Abstract
A new, widespread disease was recently observed in soybean in the United States. The disease, named Soybean vein necrosis, is manifested by intraveinal chlorosis and necrosis, and has been found in almost all of the 50 fields visited over a period of 3 years in the midwest and midsouth part of the United States. A virus was isolated from symptomatic material, and detection protocols were developed. More than 150 symptomatic specimens collected from seven US States were tested, and all were found positive for the virus unlike 75 asymptomatic samples, revealing the absolute association between virus and disease. Protein pairwise comparisons coupled with phylogenetic analyses indicate that the virus is a new member of the genus Tospovirus.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Soybean [Glycine max (L.) Merr.] is one of the most important crops in worldwide agriculture. There are several viruses reported in the crop [1], many of which cause significant losses worldwide, estimated to be well over two million metric tons annually [2], whereas new virus diseases are being identified continually [3–5]. The United States is the leading producer of soybeans accounting for about one-third of the world production with more that 30 million hectares being planted and about 90 million ton being harvested in 2009 [6]. Many viruses, including Alfalfa mosaic, Soybean mosaic and Bean pod mottle are listed as pests of great importance for the US soybean industry [7].
A disease with virus-like symptoms was observed in soybean fields in Arkansas, Kansas, Missouri, Illinois, Mississippi, Tennessee, and Kentucky. First symptoms were observed in early June in Arkansas and Illinois and were manifested as vein clearing. As the season progressed, the affected areas became chlorotic or necrotic, and in severe cases leaves died off (Fig. 1).
Immunological tests failed to identify an agent associated with the disease, and the possibility of a new virus infecting soybean was investigated. Indeed, a new virus was found in symptomatic material and detection protocols were developed. Surveys in several states demonstrated that the virus was present in all the samples showing typical disease symptoms. The importance of soybean in worldwide agriculture in combination with the high prevalence of this virus in major soybean production areas in the United States has rendered its characterization essential.
Materials and methods
Soybean virus testing
Samples showing soybean vein necrosis disease symptoms were tested by ELISA for 16 viruses or virus groups at Agdia, Inc. (Elkhart, IN). The viruses assayed were Alfalfa mosaic virus, Bean pod mottle virus, Cowpea mosaic virus, Cucumber mosaic virus, Impatiens necrotic spot virus (INSV), Peanut stunt virus, Southern bean mosaic virus, Soybean dwarf virus, Soybean mosaic virus, Tobacco mosaic virus, Tobacco ringspot virus, Tobacco streak virus, Tomato ringspot virus, Tomato spotted wilt virus (TSWV), Watermelon mosaic virus-2, and Potyvirus group.
Nucleic acid purification
Double-stranded RNA (dsRNA) isolation from symptomatic soybean leaves was performed using a lithium salt extraction protocol as described by Tzanetakis and Martin [8] with the following modification: the digested nucleic acids were bound to 20-μl silica milk (Sigma) in the presence of ethanol (instead of CF-11 cellulose) and were then eluted in 100 μl TE instead of being subjected to ethanol precipitation. For detection purposes, the total nucleic acid extraction protocol of Tzanetakis et al. [9] was used.
Detection and geographic distribution
Purified nucleic acids were subjected to reverse transcription using Maxima® reverse transcriptase (Fermentas) according to the manufacturer’s instructions using virus-specific primers, LdetR and SdetR (200 nM final concentration) (Table 1). The purified nucleic acids made to 10% of the total reaction volume. For PCR, two oligonucleotide primer sets (Table 1) that amplify regions of the L (LdetF/LdetR) and the S (SdetF/SdetR) RNAs (297 and 861 nucleotides, respectively) were optimized. The final concentration of the virus primers were 400 nM. The PCR was performed in a Mastercycler® (Eppendorf) using Taq polymerase (Genscript) at a concentration of 0.25 μl reaction consisting of 2-min incubation at 94°C followed by 40 cycles of 30-s denaturation at 94°C, 10-s extension at 55°C, and 1-min extension at 72°C. The program terminated with 10-min incubation at 72°C. The validity of the tests was confirmed after sequencing at least 30 amplicons for each primer set at the University of Arkansas Sequencing Center. The geographic distribution of the virus was studied by applying virus-specific RT-PCR tests on 150 symptomatic and 75 asymptomatic samples collected from 50 production fields and variety trials from Arkansas, Tennessee, Illinois, Missouri, Kansas, Kentucky, and Mississippi.
Genome characterization and phylogenetic analyses
DsRNA was used as template for shotgun cloning as previously described [10]. Sequences were compared with those found in Genbank using BLAST [11], and those of tospovirus origin were used to design oligonucleotide primers to acquire the complete genome of the virus as previously described [12; Table 1]. The end of the virus segments were obtained after two-tailing reactions as previously described [13]. Genome assembly was performed using CAP3 [14]. The complete virus sequences have been deposited in Genbank under accession Nos HQ728385-7.
Sequence comparisons between the new virus proteins and their orthologs were performed using MatGAT [15; Table 2]. Additional in-silico protein analysis included the identification of RNA-binding domains using BindN tool at 90% specificity [16]. The putative signal peptides, transmembrane domains, and glycosylation sites were predicted using the SignalP 3.0 [17], TMHMM 2.0 [18], NetNGlyc 1.0, and NetOGlyc 3.1 [19]. For phylogenetic analysis, the L RNA polyprotein and nucleoprotein amino acid sequences were used. Analyses were performed using ClustalW employing a neighbor-joining algorithm with Kimura’s correction and bootstrap analysis consisting of 1000 pseudoreplicates [20]. Phylograms were visualized on TreeView [21].
Results and discussion
Soybean vein necrosis symptoms were observed in all but one of the 50 fields surveyed signifying the importance of the disease and the need to study it. Although disease symptoms have not been associated with any previously described soybean viruses, testing for 16 viruses and virus groups minimized the possibility of mixed infections with synergistic effects resulting in leaf necrosis. No known virus was found being consistently associated with vein necrosis symptoms in these tests.
Shotgun cloning revealed the presence of a tospovirus and a new endornavirus, endogenous dsRNA molecules found in several plants, but never associated with disease symptoms [22]. Notwithstanding the above, testing for the endornavirus did not reveal any association between the dsRNA molecule and disease (Tzanetakis, unpublished). Detection protocols were developed and used to assess the presence of the virus in a wide geographic area, including major soybean-producing areas in the United States. All the 150 samples with typical soybean vein necrosis symptoms tested positive with two oligonucleotide primer sets, targeting different virus segments. Seventy-five asymptomatic plants from fields with the disease did not yield any amplicons. This perfect association of virus and symptoms indicates that the virus is likely the cause of the disease, and thus, the name Soybean vein necrosis-associated virus (SVNaV; Fig. 2) is proposed.
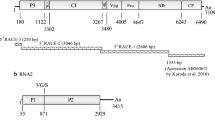
a Genome organization of Soybean vein necrosis associated virus. M RNA non-structural protein (NSm); glycoproteins (Gc/Gn); S RNA non-structural protein (NSs); and nucleoprotein (N). b Hybridization of the untranslated region termini of the genomic RNAs (L, M, S) of Soybean vein necrosis-associated virus using mfold [31]. Arrows point to mismatches found in the stems at the termini of the molecules
The SVNaV was fully sequenced, which revealed several typical and atypical characteristics for the members of the genus Tospovirus. All the SVNaV segments have the highly conserved Tospovirus 5′ terminal sequence (AGAGCA1–6) predicted to be crucial as replication and transcription signals [23]. SVNaV L RNA is 9010 nucleotides (nt), the longest among the sequenced tospoviruses. Similar to the other tospoviruses, the first and the last 19 nts are complementary, leading to the circularization of the molecule by the formation of a pan-handle structure [24; Fig. 2]. SVNaV L RNA has a single open reading frame (ORF) in the negative orientation starting at nt 8980 and terminating at nt 185 encoding for a putative polyprotein of 336 KDa. Typical RNA-dependent RNA polymerase (RdRp) domains (fingers, palm, and thumb) forming an open “U-shape” crevice and conserved sequences reported in the family Bunyaviridae, and other segmented negative-strand viruses, were identified in the protein [25]. Motif A (DXXKWS)539–544, and motif C (SDD)665-667 are present in the palm domain, being involved in divalent metal–cation binding [26]. Mutation of the Asp in the two motifs (motif A: D539 and motif C: D667) abolished polymerase activity in Bunyamwera virus [27]. The Gly528 conserved in almost all viral RdRp, is present in motif B (QGXXXXXSS)527–535 [28] and may, because of its mobility, play a crucial role in RNA binding. Lys712 (motif D) is presumably involved in catalytic activity of the enzyme because of its proximity in the tertiary structure of the protein to motif A Asp539 as determined for Lassa fever virus [26]. Motif E (EFXSE)721–725 is involved in cap scavenging and has endonuclease activity [28]. Region F, the NTP-binding site, is subdivided into three domains; F1: (KX)451–452, F2: (KXQR)459–462, and F3:(TXXDRXIY)463–470 [29]. Several RNA-binding domains participating in the formation of the RNA replication complex and maintaining nucleotides in specific positions were identified, including TSSSGSK2900–2906 and KWSKPKKKKKPKAKPKKSKKKHNK2908–2931, at the C-terminus of the protein [30].
The M RNA is 4955 nt and similar to the L segment, the end sequences are complementary; however, instead of the 19 nt in the L RNA, the first and the last 27 nts form the pan-handle structure, with two mismatches [31; Fig. 2]. The segment codes an ORF in the positive, and one in the negative orientation. ORF1 initiates at nt 58 and terminates at nt 1008 coding for a non-structural protein (NSm) of 36 kDa. The protein is probably involved in cell-to-cell movement, as the highly conserved LXDX40G motif of the “30 K movement protein superfamily” was identified in several orthologs [32–34], but SVNaV has an Ile instead of Leu at the beginning of the motif (IXDX40G165–208). The “P/D-L-X motif” and phospholipase A2 catalytic site, though present in some tospovirus NSm including TSWV, Groundnut bud necrosis virus (GBNV), Chrysanthemum stem necrosis virus (CSNV), and Tomato chlorotic spot virus (TCSV), are absent from the SVNaV ortholog. A glycosylation site was predicted at N195. ORF2 starts at nt 4863 extending to nt 1276 and codes for the precursor of the virus glycoproteins. As secretory proteins, the transportation of the mature glycoproteins from the cytosol to the endoplasmic reticulum may be directed by a signal peptide, such as the one predicted to be cleaved from the N-terminus of the protein. The 26 amino acid (aa) peptide primarily comprises basic and hydrophobic amino acids. The putative signal cleavage –or the mature proteins (Gn/Gc), predicted after alignment of tospovirus orthologs, is found between Cys378 and Ser379 yielding the 43-kDa-Gn and 91-kDa-Gc [35]. N-Glycosylation sites of Gn were predicted at N25, N229, and N343, whereas the three transmembrane domains are located between aa 6–28, 317–339, and 349–371. The RGD Gn domain, predicted to play a critical role in virion–cell attachment was found at positions 29–31 [35, 36]. A series of conserved domains present in Tospovirus Gcs are also present in the SVNaV ortholog, including a highly conserved Lys702, a motif consisting of T-X-T714-716, followed by CTGXC730–734 and TSXWGCEEXXCXAXXXGXXXGXC754–776 [36]. A transmembrane domain is found among Gc aa 77–99, leaving a long aa tail inside the virus particle. N-glycosylation modifications are predicted at three positions: N5, N20, and N171 whereas no O-glycosylation site was predicted in either of the glycoproteins.
The S RNA is 2603 nt, has the first and the last six nucleotides complementary, and similar to the other two RNAs, its untranslated region is highly structured (Fig. 2). Similar to the M RNA, the S RNA encodes one ORF each in the positive and negative orientations. ORF1 initiates at nt 59 and terminates at nt 1381 encoding a 50-kDa non-structural protein (NSs), a putative-silencing suppressor [37]. The conserved GK178–179 motif is present in the Walker motif A, where K179 is believed to interact with the ATP/ADP phosphate [38]. The SVNaV NSs Walker motif B (DEXX148–151) is located upstream motif A, similar to Watermelon silver mottle virus (WSMoV), GBNV, and Capsicum chlorosis virus. The conserved DE148–149 is presumably involved in Mg2+ binding and ATP hydrolysis. Lokesh et al. [39] have recently reported NSs to be a biofunctional enzyme: its NTPase activity hydrolyzes ATP required by Dicer to process and unwind siRNA. It also functions as phosphatase removing the phosphoryl group from the 5′ end of dsRNA, making it unidentifiable by Dicer. The issue of the SVNaV NSs employs the same strategy and functions as an RNA-silencing suppressor remains to be studied. ORF2 spans from nt 2533 to 1700, encoding for a putative nucleoprotein (N) of 31 kDa. N may be essential for RNA synthesis together with the L RNA polyprotein [30]. Similar to the TSWV ortholog, the C-terminal motif KKDGKGKKSK264–273 was predicted to bind RNA [40], whereas other discrete amino acids, including PSN7–9, RK51–52, RY54–55, and KK73–74 may interact with the virus RNAs to prevent premature termination caused by base pairing of the newly synthesized RNA strands and to protect it from degradation [30].
The SVNaV is a distinct member of the genus Tospovirus, as it shares minimal similarity with any of the established species in the genus—all SVNaV proteins exhibit less than 40% aa identity to their orthologs. Many of the characteristic tospovirus protein motifs were identified in the SVNaV orthologs, whereas certain unique features, such as the very large polyprotein and the inverted Walker motif in NSs, add to the complexity of the genus.
When SVNaV is compared to other tospoviruses using the L RNA polyprotein-RdRp, the hallmark gene for any virus evolution study and an excellent predictor of tospovirus phylogeny [41], it becomes evident that the virus occupies the phylogenetic space between the New and Old World tospoviruses, forming a new clade in the genus (Fig. 3). Because of lack of significant number of completely sequenced Tospovirus polyproteins, the nucleoprotein was also used for assessing the phylogenetic placement of SVNaV. The virus position in Tospovirus evolution stands, but more clades emerge, with the most pronounced being that of Peanut chlorotic fan-spot/Peanut yellow spot viruses. These two viruses are not fully characterized, and thus, their phylogenetic placement remains uncertain.
Unrooted phylograms using the amino acid sequences of a the L RNA polyprotein and b the nucleoprotein of Soybean necrosis-associated virus (SVNaV), and other members of the genus Tospovirus. Abbreviations and Genbank accession numbers (Polyprotein in plain text, nucleoprotein in italics): TSWV, Tomato spotted wilt virus, D10066, NP_049361; INSV, Impatiens necrotic spot virus, X93218, NP_619709; GBNV, Groundnut bud necrosis virus, AF025583, NP_619701; WSMoV, Watermelon silver mottle virus, AF133128, NP_620771; CaCV, Capsicum chlorosis virus, DQ256124, YP_717923; MYSV Melon yellow spot virus, AB061774, YP_717921; TZSV, Tomato zonate spot virus, EF552435, YP_001740044; CCSV, Calla lily chlorotic spot virus, FJ822962, AAW58115; IYSV, Iris yellow spot virus, FJ623474, ACN62253; PCFSV, Peanut chlorotic fan-spot virus, AAC99405; WBNV, Watermelon bud necrosis virus, ADD83166; ZLCV, Zucchini lethal chlorosis virus, AAF04198; PYSV, Peanut yellow spot virus, AAB94022; TCSV, Tomato chlorotic spot virus, AAL07433; GRSV, Groundnut ringspot virus, AAM47011; ANSV, Alstroemeria necrotic streak virus, ACZ18222; MSMTV, Melon severe mosaic tospovirus, ABX72231; PRTV, Polygonum ringspot tospovirus, ABO31117; and TNRV, Tomato necrotic ringspot virus, ACK99533. The bootstrap values are based on 1000 pseudoreplicates, and the support of under 70% is not shown, being considered as unreliable. Bars represent 0.1 amino acid changes per site
Tospoviruses cause significant losses and inferior quality of produce in vegetables, legumes, and ornamentals among other crops around the world. Some members of the family (i.e., TSWV and INSV) infect both dicots and monocots with host ranges that exceed several hundred species causing losses in the range of billions of US dollar [42, 43]. The prevalence of disease and virus in almost all the fields visited makes it a potential threat to the soybean industry.
While this article presents the molecular characterization of SVNaV, a virus found to be in perfect association to Soybean vein necrosis, fulfillment of Koch postulates is needed before proving that it is the causal agent of the disease. We are taking up the study on the biologic characterization and epidemiology including potential vectors, alternative hosts, and genetic diversity, aspects of the virus biology that will help us better understand the disease and assist in the implementation of disease-control strategies.
References
G.L. Hartman, J.B. Sinclair, J.C. Rupe, 4th edn. (American Phytopathological Society, St. Paul, 1999), pp. 100
A. Wrather, G. Shannon, R. Balardin, L. Carregal, R. Escobar, G.K. Gupta, Z. Ma, W. Morel, D. Ploper, A. Tenuta, Plant Health Prog. (2010). doi:https://doi.org/10.1094/PHP-2010-0125-01-RS
T. Kuroda, K. Nabata, T. Hori, K. Ishikawa, T. Natsuaki, J. Gen. Plant Pathol. 76, 382–388 (2010)
R.L. Lamprecht, G.G.F. Kasdorf, M. Stiller, S.M. Staples, L.H. Nel, G. Pietersen, Plant Dis. 94, 1348–1354 (2010)
N. Nam, S.M. Kim, L.L. Domier, S. Koh, J. Moon, H.S. Choi, H.G. Kim, J.S. Moon, S.H. Lee, Arch. Virol. 154, 1679–1684 (2009)
Anonymous, U.S. Soybean Industry: Background Statistics and Information. (2011). http://www.ers.usda.gov/news/soybeancoverage.htm
Anonymous, Soybean Checkoff Research Database. (2011). http://www.soybeancheckoffresearch.org/index.html
I.E. Tzanetakis, R.R. Martin, J. Virol. Method 149, 167–170 (2008)
I.E. Tzanetakis, A.B. Halgren, N. Mosier, R.R. Martin, Virus Res. 127, 26–33 (2007)
I.E. Tzanetakis, K.E. Keller, R.R. Martin, J. Virol. Method 124, 73–77 (2005)
S.F. Altschul, T.L. Madden, A.A. Schäffer, J. Zhang, Z. Zhang, W. Miller, D.J. Lipman, Nucleic Acids Res. 25, 3389–3402 (1997)
I.E. Tzanetakis, J.D. Postman, R.R. Martin, Plant Dis. 89(6), 654–658 (2005)
I.E. Tzanetakis, R.R. Martin, Virus Genes 28, 239–246 (2004)
X. Huang, A. Madan, Genome Res. 9, 868–877 (1999)
J.J. Campanella, L. Bitincka, J. Smalley, BMC Bioinformatics 4, 1–4 (2003)
L. Wang, S.J. Brown, Nucleic Acids Res. 34, W243–W248 (2006)
J.D. Bendtsen, H. Nielsen, G. Heijne, S. Brunak, J. Mol. Biol. 340, 783–795 (2004)
A. Krogh, B. Larsson, B. Von Heijne, E.L.L. Sonnhammer, J. Mol. Biol. 305, 567–580 (2001)
K. Julenius, A. Mølgaard, R. Gupta, S. Brunak, Glycobiology 15, 153–164 (2005)
J.D. Thompson, D.G. Higgins, T.J. Gibson, Nucleic Acids Res. 22, 4673–4680 (1994)
R.D.M. Page, Comp. Appl. Biosci. 12, 357–358 (1996)
R.A. Valverde, D.L. Gutierrez, Virus Genes 35, 399–403 (2007)
J.L. Sherwood, T.L. German, A.E. Whitfield, J.W. Moyer, D.E. Ullman, in Tomato spotted wilt, ed. by O.C.Maloy, T.D. Murray (Wiley, New York, 2000), pp. 1034–1040
P. De Haan, R. Kormelind, R. De Oliverira, F. Van Poelwijk, D. Peters, R. Goldbach, J. Gen. Virol. 72, 2207–2216 (1991)
A. Roberts, C. Rossier, D. Kolakofsky, N. Nathanson, G.-S. Franscisco, Virology 206, 742–745 (1995)
I.S. Lukashevich, M. Djavani, K. Dhapiro, A. Sanchez, E. Ravkov, S.T. Nichol, M.S. Salvato, J. Gen. Virol. 78, 547–551 (1997)
H. Jin, R.M. Elliott, Virology 67, 1396–1404 (1993)
D. Kolakofsky, D. Hacker, Curr. Top. Microb. Immunol. 169, 143–157 (1991)
J.A. Bruenn, Nucleic Acids Res. 31, 1821–1829 (2003)
S.K.J. Kukkonen, A. Vaheri, A. Plyusnin, Arch. Virol. 150, 533–556 (2005)
M. Zuker, Nucleic Acids Res. 31, 3406–3415 (2003)
M.S. Silva, C.R.F. Maertins, I.C. Bezerra, T. Nagata, A.C. De Ávila, R.O. Resendes, Arch. Virol. 146, 1267–1281 (2001)
U. Melcher, J. Gen. Virol. 80, 257–266 (2000)
A.R. Mushegian, E.V. Koonin, Arch. Virol. 133, 239–257 (1993)
R. Kormelink, P. De Haan, C. Meurs, D. Peters, R. Goldbach, J. Gen. Virol. 73, 2795–2804 (1992)
I. Cortez, A. Aires, A.M. Pereira, R. Goldbach, D. Peters, R. Kormelink, Arch. Virol. 147, 2313–2325 (2002)
A. Takeda, K. Sugiyama, H. Nagano, M. Mori, M. Kaido, K. Mise, S. Tsuda, T. Okumo, FEBS Lett. 532, 75–79 (2002)
J.M. Caruthers, D.B. McKay, Curr. Opin. Struct. Biol. 12, 123–133 (2002)
B. Lokesh, P.R. Rashmi, B.S. Amruta, D. Srisathiyanarayanan, M.R.N. Murthy, H.S. Savithri, PLoS ONE 5, e9757 (2010)
M. Kainz, P. Hilson, L. Sweeney, E. DeRose, T.L. German, Phytopathology 94, 759–765 (2004)
A.G. Bertran, A.S. Oliveira, T. Nagata, R.O. Resende, Arch. Virol. (2011). doi:https://doi.org/10.1007/s00705-011-0973-4
M.L. Daughtrey, R.K. Jones, J.W. Moyer, M.E. Daub, J.R. Baker, Plant Dis. 81, 1220–1230 (1997)
H.R. Pappu, R.A.C. Jones, R.K. Jain, Virus Res. 141, 219–236 (2009)
Acknowledgments
The authors would like to thank Drs. Sead Sabanadzovic, Tom Allen (Mississippi State University, and David Johnson (Missouri Dept. of Agriculture) for helping them with sample collection. The author also thank the Arkansas and Tennessee Soybean Promotion Board for providing funds that made this research possible.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhou, J., Kantartzi, S.K., Wen, RH. et al. Molecular characterization of a new tospovirus infecting soybean. Virus Genes 43, 289–295 (2011). https://doi.org/10.1007/s11262-011-0621-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-011-0621-9