Abstract
Sixty-three sugarcane leaf samples were collected from fifty-eight sugarcane varieties, evolved from eleven major sugarcane growing states in India, Australia, South Africa and USA. In RT-PCR, using gene specific primers for sugarcane streak mosaic virus (SCSMV)-CP, 58 of 63 sugarcane samples were found positive to the virus infection and rest of the five samples were negative. Partial CP gene sequences of 42 SCSMV isolates including an isolate from aphid colony (Melanaphis indosacchari) infested on sugarcane variety from this study were characterized after cloning and sequencing for selective isolates represented by at least one isolate from each location. The new sequences identified in the study were named as SCSMV-CB isolates. Fifty two sequences including the 10 database sequences (complete CP cds) deposited earlier from this institute were compared with each other as well as GenBank database sequences of Potyviridae members viz., Rymovirus, Potyvirus, Ipomovirus, Tritimovirus and eight sequences of SCSMV reported from elsewhere. Among the SCSMV-CB isolates sequenced in the study, 85.7–100% (nucleotide) and 89.9–100% (amino acid) sequence identities were observed and with the other data base sequences of SCSMV, the respective identities were 82.2–97.5 and 89.7–98.6%. Grouping of the isolates by the maximum likelihood with molecular clock model, distributed 60 SCSMV sequences including the eight database sequences deposited by other SCSMV working groups from India and USA in 16 different phylogenetic groups. Although the isolates of SCSMV were relatively close to Ipomovirus and Tritimovirus, they were sandwiched between Rymovirus and Ipomovirus. The sequence comparison and phylogenetic studies revealed that the relatedness of SCSMV with the potyviral related genera was comparatively low to consider it as a member of earlier described potyviral genera, hence the genus “Susmovirus” (sugarcane streak mosaic virus) has been proposed, with SCSMV as the sole species to be included. The 52 SCSMV-CB isolates from this institute were distributed in 14 phylogenetic groups and the grouping pattern revealed that the virus isolates could not be grouped based on geographical origin of the host varieties or longevity of the host variety.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sugarcane (Saccharum officinarum L.) cultivation in India perhaps antedates the known history of civilization. Its earliest records are in ‘Atharvaveda’ (1500 BC–1000 BC). It is now cultivated in several countries in the world, prominent amongst them being Australia, Brazil, Cuba, Fiji, India, Indonesia, Mauritius, Mexico, Philippines, South Africa and USA. In India, it is one of the most important commercial crops cultivated in ~4.4 mha. The crop is reported to be affected by several viral diseases, of which mosaic is most widely spread since then its first report from India by Barber [1]. Mosaic in sugarcane was attributed to a single Potyvirus called sugarcane mosaic virus (SCMV) with numerous strains [2, 3]. Currently, SCMV subgroup in the genus Potyvirus consists of seven different species viz., SCMV, Sorghum mosaic virus (SrMV), Maize dwarf mosaic virus (MDMV), Johnson grass mosaic virus (JGMV) [3–5], Zea mosaic virus (ZeMV) [6], Cocksfoot streak virus (CSV) [7] and Pennisetum mosaic virus (PenMV) [8]. Among these species, only SCMV and SrMV are known to infect sugarcane under natural conditions and are considered as the causal agents of sugarcane mosaic [9]. Strains of SCMV and SrMV infecting sugarcane were characterized earlier from United States, Australia [3, 10] and India [11, 12].
Hall et al. [13] found a new Potyvirus named as sugarcane streak mosaic virus (SCSMV) from the quarantined sugarcane germplasm material exhibiting mosaic symptoms, imported from Pakistan and characterized the virus. They proposed the new sugarcane virus as the species of the genus Whesmovirus with wheat streak mosaic virus (WSMV) as its definitive member. Later, ICTV created the genus “Tritimovirus” and assigned WSMV as the type species of the genus. Before the molecular confirmation of SCSMV, the mosaic symptoms on sugarcane were thought to be due to SCMV-F occurring in USA [14]. Later, Hema et al. [15] reported that the mosaic symptoms in sugarcane in India are due to SCSMV and not by the strains of SCMV. Subsequently, Chatenet et al. [16] reported that the mosaic symptoms in sugarcane in Asia are caused by at least by two strains of SCSMV.
We have observed numerous variations in symptom expression in mosaic infected sugarcane varieties in the country and conducted detailed studies on identifying and characterizing the viruses associated with mosaic. Initially, we have taken nine sugarcane varieties evolved from five different states of India exhibiting wide variation in symptom expression for RT-PCR studies to detect the associated virus(es). RT-PCR and subsequent sequencing of amplified products revealed association of both SCSMV and SCMV either alone or together with sugarcane mosaic in India. However, SCSMV occurrence was noticed in a higher proportion than SCMV [17]. Further studies were continued to characterize SCSMV occurring in sugarcane. Since the sequence analysis showed a wide variation between 3.1 and 12% (nt) and 0.0 and 14.8% (aa), respectively in the full length CP region of SCSMV, a new set of primers were designed using the sequences reported by various sugarcane working groups to amplify 690 bp starting from N-terminal region of CP gene and the same primer pair was used to screen 63 sugarcane samples and sugarcane aphid (Melanaphis indosacchari David).
The virus systemically infects the crop and once the plant is infected, the virus is transmitted through vegetative cuttings. Hence we observe disease symptoms in all the plants in the field. Sugarcane Breeding Institute at Coimbatore in Tropical India maintains one of the two world collections for sugarcane germplasm recognized by International Society of Sugar Cane Technologists (ISSCT) [18] and the institute developed more than 2,800 varieties named ‘Co’ canes in the past 95 years. These varieties along with varieties developed in other states are being maintained in the institute’s germplasm collections. Information on the intensity of SCSMV infections and prevailing viral strains in the field as well as varietal collections are not available. Hence detailed studies were conducted on the detection of SCSMV from 63 sugarcane samples evolved from India and in other countries and in an aphid colony. The SCSMV sequences were profusely compared with the members of Potyviridae and among the SCSMV isolates with special reference to their place and year of origin of the varieties/genotype, finally the genus “Susmovirus” (sugarcane streak mosaic virus) has been proposed, with SCSMV as its sole species at present.
Materials and methods
Virus isolates
The first unfurled leaves in the whorl from 58 varieties were collected during 2006–2007 crop season maintained at the germplasm collections of the Institute. The sampled varieties represented 11 major sugarcane growing states of India; two from USA, one each from Australia and South Africa. Additionally, in some prominent varieties viz., Co 6304, Co 86032, CoC 671 and CoV 94101 which are under large scale cultivation samples were also collected from grower’s fields. A sugarcane aphid colony (Melanaphis indosacchari David) infested on sugarcane variety at the institute farm was also included in the study. The details on the period of varietal evolution, place of collection and information on their respective virus isolates which have been subjected to phylogenetic analysis were given in the Table 1.
RNA extraction and RT-PCR
Total RNA was extracted from the newly unfurled leaf using TRI Reagent (Sigma, USA) following the manufacturer’s protocol. The total RNA was re-dissolved in a final volume of 30 μl of RNA re-suspension buffer (Ambion, USA) and stored at -80°C. The quality of the RNA was checked using 1% agarose gel. Based on the published sequence information ([13, 15, 17]; Acc. nos. EF088797; EF079666; EF079667; EF088798; EF079665; EF088799), a pair of oligonucleotides, the forward primer SCSMV-F690 (F5′ GGACAAGGAACGCAGCCACCTC 3′) and the reverse primer SCSMV-R690 (R5′ CGCACGTCGATTTCTGCTGGTC 3′) were designed to amplify ~690 bp starting from N-terminal region of the coat protein. Subsequently, the total RNA from all the 63 sugarcane samples and one aphid colony sample were reverse transcribed using RevertAid H Minus first strand cDNA synthesis kit (MBI Fermentas, USA) using oligo (dT) as primer by following the manufacturer’s protocol in a PCR machine (Mastercycler gradient; Eppendorf, Germany). The PCR reaction was performed in a total volume of 25 μl containing 2 μl cDNA, 2.5 μl of 10× PCR buffer containing 15 mM MgCl2, 0.5 μl of 10 mM dNTP mix, 20 pmol each of SCSMV-F690 and SCSMV-R690, 1.25 units of Taq polymerase (Intron, South Korea), and sterile milliQ water to a final volume. The PCR reaction was performed with initial denaturation at 94°C for 3 min, 30 cycles of 94°C for 30 s, 65°C for 30 s and 72°C for 1 min and a final extension of 72°C for 10 min in a PCR machine. A 10 μl aliquot of each amplified product was analyzed by electrophoresis on 1.5% agarose gels stained with ethidium bromide.
Purification of RT-PCR products, cloning and sequencing
Although 58 sugarcane samples and an aphid colony were found positive to SCSMV infection, only the amplicons (~690 bp) from 37 sugarcane samples were selected based on the host variety evolution and place of origin for further studies. The only virus isolate from the aphid colony was also included in the study. The amplicons were excised from the low melting agarose gel and eluted using GenElute Gel Extraction kit (Sigma, USA), ligated in to pTZ57R/T vector (MBI Fermentas, USA) and then cloned in E. coli DH5α. The plasmids with the inserts of expected size were purified using Fastplasmid Mini Prep Kit (Eppendorf, Germany). For each virus isolate, four cloned inserts were completely sequenced on both strands in ABI PRISM 377 DNA sequencer, using BigDye Terminator Cycle Sequencing Kit v3.0/v3.1 (Perkin Elmer). The primer pair used for sequencing were M13F (20) and M13R puc (26). Nucleotide sequencing was performed at the DNA sequencing facility of 1st base, Selangor Darul Ehsan, Malaysia. RT-PCR errors were eliminated by compiling consensus sequences derived from the independent clones of each virus isolate. In few cases, sequences derived from the same variety with less than 99.5% identity were treated as separate isolates, hence we had 42 SCSMV isolates for comparison. The newly identified virus isolates in this study were named as SCSMV-CB isolates.
Similarity and phylogenetic analysis
Since limited information is available worldwide on SCSMV genome, the unclassified member of the Potyviridae, taxonomic affiliation of SCSMV is not well established at present. Hence with a view to identify the correct taxonomical position among Potyviridae members, the SCSMV sequences characterized in this study were compared with rest of the Potyviridae members viz., Wheat yellow mosaic virus (WYMV), a species of Bymovirus, SCMV, Potato virus-Y (PVY), Tobacco etch virus (TEV) (Potyvirus), Ryegrass mosaic virus (RgMV), Agropyron mosaic virus (AgMV) and Hordeum mosaic virus (HoMV) (Rymovirus), Sweet potato mild mottle virus (SpMMV) (Ipomovirus), WSMV, Oat necrotic mottle virus (ONMV) and Brome streak mosaic virus (BrSMV) (Tritimovirus) and the unclassified member, SCSMV.
Totally 60 SCSMV isolates including 18 database isolates (Table 1) were compared among themselves and Potyviridae members. The nucleotide (nt) and amino acid (aa) sequences were aligned separately using clustal W [19]. The 5′-terminal and 3′-terminal regions were trimmed uniformly in all the taxa such that the final alignment compared at 1–689 nt and 1–229 aa (coordinates relative to SCSMV-CB285) and DNAmlk maximum likelihood with molecular clock (Maximum Likelihood method with molecular clock, version 3.6a2.1), DNApars DNA parsimony method (DNA parsimony algorithm, version 3.6a2.1) and DNAml DNA Maximum Likelihood programme (Maximum Likelihood method, version 3.6a2.1) phylograms were generated and visualized using Treeview (Win32), a tree visualizing software inbuilt with BioEdit version 7.0.4.1 [20] and compared the grouping pattern. Using the same alignment, similarity index was generated and the output was converted in to per cent sequence similarity, from which per cent identities were calculated between different genera of Potyviridae and among the SCSMV isolates characterized from different varieties evolved from the length and breadth of the country. Subsequently, using the same alignment after deleting the other Potyviridae members from the alignment, DNAmlk maximum likelihood with molecular clock phylogram, DNApars DNA parsimony phylogram, centre and state wise sequence similarity index were generated and the similarity index was converted in to per cent sequence similarity.
Results
Detection of SCSMV in sugarcane varieties
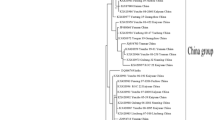
We have amplified SCSMV-CP with expected amplicon size of ~690 bp in 59 of 64 samples. The five samples from the following varieties, Co 89003, CoS 94270, Co 94006, B38192 and Co 91010 were negative to SCSMV infection (Fig. 1). The latter three varieties were asymptomatic in which sugarcane yellow leaf virus (SCYLV) was detected and the former two were observed with characteristic mosaic symptoms from which SCMV was detected using SCMV-CP gene specific primers (data not shown).
Detection of SCSMV in sugarcane varieties evolved from 1924 to 2000 at different states of India and representative varieties evolved from Australia, South Africa and USA by RT-PCR. (A) Lanes M. 100 bp DNA marker; 1–33 corresponds to varieties/genotypes 93A53, CoA 7701, CoA 8201, CoV 94101(AP), CoV 94101 (TN), CoV 92101, CoBln 94063, BO 47, BO 54, BO 72, BO 92, CoH 76, CoJ 64, CoJ 65, CoJ 77, CoJ 86, CoPant 84213, CoPant 84211, KMS 2095, CoSi(S) 6, CoSi 86071, CoC 671 (NHG), CoC 671 (ECC), CoC 92061, CP 44-101, CP 52-68, Q63, CoJn 80151, CoJn 862035, CoS 611, CoLk 97023, CoLk 97154 and CoS 94270, respectively. (B) Lanes M. 100 bp DNA marker; 34–63 CoM 9217, Co 94006, Co 285, Co 312, Co 419, Co 453, Co 617, Co 740, Co 976, Co 1148, Co 6304 (ECC), Co 7219, Co 8208, Co 62198, Co 85019, Co 89003, Co 86010, Co 86032 (NHG), Co 86032 (ECC), Co 86032 (Erode), Co 91010, Co 94003, Co 94005, B 38192, Co 6304 (NHG), Co 94008, Co 99016, Co 2000-10, ISH 69, NCo 310, respectively; 64. M. indosacchari; 65 & 66. PCR product from SCSMV-CB671 cDNA clone (positive control); 67. healthy sugarcane
Cloning and sequencing of SCSMV-CP
The RT-PCR amplicons derived from 37 sugarcane samples and aphid were cloned and sequenced, and the isolates were named as SCSMV-CB. The sequences obtained from the cloned insert were consistent with expected length of 689 nucleotides (nt), except the isolates CB86071 and CB94101 (AP) (683 nt); CB97154-2 and CB92061-3 (688 nt) and CBKMS2095 and CB52-68 (690 nt). Of the four clones sequenced, each from CoLk 97154 and CoC 92061, two clones from each variety were consistent with 688 nt and other two with 689 nt (Table 1), with 0.5% (nt) and 2.7% (aa) sequence variation between the two differential length sequences obtained from CoLk 97154 showed 1.5% (nt) and 4.9% (aa) sequence variation between them. Similarly two sequences characterized from CoC 92061 (data not shown).
Comparison of SCSMV-CP nucleotide and amino acid sequences with Potyviridae members
Comparisons of the SCSMV sequences characterized in this study with rest of the Potyviridae members revealed that the unclassified SCSMV taxa shared the mean sequence homologies of 36.0, 36.1, 35.7, 39.2 and 45% at nt level and 15.1, 17.2, 18.6, 23.4 and 30.1% at aa level, respectively, with Bymovirus, Potyvirus, Rymovirus, Ipomovirus and Tritimovirus. Similarly the mean sequence identity of SCSMV with the other sugarcane mosaic causing virus SCMV was 36.1 and 16.6%, respectively at nt and aa level (Table 2).
Among the three Potyvirus members included in the comparison, 58.6–61.2 (nt) and 46.3–64.8% (aa) sequence similarities have been observed. The sequence identity of Bymovirus with the rest of the members of the Potyviridae is very minimum as compared to other members and was equally distant with the members of Potyviridae. The species of Tritimovirus, WSMV, ONMV and BrSMV showed identities of 55–74.5 (nt) and 46.8–82.2% (aa) in their CP sequences. Among the Tritimovirus, the maximum nt (73.8–74.5%) and aa (81–82.2%) sequence identity was observed between ONMV and WSMV. However, the sequence identities of BrSMV with WSMV and ONMV was comparatively low (55–58.1 and 46.8–48% respectively, at nt and aa level). Among the species of Rymovirus, AgMV and HoMV showed maximum sequence identities of 75.8 (nt) and 68.9% (aa), however their identities with the third species RgMV, were comparatively less both at nt (51.4%) and aa (56.8%) sequence level (data not shown).
Among SCSMV-CB isolates, 85.7–100 and 89.9–100% sequence identities have been observed respectively at nt and aa level. The nt and aa sequence identities of SCSMV-CB isolates with the database SCSMV sequences were 82.2–97.5 and 89.7–98.6%, respectively. The identities of the SCSMV-CB isolates with the only reported strain SCSMV-PAK had ranged from 83.8% with CBH76 to 93.6% with CB671 at nt level and 90.4% with CB86071 and CB9217-2 to 95.6% with CB94063 at aa sequence level. The most diverged isolate CB86071 shared wide range of sequence identities with rest of the SCSMV-CB isolates sharing 85.7% (CB740) to 90.5% (CB94063) and 90.3% (CB92061-3 and CB52-68) to 95.1% (CB84211) respectively at nt and aa level. The predominant isolate SCSMV-CB671 shared 86.3–94.7 (nt) and 93–97.8% (aa) sequence identities with rest of the SCSMV-CB isolates and its relation was comparatively low with other earlier characterized SCSMV isolates. The earlier characterized South Indian isolates, TN, TA and KA shared 98.7–98.9 (nt) and 98.9–98.6% (aa) sequence identities among each other; while the identities of these three isolates with rest of the isolates ranged from 83.2–91.9 to 83.6–93.6% respectively at nt and aa level. The sequences characterized from aphid viz., CBAPHIDS shared 97.2–99.5% (aa) with AP, CB6304, CBQ63-1, CBQ63-2, CB62198, NCO310, CBH76, CBA8201, MR-CoC671, CBECC86032 and CB2000-10 and at nucleotide level, it varied from 96.7 to 100% (data not shown).
Two way comparisons of SCSMV-CB isolates i.e. period of host variety origin and place of the sugarcane varieties origin were made to understand the significance of these parameters on SCSMV variation. There is no homogenous population existed among the representative varieties from different states. However, 100% nt and aa sequence similarities were observed among the isolates of CB312, CB85019 and CB99016 respectively, from cvs. Co 312, Co 85019 and Co 99016. Similarly, 100% aa sequence similarity was observed among CB85019 and CBNHG86032; CB62198, CB419-1 and, CB419-4; and CB312, CB740 and CB2000-10 (Table 3).
The overall mean nt and aa sequence similarities of SCSMV-CP characterized in India were 92.1 and 95.3%, respectively. The respective overall mean identities of 94.3 and 97.0% were observed among the isolates characterized from Coimbatore varieties, which were higher than the overall India mean sequence identity, except CB671 and CB86071 characterized, respectively originated from Cuddalore and Sirugamani in Tamil Nadu state. However, the isolates CB92061-3 (94.5%) and CB740-1 (94.6%) shared relatively lower overall mean aa sequence identities (Table 3). Similarly, the SCSMV sequences characterized from Andhra Pradesh, Punjab and Uttar Pradesh varieties, showed sequence identities higher than the overall mean of India, except CB94101 (AP) and CB77-2, respectively characterized from Andhra Pradesh and Punjab. The aa sequence similarities of the isolates characterized from Bihar varieties shared below (94.8%) the overall mean sequence similarity. The overall mean sequence identities of the isolates characterized from Australia, South Africa and USA were higher than the overall mean of India. However, the PAK isolate originated from Pakistan shared below overall mean aa sequence identity. The earlier characterized South Indian isolates viz., KA, TA and TN for which the source varieties were not mentioned, shared below the overall mean nt and aa sequence identity (data not shown).
Phylogenetic relationship of SCSMV with other Potyviridae members
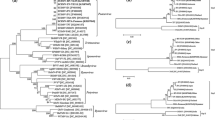
The phylogram constructed with DNAmlk DNA Maximum Likelihood program with molecular clock algorithm, distributed the Potyviridae members in four clusters (Fig. 2). The Bymovirus alone in a cluster, from which the aphid transmitted taxa of Potyvirus and mite transmitted taxa of Rymovirus were separated in a cluster, and then the unclassified potyviral member SCSMV, the members of Ipomovirus and Tritimovirus share the common ancestor. The taxa of the SCSMV were sandwiched between RgMV (Rymovirus) and SpMMV (Ipomovirus). Further, the taxa of the SCSMV, distributed in three sub clusters. The most diverged isolates KA, TA and TN reported earlier in a sub cluster (sub cluster 1); the other diverged SCSMV-CB isolates viz., CB671, CB86071, CB94003, CB94063, CBKMS2095, and the recognized strain SCSMV-PAK alone in another sub-cluster (sub-cluster 3); and rest of the 51 sequences were distributed in a sub-cluster (sub-cluster 2) sandwiched between relatively diverged isolates containing sub-cluster 1 and sub-cluster 3. The phylogram constructed with DNAPars DNA Parsimony algorithm had yielded a similar grouping pattern, except the isolates SCSMV-PAK and SCSMV-AP which were inconsistent in grouping pattern. Similar phylograms (DNAmlk DNA Maximum Likelihood program with molecular clock, DNAPars DNA Parsimony and DNAml DNA Maximum Likelihood programme) generated with SCSMV isolates alone after deleting the other Potyviridae members from the alignment, showed distribution of SCSMV isolates in 16 groups under three clusters (Fig. 3). In both the methods, the placement of the isolates SCSMV-PAK, SCSMV-CB671 and SCSMV-AP were inconsistent and rest of the sequences were consistently placed in both the phylograms. The phylogram constructed with maximum likelihood program with molecular clock placed SCSMV-PAK isolate in a separate cluster from which the other SCSMV sequences were separated. However in the phylograms generated using DNA parsimony method and DNA Maximum Likelihood programme, the SCSMV-CB671 isolate was placed in a separate cluster and the SCSMV-PAK isolate shared the common clade with South Indian isolates viz., SCSMV-KA, TA and TN. Similar grouping pattern was observed among the remaining isolates in all the three phylograms. The SCSMV isolates, PAK, CB94101 (TN), CB453 and CBSIS6, CB94003 and CB671 each represented a separate group showing their diversity with the rest of the isolates.
Phylogenetic relationships of the unclassified member of Potyviridae, SCSMV with the recognized potyviral members. Presented is phylogram derived from DNAmlk DNA Maximum Likelihood program with molecular clock based on a nucleotide sequence alignment corresponding to 689 nt of SCSMV-CB285. Proposed genus of the taxa SCSMV is given in bold letter in right side of the phylogram. Abbreviations and accession number in Genbank: WYMV (D86634), AgMV (U30615), HoMV (U30616), SCMVCBC92061-1 (EF655896), SCMVCB92061-2 (EF655897), SCMVCB1148 (EF655898), SCMV-CB44-101 (EF655894), PVY-N (X12456), TEV (M11458), RgMV (U27383), RgMV (Y09854), SpMMV (Z73124), CVYV (AF233429), BrMV (NC003501), ONMV Type (AF454460), WSMV-I (AF454458), WSMV-H (AF454456) and WSMV-R (AF454459) and see Table 1 for other sequences
Phylogenetic relatedness of SCSMV-CB isolates and data base SCSMV isolates. Presented is phylogram derived from DNAmlk DNA Maximum Likelihood program with molecular clock based on a nucleotide sequence alignment corresponding to 689 nt of SCSMV-CB285. The 60 SCSMV taxa were distributed in three clusters and the placements of the isolates viz., SCSMV-PAK, CSMV-CB671 and SCSMV-AP in DNAmlk DNA Maximum Likelihood program with molecular clock and DNAPars DNA Parsimony method were inconsistent. The details of the sequences and their accession numbers were given Table 1
Discussion
The viruses belonging to Potyviridae have been classified into genera that coincide with vector taxa [21]. The numerous economically important aphid transmitted potyviruses share evolutionary relationship with fungus transmitted bymoviruses [22, 23]. The studies on molecular characterization and virus vector relationships resulted many revisions and there were proposals to assign genus name to the unassigned Potyviridae members viz., whitefly transmitted SpMMV of Ipomovirus [24, 25], mite transmitted RGMV [26], AgMV and HoMV [27] of Rymovirus [28], BrSMV [29], WSMV [30, 31] of Tritimovirus [25]. The revision and proposals were reflected in the subsequent ICTV report [32]. Before 1998, the mosaic in sugarcane was reported to be associated with different strains of SCMV and SrMV under natural conditions. Studies of Hall et al. [13] clearly established the association of SCSMV with the disease and molecular characterization revealed it is a new virus species in Potyviridae. Subsequently, the SCSMV was reported from Bangladesh, Pakistan, Sri Lanka, Thailand and Vietnam [16] and in India [15–17, 33].
Hema et al. [15] reported that sugarcane mosaic in India is due to SCSMV and not by the SCMV subgroups. Later, Chatenet et al. [16] reported that sugarcane mosaic is caused by at least by two strains of SCSMV in most of the sugarcane growing Asian countries. We strongly established that mosaic in sugarcane is caused either by SCMV or SCSMV alone or both in combinations [17]. The association of SCMV strains viz., SCMV-C, G, K and L from USA [3] and SCMV-N from India [12] were reported based on serology and differential host reaction are not available at present. By nucleotide sequencing of these strains we may likely to identify new virus strains diverged from SCMV or SCSMV, probably responsible for the mosaic in the crop as like strain SCMV-F, presumed as the casual agent of mosaic [14] reported later as a distinct Potyvirus based on nucleotide sequencing, named as SCSMV [13]. Grisham and Pan [34] failed to detect SCMV or SrMV in 8% of the symptomatic sugarcane plants in Louisiana. Similarly primers for these two viruses failed to detect the viruses in the mosaic infected sugarcane in Argentina [35]. These scenarios suggest that probably SCSMV might be associated with mosaic there also and possibly in other sugarcane growing countries. The sugarcane varieties under cultivation in India are widely infected with SCSMV as evidenced by more than 92% infection in our samples (Fig. 1).
The comparison of N-terminal and core region of the SCSMV-CP revealed a wide range (89.9–100%) of aa sequence similarity. However we found greatly varied isolates like KA, TA, TN, CB86071, CB94003 and CBKMS2095 and existence of more than one isolates in three sugarcane varieties. The SCSMV population structure greatly varied in the samples (CoC 671) collected from Tamil Nadu and Maharashtra and also the two isolates from Co 86032 sampled at different fields of the same place were relatively heterogeneous. The heterogeneous populations in these two samples support the earlier report made by Garcia-Arenal et al. [36]. In contrast, we also found highly homogeneous virus population from sugarcane varieties Co 312, Co 85019 and Co 99016. These findings suggest that the viral population structure is varied with field to field, region to region and even in a single leaf. The variants in these varieties might be due to the existence of mixture of populations possibly generated by high mutation rate during RNA replication as it is a common weapons hold by RNA virus that is needed for rapid replication of their chemically unstable RNA genome rather than being an evolutionary strategy [37].
Distribution of the 60 SCSMV sequences in 16 phylogenetic groups clearly explains their heterogeneity. In DNA maximum likelihood method with molecular clock phylogram constructed with all the Potyviridae members, the isolates, SCSMV-PAK and CB671 distributed in a sub-cluster, meanwhile in the phylogram constructed with SCSMV alone they were distributed in different clusters and are inconsistently placed in the phylogram constructed with other methods (Fig. 3). Moreover, 14 amino acid variations have been observed between these two isolates from 1 to 45th amino acid residues (data not shown). The aa comparison revealed that none of the Indian isolates are belonging to SCSMV-PAK strain as evidenced from 10 to 19 aa variation observed between SCSMV-PAK and other isolates, sharing 90.4–100% identity. However, Hema et al. [38] reported that SCSMV-AP isolate belongs to SCSMV-PAK strain. The strains of the Potyvirus generally exhibit amino acid similarities in the CP from 90 to 99% [39, 40]. Van Regenmortel et al. [41] demarcated the virus isolates showing less than 90% identity in CP sequence as distinct species in the family Potyviridae. However, the most recent criterion [42] points that the aa sequence identity of the CP should be lesser than 80% before new virus species is designated. Hence, the mosaic disease of sugarcane in India is caused by more than one strains of SCSMV. Further we found SCSMV groupings do not coincide with either place or year of varietal evolution or both. Probably this may be due to maintaining the varieties at our collections for varying periods, which enabled them to exchange viral populations immensely through vector or human interventions. Further, among the mixed population, survival of the fittest theory might have operated, so as to adapt and encounter the selection pressure. To meet out this requirement, evolutionary process viz., mutation and genetic drift would have switched on among the viral populations leading to the existence of heterogeneous populations irrespective of the examined parameters. In this regard, further studies are needed to establish the evidences of the evolutionary strategy undergone by this virus. The distribution of SCSMV in a separate cluster and not within the cluster containing related genera Ipomovirus and Tritimovirus clearly demonstrates the uniqueness of the virus to place under a separate genus, for which the genus “Susmovirus” has been suggested through this study. We presumed that evolution of the virus is still continuing as it was evidenced from the huge variations in the hyper variable regions which ranged from 0.0 to 40.9% variation at aa level (data not shown). The divergence of RNA viruses accounts for the sequence drift of various RNA viruses under certain selection pressures [43, 44]. Further, this extreme variation in the N-terminal region might be associated with its successful wide spread occurrence as major causative agent for mosaic of sugarcane, in India.
Present study reveals the existence of high levels of variation in SCSMV infecting different sugarcane varieties evolved over eight decades in India. It also demonstrates, for the first time that very higher level of variation in SCSMV populations in a geographical region. In world scenario, genomic information on SCSMV is totally lacking except a few and there are chances to get even higher variation if sequence information is made available from other countries. Almost all of the sequences except SCSMV-PAK, discussed in this paper are characterized from India.
Even though the Potyvirus and Rymovirus species are transmitted by aphids and mites, respectively, both of them shared the common clade. However, Rymoviruses and Tritimoviruses are distributed in separate clusters that explain the divergence among the members of the two genera. The taxa of the Ipomovirus and Tritimovirus were distributed in a cluster that shared the common ancestor with unclassified SCSMV taxa. The SCSMV sequence relatedness is progressively increasing from Bymovirus to Tritimovirus and it showed that the latter is closer than other genus. In this regard, detailed studies are required on the complete genome characterization, genomic scanning of Ipomovirus, Tritimovirus and the unclassified SCSMV taxa with special reference to genetic exchange through recombination, mutation and genetic drift among the taxa of the genera.
Practically it is highly complicated to control the disease even with genetically engineered resistance, since most of the genetically engineered resistance relies on low sequence variation within the countries/regions being targeted [45]. Hence, the complex scenarios i.e. existence of more than a strain in a single variety and high levels of divergence among the SCSMV populations will pose a significant challenge among the Scientific community to control the virus even with genetic engineering. It is now clearly established that more than one strains of Susmovirus cause sugarcane mosaic in India in higher levels and SCMV does occur in relatively lower levels. Further studies are required to characterize SCSMV in different continents and to assess relative proportion of SCSMV and SCMV causing sugarcane mosaic.
References
C.A. Barber, Int. Sugar J 23, 12–19 (1921)
H. Koike, A.G. Gillespie, in Disease of Sugarcane—Major Diseases, ed. by C. Ricaud, B.T. Egan, A.G. Gillespie, C.G. Hughes (Elsevier, Amsterdam, 1989), pp. 301–322
D.D. Shukla, C.W. Ward, A.A. Brunt, The Potyviridae (CAB International, Cambridge University Press, Cambridge, 1994), p. 516
N.M. Mckern, D.D. Shukla, R.D. Toler, S.G. Jensen, M. Tosic, R.E. Ford, O. Leon, C.W. Ward, Phytopathology 81, 1025–1029 (1991)
D.D. Shukla, M. Tosic, J. Jilka, R.E. Ford, R.W. Taker, M.A.C. Langham, Phytopathology 79, 223–229 (1989)
D.L. Seifers, R. Salomon, V. Marie-Jeanne, B. Alliot, P. Signoret, S. Harber, A. Loboda, W. Ens, Y.M. She, K.G. Standing, Phytopathology 90, 505–513 (2000)
R. Gotz, E. Maiss, Arch. Virol. 147, 1573–1583 (2002)
Z. Fan, H. Chen, S. Cai, C. Deng, W. Wang, X. Liang, H. Li, Arch. Virol. 148, 1219–1224 (2003)
M.P. Grisham, in A Guide to Sugarcane Diseases, ed. by P. Rott, R.A. Bailey, J.C. Comstock, B.J. Croft, A.S. Saumtally (CIRAD-ISSCT, CIRAD Publication Services, Montepellier, France, 2000), pp. 249–254
D.D. Shukla, M.J. Frenkel, N.M. Mckern, C.W. Ward, J. Jilka, M. Tosic, R.E. Ford, Arch. Virol. [Suppl.] 5, 363–373 (1992)
E. Kondaiah, M.V. Nayudu, Curr. Sci. 53, 273–275 (1984)
E. Kondaiah, M.V. Nayudu, Sugar Cane 4, 11–14 (1985)
J.S. Hall, B. Adams, T.J. Parsons, R. French, L.C. Lane, S.G. Jensen, Mol. Phylogenet. Evol. 10, 323–332 (1998)
A.G. Gillaspie Jr., R.G. Mock, F.F. Smith, Proc. Int. Soc. Sugar Cane Technol. 16, 347–355 (1978)
M. Hema, J. Joseph, K. Gobinath, P. Sreenivasulu, H.S. Savithri, Arch. Virol. 144, 479–490 (1999)
M. Chatenet, C. Mazarin, J.C. Girard, E. Fernandez, D. Gargani, G.P. Rao, M. Royer, B. Lockhart, P. Rott, Sugarcane Int. 23, 12–15, 28 (2005)
R. Viswanathan, M. Balamuralikrishnan, R. Karuppaiah, Sugarcane Int. 25, 6–14 (2007)
D.J. Heinz, in Sugarcane Varietal Improvement, ed. by M.K. Naidu, T.V. Sreenivasan, M.N. Premachandran (The Director, Sugarcane Breeding Institute, Coimbatore, 1989), p. 364
J.D. Thompson, D.G. Higgins, T.J. Gibson, Nucleic Acid Res. 22, 4673–4680 (1994)
T.A. Hall, Nucl. Acids Symp. Ser. 41, 95–98 (1999)
C.W. Ward, G.F. Willer, D.D. Shukla, A. Gibbs, in Molecular Basis of Virus Evolution, ed. by A.J. Gibbs, C.H. Calisher, G. Arenal (Press Syndicate of the University of Cambridge, Cambridge, 1995), pp. 447–500
S. Kashiwazaki, Y. Minobe, T. Omura, H. Hibino, J. Gen. Virol. 71, 2781–2790 (1990)
E. Peerenboom, M. Prols, J. Schell, H.H. Steinbiss, A.D. Davidson, J. Gen. Virol. 73, 1303–1308 (1992)
D. Colinet, J. Krummert, P. Lepoivre, Arch. Virol. 141, 125–135 (1996)
D.C. Stenger, J.S. Hall, I.R. Choi, R. French, Phytopathology 88, 782–787 (1998)
J. Schubert, F. Rabenstein, E. Proll, Agronomie 15, 447–452 (1995)
S.N. Salm, M.E.C. Rey, N.L. Robertson, R. French, F. Rabenstein, J. Schubert, Arch. Virol. 141, 2115–2127 (1996)
S.N. Salm, M.E.C. Rey, E.P. Rybicki, Arch. Virol. 141, 2237–2242 (1996)
R. Gotz, E. Maiss, J. Gen. Virol. 76, 2035–2042 (1995)
K.D. Chenault, R.M. Hunger, J.L. Sherwood, Virus Genes 13, 187–188 (1996)
C.L. Niblett, K.R. Zagula, L.A. Calvert, T.L. Kendall, D.M. Stark, C.E. Smith, R.N. Beachy, S.A. Lommel, J. Gen. Virol. 72, 499–504 (1991)
C.M. Fauquet, M.A. Mayo, J. Maniloff, U. Desselberger, L.A. Ball (eds.), Virus Taxonomy. Classification and Nomenclature of Viruses (8th ICTV Report, Academic Press, Elsevier, 2005), p. 1217
R.K. Gaur, M. Singh, A. Singh, L.C. Mishra, G.P. Rao, Sugarcane Int. 24, 13–17 (2006)
M.P. Grisham, Y.P. Pan, Plant Dis. 91, 453–458 (2007)
M.F. Perera, M.P. Filippone, J. Ramallo, M.I. Cuenya, M.L. Garcia, A.P. Castagnaro, Proc. Int. Soc. Sugar Cane Technol. 26, 988–992 (2007)
F. Garcia-Arenal, A. Fraile, J.M. Malpica, Int. Microbiol. 6, 225–232 (2003)
J.W. Drake, J.J. Holland, Proc. Natl. Acad. Sci. USA 96, 13910–13913 (1999)
M. Hema, P. Sreenivasulu, H.S. Savithri, Arch. Virol. 147, 1997–2007 (2002)
D.D. Shukla, C.W. Ward, J. Gen. Virol. 69, 2703–2710 (1988)
C.W. Ward, N.M. Mckern, M.J. Frenkel, D.D. Shukla, Arch. Virol. [Suppl.] 5, 283–297 (1992)
M.H.V. Van Regenmortel, D.H.L. Bishop, C.M. Fauquet, M.A. Mayo, J. Maniloff, C.H. Calisher, Arch. Virol. 142, 1505–1518 (1997)
P.H. Berger, O.W. Barnett, A.A. Brunt, D. Colinet, J.R. Edwardson, J. Hammond, J.H. Hill, R.L. Jordon, W. Kashiwazaki, K. Makkouk, G. Morales, E.P. Rybicki, N. Spence, S.T. Ohki, I. Uyeda, A. van Zaayan, H.J. Vetten, in Virus Taxonomy: Seventh Report of the International Committee on Taxonomy of Viruses, ed. by M.H.V. van Regenmortel, C.M. Fauquet, D.H.L. Bishop, E.B. Carstens, M.K. Estes, S.M. Lemons, J. Maniloff, M.A. Mayo, C.R. Pringle, R.B. Wickner (Academic Press, New York, 2000)
D.A. Steinhauer, J.C. De la Torre, J.J. Holland, J. Virol. 63, 2063–2071 (1989)
D.A. Steinhauer, J.C. De la Torre, E. Meier, J.J. Holland, J. Virol. 63, 2072–2080 (1989)
P.F. Tennant, C. Gonsalves, K.S. Ling, M. Fitch, R. Manshardt, J.L. Slightom, D. Gonslaves, Phytopathology 84, 1359–1366 (1994)
Acknowledgements
The authors wish to express sincere thanks to Dr. N. Vijayan Nair, Director of the Institute for the encouragement and facilities. Financial support received from ICAR, New Delhi as part of Network Project on Developing Diagnostics of Emerging Plant Viruses is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Viswanathan, R., Balamuralikrishnan, M. & Karuppaiah, R. Characterization and genetic diversity of sugarcane streak mosaic virus causing mosaic in sugarcane. Virus Genes 36, 553–564 (2008). https://doi.org/10.1007/s11262-008-0228-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-008-0228-y