2). However, DNA-Lambda does not enhance core-specific antibody values than that in the mice with Lambda–DNA and DNA–DNA groups.
Antibody isotype responses
Immunoglobulin isotype-specific anti-HCV Core values were quantified by ELISA. The production of IgG1 is primarily induced by Th2-type cytokines, while IgG2c is produced through Th1-type cytokines. The IgG2c/IgG1 ratio can also help to define the T cell phenotype induced by vaccination. Thus, IgG2c/IgG1 ratios were used as indicators of Th1 or Th2 biased responses induced by different prime–boost strategies. Therefore, the isotype of the specific IgG was measured to investigate balance of immune responses. Specific IgG1 and IgG2c antibody subtypes were measured using specific secondary antibodies.
As shown in Fig. 3a, mice immunized with Lambda–Lambda regime elicited higher levels of IgG1 as compared to Lambda–DNA, DNA–Lambda, and DNA–DNA (P < 0.01). Lambda–Lambda and DNA–Lambda regimes showed significant levels of IgG2c as compared to Lambda–DNA and DNA–DNA groups (P < 0.01) (Fig. 3b).
Determination of specific IgG isotypes in mouse sera. Analysis of isotype-specific IgG antibody levels in mice sera. IgG1 (a) and IgG2c (b) values have been represented in two distinct graphs. Each mouse (n = 5) sera analyzed in triplicate by direct ELISA using IgG isotype-specific HRP-labeled detection antibodies. Values for individual isotypes are expressed in OD 450 nm (mean ± SD) of mice in each group. ***Indicates statistically significant difference between the DNA–Lambda group with that of Lambda–DNA, DNA–Lambda, and DNA–DNA (P < 0.001) (a). *** Also indicates statistically significant difference between the Lambda–Lambda and DNA-Lambda groups with that of Lambda-DNA and DNA–DNA groups (P < 0.001) (b). The graphs also shows the statistical significant differences between all treatments and control groups (P < 0.05). The IgG2c/IgG1 ratio, indicative of a Th1/Th2 response, was calculated based on subclass tittering (c)
The induced anti-HCV Core antibodies by the DNA–Lambda regime were predominantly of the IgG2c isotype, implying that DNA vaccine with a recombinant Lambda booster favored a Th1 prone immune response. Whereas vaccination with Lambda–Lambda resulted in a balanced increase of both IgG1 and IgG2c subtypes. There were no significant differences in IgG1 and IgG2c values between the groups receiving Lambda-DNA and DNA–DNA.
Low levels of both IgG1 and IgG2c were found in wild phage, PBS and pcDNA3 control sera. The results are expressed at a serum dilution of 1/100.
We further characterized specific IgG2c/IgG1 antibodies against the HCV core peptide in serum. The IgG2c/IgG1 ratios were calculated as 4 (DNA-Lambda), 2.11 (Lambda–Lambda), 1.25 (Lambda-DNA), 0.8 (DNA–DNA), 0.85 (Wild Lambda), and 1.2 (PBS and pcDNA3) (Fig. 3c).
The results showed that the ratio of IgG2c/IgG1 for the mice immunized with DNA-Lambda was higher than that for the mice immunized with Lambda–Lambda, Lambda–DNA, and DNA–DNA. This analysis suggests that DNA–Lambda drive humoral immune responses toward a Th1 phenotype in vivo.
Lymphocyte proliferation assay
To determine whether splenocyte proliferation response to the DNA vaccine encoding HCV Core may be boosted by Lambda phage nanoparticles, 1 week after last administration, splenocytes from the vaccinated mice were examined for proliferation. As shown in Fig. 4, Lambda–Lambda, DNA–Lambda, Lambda–DNA, and DNA–DNA produced significant lymphocyte responses against HCV Core antigen compared to wild lambda or pcDNA3 groups (P < 0.05).
Splenocyte proliferation levels after in vitro stimulation with HCV Core antigen. The mice were injected sub-cutaneously (s.c.) three times at 2-weeks intervals with different prime–boost regimes of DNA and Lambda phage nanoparticles expressing the HCV Core. One week after final immunization, spleens of individual mice (five per group) were removed and splenocytes from the immunized mice were incubated with HCV Core protein (1 μg/ml) for 2 days and then measured by the MTT assay to calculate the SI. Values are the mean ± standard error of the mean for the experiments. All tests were performed in triplicate for each mouse.***Indicates statistically significant difference between the DNA-Lambda group with that of Lambda–Lambda, Lambda–DNA, and DNA–Lambda groups (P < 0.001). The graph also shows the statistical significant differences between all treatments and control groups (P < 0.05)
Splenocyte proliferation response to the DNA-Lambda nanoparticles was about twofold higher than that to the Lambda–Lambda vaccine. DNA–Lambda immunization significantly enhanced proliferation response, which implied an antigen-specific boosting effect by the recombinant Lambda nanoparticles. The results also demonstrated that heterologous immunization of DNA–Lambda induced statistically the most significant proliferation response than homologous (Lambda–Lambda and DNA–DNA) and heterologous (Lambda–DNA) prime- boost groups.
LDH cytolytic activity
In order to analyze the capacity of recombinant HCV Core to enhance the Core-specific cytotoxic T lymphocyte (CTL) response, splenic cells, derived from the immunized mice 1 week after the last administration, were restimulated specifically by naive El4 (target cells) pulsed with Core-specific antigen in vitro. The cytotoxic activity was tested by non-radioactive LDH release assay. As shown in Fig. 5, all the treatment groups (except DNA–DNA group) given Lambda–Lambda, DNA–Lambda, and Lambda–DNA regimes produced significant cytolytic activity compared to wild lambda or pcDNA3 control groups. DNA-Lambda immunization resulted in strong lytic activity, with almost 60 % specific lysis at 100:1 E/T ratio, versus DNA–DNA (∼35 % at100:1 E/T ratio) and Lambda-DNA (∼42 % at 100:1 E/T ratio). CTL responses induced by the Lambda–Lambda vaccine (∼62 % at 100:1 E/T ratio) appeared to be slightly higher than those induced by DNA–Lambda, though this was not statistically significant.
Analysis of the cytotoxic activity of CTLs induced by prime–boost combinations of DNA and Lambda phage nanoparticles expressing the HCV core (Five mice/group). CTL activity of the lymphocytes from immunized mice was measured by LDH release assaying kit (Takara) as described in “Materials and methods” section. Target cells were stimulated with the corresponding antigens, and mean percentages of specific lysis from five mice were determined at different effector to target (E:T) ratios. Specific lysis of target cells is shown with nonspecific background lysis subtracted. ***indicates statistically significant difference between the Lambda–Lambda and DNA–Lambda groups as determined by one-way ANOVA with that of Lambda–DNA and DNA–DNA groups (P < 0.001). The graph also shows the statistical significant differences between all treatments and control groups (P < 0.05)
Collectively, our results indicate that the strengths of the CTL responses induced by the DNA–Lambda and Lambda–Lambda groups are similar.
Cytokine assay
Th1 cytokines (IFN-γ) and Th2 cytokines (IL-4) are major parameters in our understanding of the polarization of immune responses. Th1 immune responses are thought to drive induction of cellular immunity, whereas Th2 immune responses preferentially drive humoral immunity. Therefore, we examined the cytokine secretion profiles 1 week after the last immunization. As demonstrated in Fig. 6a, b, in mice immunized by prime–boost strategy, the levels of IFN-γ and IL-4 were significantly higher (P < 0.05), compared with the negative groups.
Concentration of IFN-γ and IL-4 in supernatant following stimulation of splenocytes with HCV Core protein (1 μg/ml) (Five mice/group). The concentration of IFN-γ and IL-4 in the supernatants was estimated using a commercial ELISA kit. Data presented as mean ± standard deviation for five mice per group. **indicate statistically significant difference between the Lambda–Lambda and DNA–Lambda groups as determined by one-way ANOVA with that of Lambda–DNA and DNA–DNA groups (P < 0.01) (a). ** Also indicates statistically significant difference between the Lambda–Lambda group as determined by one-way ANOVA with that with DNA–Lambda, Lambda–DNA, and DNA–DNA groups (P < 0.01) (b). The graphs also shows the statistical significant differences between all treatments and control groups (P < 0.05)
As shown in Fig. 6a, the level of IFN-γ was significantly increased in DNA–Lambda and Lambda–Lambda groups, compared to Lambda–DNA and DNA–DNA vaccination groups (P < 0.01). However, there was no significant difference between DNA–Lambda and Lambda–Lambda (P > 0.05).
As demonstrated in Fig. 6b, in mice immunized by prime–boost strategy, the levels of IL-4 were increased (P < 0.05), compared with the negative groups. Lambda–Lambda significantly increased the secretion of and IL-4 compared with other prime–boost strategy (P < 0.01). In contrast, there was no significant difference in IL-4 levels between DNA–Lambda, Lambda–DNA, and DNA–DNA groups (P > 0.05). This profile of cytokine secretion suggests that the cellular immune response induced by the heterologous DNA prime–Lambda boost strategy was skewed to the Th1-type.
Discussion
It is known that individuals who recover from acute HCV infection appear to have quantitatively vigorous CD4+ proliferative and CD8+ cytotoxic T cell responses against one or more HCV proteins compared with those individuals who develop chronic disease [23]. Specifically, a strong T cell response against the structural proteins of the virus, in particular the Core protein, has been reported to be related to the clinical outcome and the kinetics of viral replication in HCV infection [24]. Many of the studies with DNA vaccines have focused on using plasmids expressing the HCV Core antigen, the results indicated generally low helper T (Th) cell and humoral immune responses against the Core protein in mice [25, 26]. Thus, DNA vaccines need to be made much more potent to be candidates for immunization. DNA vaccines are good at priming immune responses, but are inefficient at boosting immune response, whereas eukaryotic viral vectors, in particular adenoviruses, are good at boosting but suffer from the imitation of low clinical efficacy, because of viral vector-specific immunity and neutralization.
These considerations recommend that the ideal boosting vaccine might consist of a safe and non-pathogenic vector that can stimulate potent immune responses. Recently, Lambda bacteriophages have been used as vehicles for the delivery of vaccines in our previous studies [18]. Bacteriophage-based vectors have many of the desirable properties of both animal viral and non-viral systems. However, it is unclear if this vector is also capable of boosting DNA vaccine-induced-immune response as a booster immunogen efficiently.
In the current study, we have evaluated the homologous and heterologous prime–boost immunizations consisting of various combinations of DNA vaccine and recombinant Lambda phage nanoparticles encoding HCV Core for the induction of HCV-Core-specific cellular and humoral immunity. We demonstrated for the first time that a heterologous prime–boost strategy, DNA-Lambda nanoparticles, which encode HCV Core antigen, can efficiently induce strong antigen-specific splenocyte proliferation and CTL cytotoxic responses.
It will be interesting to determine the underlying mechanism as to how DNA prime–Lambda markedly boosts Th1 responses.
The use of the bacteriophage as a delivery vehicle is a novel approach in which a eukaryotic expression cassette is cloned into a bacteriophage genome and whole phage particles are packaged and injected as vaccine. The strategy have several features including expression of vaccine antigen within host cells so that produced proteins exhibit fully eukaryotic post-translational modifications along with induction of strong immune response.
March and colleagues have shown that multiple injections of whole Lambda phages containing hepatitis B surface antigen (HBs Ag) results in a strong antigen-specific humoral immune response in mice and rabbits [27]. In our previous study, we demonstrated that subcutaneous administration of Lambda-ZAP bacteriophage encoding HPV-16 E7 gene as therapeutic cancer vaccine protected against TC-1 tumor [19]. The study showed significant secretion of IFN-γ and granzyme B along with induction of cytotoxic T lymphocytes. The results of recombinant Lambda phage nanoparticles provided better tumor protection than DNA vaccine employing the same gene. Together, our results revealed several surprising aspects of Lambda phage and have interesting implications for designing a new prime/boost strategy involving Lambda vaccine.
In our study, the Lambda–Lambda and DNA prime–Lambda boost regime elicited very similar cell-mediated responses, both inducing a cellular immune response characterized by a predominance of IFN-γ (Fig. 6a) and high CD8+ cytotoxicity (Fig. 5).
Other results also demonstrated that the mice immunized with heterologous DNA prime–Lambda boost induce strong lymphocyte proliferation response (Fig 4) in C57BL/6 mice, whereas homologous Lambda–Lambda prime–boost elicited antibody (Figs. 2 and 3a), but moderate lymphocyte proliferation responses compared to other groups.
Antibody isotype analysis showed that Lambda–Lambda induced balanced amounts of IgG1 and IgG2c antibodies, whereas the anti-Core antibodies elicited by DNA-Lambda were predominantly of the IgG2c isotype (Fig 3b), indicating a highly polarized Th1-type response. The Th1 cytokine IFN-γ is important in vivo for enhancement of IgG2c secretion, and the secretion of IgG2c isotype showed a strong inverse correlation with the concentration of IL-4 in DNA-Lambda immunized mice.
It is noteworthy that the DNA prime–Lambda boost was absolutely superior to the reverse sequence (Lambda-DNA) in the induction of HCV Core-specific IFN-γ, lymphocyte proliferation, CTL responses, and IgG2c level. However, no significant difference was found between two groups for IgG1 subtype levels. Our findings are in accordance with previous studies showing that DNA prime/adenoviral vector boost elicited the highest level of Th1 CD4+ T cell responses when compared to the reversed adenoviral prime–DNA boost with the same vaccines [28]. Fournillier et al. [29] also demonstrated that heterologous prime–boost immunizations with a DNA and a MVA vaccine successfully enhanced the number of multifunctional T cells compared with each vaccine alone.
So its possible that Lambda phage nanoparticles are very good boosting vaccine candidate just as DNA is quite effective as an initial immunogen in a bimodal vaccine strategy.
The capacity of our Lambda nanoparticles (with or without DNA vaccine priming) to induce HCV-specific CD8+ T lymphocyte (CTL) together with specific lymphocyte proliferation responses and the fact that overall induced responses were capable to control splenic or hepatic expression of a HCV antigen, are extremely encouraging [30].
In the chronic HCV infection, frequency of HCV-specific CTLs in the peripheral blood appears to be quite low. Therefore, it is believed that T cell responses, including both CTLs and helper T lymphocytes, are critical in determining the outcome. In addition to quantity of T cell responses, it has been described that HCV-specific CD8+ T cells in the chronic HCV infection display less HCV-specific cytotoxicity, impaired lymphocyte proliferation and less IFN-γ secretion [31, 32]. However, Lambda phage nanoparticles as a vaccine and booster vaccine may induce the normal development of cellular immunity in patients with chronic HCV infection, thereby causing improved induction of HCV-specific CTLs.
In conclusion, we have for the first time designed and generated a safe HCV candidate vaccine based on a recombinant Lambda phage nanoparticle vaccine encoding HCV Core antigen. We showed that the nanoparticle in the prime–boost immunization enhanced the induction of HCV-specific cellular immunity with a Th1 profile in mice. Further analysis of the mechanisms of the improved immunogenicity of the Lambda phage nanoparticles would facilitate the design of universally efficacious vaccines against HCV infections.
Abbreviations
- HCV:
-
Hepatitis C virus
- PHA:
-
Phytohemaglotinin
- APC:
-
Antigen-presenting cell
- CTL:
-
Cytolytic T lymphocyte
- IFN-γ:
-
Interferon γ
- IL-4:
-
Interleukin 4
- PMSF:
-
Phenylmethanesulfonyl fluoride
- LDH:
-
Lactate dehydrogenase
- MTT:
-
3[4,5-dimethylthiazol-2-ll]-2,5-diphenyltetrazolium bromide, thiazolyl-blue
- CHO:
-
Chinese hamster ovary
- OD:
-
Optical density
- FCS:
-
Fetal calf serum
- RPMI:
-
1640 Roswell Park Memorial Institute (name of the medium)
- Th:
-
T helper
References
L. Zhang, M. Gwinn, D.J. Hu, Public Health Genomics 16, 192–197 (2013)
L. Mailly, E. Robinet, P. Meuleman, T.F. Baumert, M.B. Zeisel, Front. Microbiol. 4, 213 (2013)
F. Poordad, D. Dieterich, J. Viral Hepat. 19, 449–464 (2012)
L.G. Guidotti, F.V. Chisari, Annu. Rev. Pathol. 1, 23–61 (2006)
G.M. Lauer, E. Barnes, M. Lucas, J. Timm, K. Ouchi, A.Y. Kim, C.L. Day, G.K. Robbins, D.R. Casson, M. Reiser, G. Dusheiko, T.M. Allen, R.T. Chung, B.D. Walker, P. Klenerman, Gastroenterology 127, 924–936 (2004)
J.M. Pestka, M.B. Zeisel, E. Blaser, P. Schurmann, B. Bartosch, F.L. Cosset, A.H. Patel, H. Meisel, J. Baumert, S. Viazov, K. Rispeter, H.E. Blum, M. Roggendorf, T.F. Baumert, Proc. Natl. Acad. Sci. USA 104, 6025–6030 (2007)
M. Houghton, Immunol. Rev. 239, 99–108 (2011)
J. Torresi, D. Johnson, H. Wedemeyer, J. Hepatol. 54, 1273–1285 (2011)
J.M. Pawlotsky, Nat. Rev. Gastroenterol. Hepatol. 6, 383–385 (2009)
Q.L. Choo, G. Kuo, R. Ralston, A. Weiner, D. Chien, G. Van Nest, J. Han, K. Berger, K. Thudium, C. Kuo et al., Proc. Natl. Acad. Sci. USA 91, 1294–1298 (1994)
C.S. Klade, H. Wedemeyer, T. Berg, H. Hinrichsen, G. Cholewinska, S. Zeuzem, H. Blum, M. Buschle, S. Jelovcan, V. Buerger, E. Tauber, J. Frisch, M.P. Manns, Gastroenterology 134, 1385–1395 (2008)
M. Puig, K. Mihalik, J.C. Tilton, O. Williams, M. Merchlinsky, M. Connors, S.M. Feinstone, M.E. Major, Hepatology (Baltimore, Md) 44, 736–745 (2006)
A. Folgori, S. Capone, L. Ruggeri, A. Meola, E. Sporeno, B.B. Ercole, M. Pezzanera, R. Tafi, M. Arcuri, E. Fattori, A. Lahm, A. Luzzago, A. Vitelli, S. Colloca, R. Cortese, A. Nicosia, Nat. Med. 12, 190–197 (2006)
R.R. Amara, J.M. Smith, S.I. Staprans, D.C. Montefiori, F. Villinger, J.D. Altman, S.P. O’Neil, N.L. Kozyr, Y. Xu, L.S. Wyatt, P.L. Earl, J.G. Herndon, J.M. McNicholl, H.M. McClure, B. Moss, H.L. Robinson, J. Virol. 76, 6138–6146 (2002)
D. Ferber, Science (New York, NY) 294, 1638–1642 (2001)
N. Somia, I.M. Verma, Nat. Rev. Genet. 1, 91–99 (2000)
C.D. Jepson, J.B. March, Vaccine 22, 2413–2419 (2004)
A. Ghaemi, H. Soleimanjahi, P. Gill, Z. Hassan, S.R. Jahromi, F. Roohvand, Genet. Vaccines Ther. 8, 3 (2010)
A. Ghaemi, H. Soleimanjahi, P. Gill, Z.M. Hassan, S. Razeghi, M. Fazeli, S.M. Razavinikoo, Intervirology 54, 105–112 (2011)
J.R. Clark, K. Bartley, C.D. Jepson, V. Craik, J.B. March, FEMS Immunol. Med. Microbiol. 61, 197–204 (2011)
J.R. Clark, J.B. March, FEMS Immunol. Med. Microbiol. 40, 21–26 (2004)
T. Akuta, A. Eguchi, H. Okuyama, T. Senda, H. Inokuchi, Y. Suzuki, E. Nagoshi, H. Mizuguchi, T. Hayakawa, K. Takeda, M. Hasegawa, M. Nakanishi, Biochemical and biophysical research communications 297, 779–786 (2002)
A. Grakoui, N.H. Shoukry, D.J. Woollard, J.H. Han, H.L. Hanson, J. Ghrayeb, K.K. Murthy, C.M. Rice, C.M. Walker, Science (New York, NY) 302, 659–662 (2003)
G.A. Elmowalid, M. Qiao, S.H. Jeong, B.B. Borg, T.F. Baumert, R.K. Sapp, Z. Hu, K. Murthy, T.J. Liang, Proc. Natl. Acad. Sci. USA 104, 8427–8432 (2007)
J. Cao, Z. Chen, Y. Ren, Y. Luo, M. Cao, W. Lu, P. Zhao, Z. Qi, Vaccine 29, 3714–3723 (2011)
M.G. Isaguliants, N.V. Petrakova, E.V. Kashuba, Y.G. Suzdaltzeva, S.V. Belikov, V.V. Mokhonov, A.G. Prilipov, L. Matskova, I.S. Smirnova, C. Jolivet-Reynaud, E. Nordenfelt, Vaccine 22, 1656–1665 (2004)
J.B. March, J.R. Clark, C.D. Jepson, Vaccine 22, 1666–1671 (2004)
S.H. Park, S.H. Yang, C.G. Lee, J.W. Youn, J. Chang, Y.C. Sung, Vaccine 21, 4555–4564 (2003)
A. Fournillier, L. Frelin, E. Jacquier, G. Ahlen, A. Brass, E. Gerossier, F. Holmstrom, K.E. Broderick, N.Y. Sardesai, J.Y. Bonnefoy, G. Inchauspe, M. Sallberg, J. Infect. Dis. 208, 1008–1019 (2013)
A. Fournillier, E. Gerossier, A. Evlashev, D. Schmitt, B. Simon, L. Chatel, P. Martin, N. Silvestre, J.M. Balloul, R. Barry, G. Inchauspe, Vaccine 25, 7339–7353 (2007)
M. Matsui, O. Moriya, M.L. Belladonna, S. Kamiya, F.A. Lemonnier, T. Yoshimoto, T. Akatsuka, J. Virol. 78, 9093–9104 (2004)
Spangenberg H.C., Viazov S., Kersting N., Neumann-Haefelin C., McKinney D., Roggendorf M., von Weizsacker F., Blum H.E., and Thimme R., Hepatology (Baltimore, Md) 42, 828-837, 2005
Acknowledgments
The authors appreciate the financial support of the Research Deputy at Golestan Medical University. This project was extracted from a MSC thesis.
Conflict of interest
All the authors have no conflicting interests.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Saeedi, A., Ghaemi, A., Tabarraei, A. et al. Enhanced cell immune responses to hepatitis c virus core by novel heterologous DNA prime/lambda nanoparticles boost in mice. Virus Genes 49, 11–21 (2014). https://doi.org/10.1007/s11262-014-1070-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-014-1070-z