Abstract
The resistance of 220 coagulase-negative Staphylococci (CNS) (associated with animal disease) to 13 antibiotics were determined using the disk diffusion method. 35.9% of multidrug-resistant coagulase-negative Staphylococci (MR-CNS) exhibited resistance to five or more than five antibiotics; all of these bacteria were resistant to methicillin too. The new Streptomyces sp. ABRIINW111 was isolated from the Zagros Mountains Hamadan, Iran. The 16S rDNA sequence of the isolate indicated that it has 98% similarity to S. levis, but some mutations in the alpha and gamma regions of the 16S rDNA sequence emphasize the probability of the existence of a new species. Preliminary and secondary antibacterial screenings revealed that the isolate is active against gram negative and positive bacteria. The diethyl ether extracted metabolite of the Streptomyces sp. ABRIINW111 showed an effective antibacterial activity against MR-CNS. So the diethyl ether extract of the new Streptomyces sp. strain ABRIINW111 can inhibit the MR-CNS in vitro, and it can offer a new approach to treat MR-CNS infectious patients.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Staphylococcus genus contains 41 validly described Species. These are traditionally grouped into coagulase positive (CPS) and coagulase-negative Staphylococci (CNS) (Resch et al. 2008). Main habitats of these bacteria are skin, skin glands and mucous membranes of humans and animals (Sudagidan et al. 2005; Lis et al. 2009; Koksal et al. 2007). CNS have become the most frequently isolated organisms from bovine intra mammary infections in several countries and their prevalence is generally highest at calving (Katsuda et al. 2005; Schultz et al. 2009). CNS may behave as opportunistic pathogens, often introduced by medical devices or colonizing exposed wounds (Faria et al. 2009).
Due to the intensive using of antibiotics in public health and animal husbandry, antibiotic resistance in pathogens including the genus Staphylococcus has been an increasing medical problem during the last decades (Schultz et al. 2009; Resch et al. 2008).
Streptomyces, and related Actinomycetales, continue to be prolific sources of novel secondary metabolites with a range of biological activities that may ultimately find application as anti-infective, anti-cancer agents or other pharmaceutically useful compounds (Bibb 2005). These genera are widely recognized as industrially important microorganisms, and produce many commercially and medically useful antibiotics (Baltz 2008).
It is generally accepted that the ‘golden age’ of antibiotic discovery from Actinomycetales has passed (Higginbotham and Murphy 2010). Nonetheless, these bacteria are source for the discovery of novel bioactive secondary metabolites (Lam 2006); Nowadays 75-80% of the commercially and medicinally useful antibiotics have been derived from this genus (Thakur et al. 2007), thus many more antibiotics remain to be identified.
At a time when antibiotic resistance amongst pathogens is a chronic clinical problem, the diminishing numbers of new antibiotics being discovered is highly worrying (Higginbotham and Murphy 2010). In this paper we describe the isolation and characterization of a bacterium that exhibits strong antibacterial activity, notably against multidrug-resistant coagulase-negative Staphylococci (MR-CNS), and describe some of the antibiotic’s properties.
Material and methods
Isolation and maintenance
Streptomyces sp. isolate ABRIINW111 (Agriculture Biotechnology Research Institute of Iran, North West Branch) has been derived from the Zagros Mountains Hamadan, Iran. It was grown on starch casein agar medium at 28°C for ten days. Plates containing the isolate were stored at 4°C. For long storage, it was grown in International Streptomyces Project medium 2 (ISP-2) broth for seven days and glycerol was added to make the final concentration 15%. Storage was at -70 C.
Screening of Actinomycetales for antimicrobial activity
For the preliminary screening, Escherichia coli ATCC 1399, Klebsiella pneumoniae ATCC 1290, Shigella flexneri ATCC 1234, Listeria monocytogenes ATCC 33090, Bacillus cereus ATCC 1431, Yersinia enterocolitica ATCC 35669, Staphylococcus aureus ATCC 29213 were used as test organisms.
The bacteria were grown overnight at 37°C in Mueller-Hinton agar, and inocula for the assays was prepared by diluting cell mass in 0.85% NaCl solution, adjusted to McFarland scale of 0.5 and confirmed by spectrophotometrical reading at 620 nm (0.08 O.D.). Cell suspensions were finally diluted to 108 cfu mL-1 for being used in the antibacterial assays.
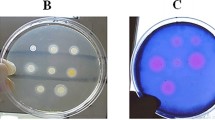
Mueller-Hinton agar was used as an assay medium. The medium at 45°C was mixed with the pathogen bacterial suspension containing approximately 108 cfu ml-1. Next, the mixture was poured on 9 cm Petri dishes and mixed with 108 cfu ml-1 Streptomyces sp. ABRIINW111 in same volume and allowed to solidify, then incubated at 37°C for 10 days. At the end all plates that had a Streptomyces culture without pathogenic bacteria, were assessed as a positive result.
Extraction of antimicrobial metabolites
ISP-2 was used for antimicrobial compound production (for in the previous step active isolates). The medium (200 ml in 500 ml Erlenmeyer flask) was inoculated with 5 ml of homogenous bacteria suspension (620 nm, 0.08 O.D.) and incubated at 30°C on a rotary shaker (120 rpm) for five days (Taechowisan et al. 2005). The Streptomyces sp. ABRIINW111 culture broth was centrifuged at 8000 g for 10 min. The antimicrobial compound containing supernatant was extracted using an equal volume of 7 different polar and non-polar solvents such as hexane, chloroform, ethyl acetate, diethyl ether, dichloromethane, methanol and water. Solvents were concentrated to 1 ml by evaporator at 40°C.
Secondary screening antimicrobial test
Antibacterial tests were carried out by the disk diffusion method, using 108 cfu ml-1 of inoculums (E. coli ATCC 1399, B. cereus ATCC 1431, Y. enterocolitica ATCC 35669, S. aureus ATCC 29213) (620 nm, 0.08 O.D.). The sterile disks (Whatman No. 1, 6 mm in diameter) were impregnated with 200 μl/disc of each metabolite, then were dried and placed on the inoculated agar. Negative controls were prepared using sterile water. The inoculated plates were incubated at 37°C for 24 h. Antibacterial activity was evaluated by measuring the zone of inhibition against the test organism.
Clinical Staphylococci isolates
CNS (n = 220) were obtained from the strain collection of the veterinary microbiological diagnostic center (veterinary medicine microbiology laboratory, Azad University). CNS were detected based on colony morphology, Gram staining, biochemical reactions and the absence of coagulase activity (Quinn et al. 1994).
This strain collection contains CNS isolated from clinical infections of dogs, cats, horses, cattle and other animal species. All these isolates were from different patients. The isolates were cultured from various clinical infectious disease processes including mastitis, pyoderma, cystitis, arthritis, and infections of the respiratory and genital tract. CNS isolates used in this study were collected in the period 2007–2009.
Antibacterial susceptibility testing
Antibiotic resistances were tested by applying the disk diffusion assay according to the guidelines of the National Committee for Clinical Laboratory Standards (NCCLS, 2003) using Mueller Hinton agar. The tested antibiotics were Ciprofloxacin, Oxacillin, Gentamicin, Tetracycline, Erythromycin, Co-Trimoxazole, Rifampicin, Vancomycin, Penicillin G, Cephalothin, Cloxacillin, Methicillin and Azlocillin. Staphylococcus aureus ATCC 25923 and Escherichia coli ATCC 35218 served as reference strains for disk diffusion testing.
The categories sensitive (S), intermediate (I) or resistant (R) were assigned on the basis of breakpoints recommended by the NCCLS. In this step MR-CNS (resistant to 5 or more antimicrobials) were selected (Higuchi et al. 2007).
Active metabolite test on MR-CNS
In this step, active metabolites of "Secondary screening antimicrobial test" were tested on MR-CNS by the disk diffusion as described.
Spectroscopic studies with active metabolites
The absorption spectrum of the active metabolites was determined by a Park Elmer Vis/UV spectrometer at 200-400 nm.
The infrared spectrum was obtained by dispersing the material to be analyzed in potassium bromide and working with a Shimadzu FTIR-8400 S spectrometer at 400-4000 cm-1.
Molecular identification of the Streptomyces strain
The isolate ABRIINW 111 was grown during four days at 28°C with agitation in 500 ml flasks containing 100 ml of ISP-2 medium. The biomass was harvested by centrifugation at 8000 g for 10 min and washed twice with double-distilled water. About 200 mg of mycelia was used for DNA extraction as follows: the sample was dispersed in 800 ml of the aqueous lysis solution (100 mM Tris-HCl, pH 7; 20 mM EDTA; 250 mM NaCl; 2% SDS; 1 mg mL-1 lysozyme). About 5 μl of a 50 mg ml-L RNase solution was added, and the suspension incubated at 37°C for 60 min. About 10 μl of a proteinase K solution (20 mg ml-L) was added, and the lysis solution was re-incubated at 65°C for 30 min. The lysate was extracted with an equal volume of phenol and centrifuged at 7000 g for 10 min. The aqueous layer was re-extracted with phenol (50-50%, v/v), then by chloroform (50-50%, v/v). DNA was recovered from the aqueous phase by the addition of NaCl (150 mM final concentration) and two volumes of cool 95% (v/v) ethanol prior to centrifugation. The precipitated DNA was cleaned with 50 ml of 70% (v/v) ethanol, centrifuged at 7000 g for 10 min, re-suspended in 50 ml of TE buffer (10 mM Tris-HCl, pH 7.4; 1 mM EDTA, pH 8) and stored at -20°C. The purity of DNA solutions was checked spectrophotometrically at 260 and 280 nm, and the quantities of DNA were measured at 260 nm.
Identification was done with 16S rDNA gene sequencing. The 16 S ribosomal RNA gene was amplified by using the PCR method with Taq DNA polymerase and primers AF (5' AGAGTTTGATCCTGGCTCA 3') and AR (5' AAGGAGGTGATCCAGCCGC 3'). The conditions for thermal cycling were as follows: denaturation of the target DNA at 96°C for 5 minutes followed by 30 cycles at 94°C for one minute, primer annealing at 55°C for one minute and primer extension at 72°C for 2 minutes. At the end of the cycling, the reaction mixture was held at 72°C for 10 min and then cooled to 4°C. PCR amplification was detected by agarose gel (1.5%) electrophoresis and was visualized by ultraviolet (UV) after ethidium bromide staining. The amplified products (~1500 bp) were purified individually using the high pure PCR product purification kit (Roche) according to manufacturer’s instruction. The 16S rDNA amplicon was cloned in pTZ57R/T vector according to the manufacturer’s instruction (T/A clone TM PCR product cloning kit, Fermentas). Sequencing of the 16S rDNA gene (~1500 bp) of the isolate was carried out in Macrogen (Seoul, Korea). The obtained sequences were assessed, analyzed and manually edited by using of the Choromas lite software package and compared with sequences with in the NCBI database (http://www/ncbi.nlm.nih.gov/) using the basic local alignment search tool (BLAST). Multiple sequence alignment was carried out using CLUSTALX. Bootstrap analysis was performed to validate the reproducibility of the branching pattern with Mega 4 software.
Statistical analyses
Data were examined using a commercially available statistical package (SPSS version 17 for Windows), and comparisons were made using the descriptive statistics.
Results
Properties of various isolated soil samples were initially screened for antibiotic activity. 110 different Streptomyces isolates were recovered, but antibacterial activity was exhibited in 12.72% of all isolates and only one isolate had antibacterial effects on CNS. The isolate that displayed strong antibacterial activities was identified by sequence comparisons of its 16S rDNA. Comparison of the 16S rDNA gene ABRIINW111 (GenBank accession number GU433228) (~1500 bp) with sequences in the GenBank data base revealed that the bacterium was a Streptomyces sp. with a 98% similarity to the S. levis strain NRRL B-16370 (Fig. 1).
Phylogenetic dendrogram based on 16S rDNA gene sequence analysis, reconstructed from evolutionary distances by using the neighbour-joining method, showing the phylogenetic position of strain ABRIINW111 (GenBank accession number GU433228, pointed by ■) with the most similar species. Bootstrap values (>50%, value above branch: neighbor joining) are indicated at the relevant branching points
Permissive temperature ranges for growth of the strain ABRIINW111 were 22 to 40 ◦C with an optimum at 28 ◦C. Culture characteristics of strain ABRIINW111 were followed on the basis of observations made after 7, 14 and 21 days of incubation on nutrient agar and starch casein agar. Generally, strain ABRIINW111 grew well; the colonies were elevated and spreading. The colors of the vegetative and aerial mycelia were light gray and grayish respectively. The spore chains were white and moderately developed. The characteristics of strain ABRIINW111 were compared with those of the known species of Actinomycetales described in Bergey’s manual of systematic bacteriology (Brenner et al. 2004), and the obtained morphological properties suggested strongly that strain ABRIINW111 belonged to the genus Streptomyces.
The metabolites of strain ABRIINW111 exhibited significant antibiotic activity against gram-positive and gram-negative bacteria. Table 1 shows secondary screening antibacterial activities of metabolites of Streptomyces sp. ABRIINW111. Most interestingly, some of the cultured metabolites were highly active against clinical isolates of MR-CNS. Table 2 shows the percentages of antibiotic resistance in CNS.
35.9% of MR-CNS have exhibited resistances against five or more than five antibiotics, all of these bacteria were resistant to methicillin too.
The UV spectrum of the compound showed that the maximum absorption was at 275 nm in diethyl ether.
Discussion
The presence of antibiotic resistant bacteria or resistance determinants has been reported sporadically for more than 20 years in different countries (Faria et al. 2009; Resch et al. 2008; Ruscher et al. 2009; Lis et al. 2009; Seyler et al. 2004). CNS were the most frequently isolated organisms in animals, in agreement with several studies too (Luthje and Schwarz 2007; Schultz et al. 2009; Koksal et al. 2007; Malik et al. 2007).
According to Moodley and Guardabassi (2009) occurrence and distribution of methicillin-resistant CNS among horses, personnel and environmental sites at equine facilities was 69%. In another study some strains of CNS were to 95% resistant against up to seven antibiotics (Resch et al. 2008). Resistance to methicillin was detected in 67.5% of CNS isolates of septicemic patients in Turkey (Koksal et al. 2007). However, Higuchi et al. (2007) found 26.8% MR-CNS isolates from medical students in Japan. This fact indicates that the prevalence of resistant CNS may differ considerably between countries and host (Duijkeren, et al. 2004).
In this study the prevalence of MR-CNS in clinical specimens of different animal species was defined by investigating a total of 220 clinical samples. Of all samples examined, 79 were positive for MR-CNS, giving an overall prevalence of 35.9%.
The percentage of resistance in CNS isolates is given in Table 2.
According to Koksal et al. (2007) methicillin resistant Staphylococci are resistant to all other penicillins, carbapenems, cephems and beta-lactam/beta-lactamase inhibitor combinations. It is important that 100% of this study MR-CNS were resistant to methicillin.
Nowadays morbidity and mortality rates due to MR-CNS infections remain high, and determining the best therapeutic agent for treatment is an ongoing challenge.
On the other hand we are in the early stages of a "renaissance" in antibiotic discovery from Actinomycetales (Baltz 2008) and in recent year's investigation of resources for isolation of new active bacterial strains were resumed (Higginbotham and Murphy 2010; Nithya and Pandian 2009).
In one study, from 10 farming soil samples collected from the Manisa province, 50 isolates of Actinomycetales were obtained. Approximately 34% of the isolates produced antibiotics against gram-positive and gram-negative bacteria (Oskay, et al. 2004). In another study a total of 20 different Streptomyces isolates were recovered from 33 soil samples. Antibacterial activity was exhibited in 44.5% of all isolates (Ilic, et al. 2005). In the work of Thakur et al. (2007), among the 110 isolates, 65 (59.09%) strains showed antibacterial effects.
In the present study, 110 different Streptomyces isolates were recovered but antibacterial activity was only exhibited in 12 (12.72%) of all isolates. We have described the isolation and identification of a new Streptomyces sp., strain ABRIINW111 that produces highly active secondary metabolites that have a broad spectrum of activity and are particularly active against clinical isolates of MR-CNS. Table 1 shows that the diethyl ether extracted metabolite of Streptomyces sp. strain ABRIINW111 exhibits a 15 ± 0.8 mm inhibitory zone in MR-CNS. All these MR-CNS strains were resistant to most beta lactam and non-beta lactam antibiotics.
According to the 16S rDNA, strain ABRIINW111 has 98% similarity to the S. levis strain NRRL B-16370 but this strain has some mutations in the alpha and gamma regions of the 16S rDNA sequence (Stackebrandt et al. 1991), that have a high potency to define species. So this bacterium is probably a novel species of the Streptomyces genus (Fig. 1).
The UV visible spectrum exhibited maxima at 275 nm, suggesting the presence of chromophores or other kinds of conjugated pi-electron systems such as unsaturated aldehydes, ketones and aromatic ring compounds (Mellouli et al. 2003).
The FTIR spectrum indicates that the compound had an O-H group (2927 cm-1). The absence of carboxylic acid (COOH), ester (COOR), and alkyne (C = C-), was confirmed by the lack of a band in the region of 1670-1740, 1700-1750 and 2100-2260 cm-1, respectively (Dhanasekaran et al. 2008).
More investigations are needed to discover the structure of this antibacterial compound and additional in vivo studies are necessary for clinical evaluation. But generally, the diethyl ether extract of the new Streptomyces sp. strain ABRIINW111 can inhibit the MR-CNS in vitro, and it can offer a new approach to treatment of MR-CNS infected patients in future.
References
Baltz RH (2008) Renaissance in antibacterial discovery from Actinomycetes. Curr Opin Pharmacol 8:557–563
Bibb MJ (2005) Regulation of secondary metabolism in streptomycetes. Curr Opin Pharmacol 8:208–215
Brenner DJ, Krieg NR, Staley JT (2004) Bergey’s manual of systematic bacteriology, 2nd edn. Springerlink, Michigan, pp 95–100
Dhanasekaran D, Thajuddin N, Panneerselvam A (2008) An antifungal compound: 4' PHENYL -1-NAPTHYL –PHENYL ACETAMIDE From Stereptomyces sp. DPTB16. FACTA Universitatis: Medicine and Biology 15:7–12
Duijkeren EV, Box AT, Heck ME, Wannet WJ, Fluit AC (2004) Methicillin-resistant staphylococci isolated from animals. Vet Microbiol 103:91–97
Faria C, Vaz-Moreira I, Serapicos E, Nunes OC, Manaia CM (2009) Antibiotic resistance in coagulase negative staphylococci isolated from wastewater and drinking water. Sci Total Environ. doi:10.1016/j.scitotenv.2009.02.034
Higginbotham SJ, Murphy CD (2010) Identification and characterization of a Streptomyces sp. isolate exhibiting activity against methicillin resistant Staphylococcus aureus. Microbiol Res 165:82–86
Higuchi W, Isobe H, Iwao Y, Dohmae S, Saito K, Takano T, Otsuka T, Baranovich T, Endo C, Suzuki N, Tomiyama Y, Yamamoto T (2007) Extensive multidrug resistance of coagulase-negative staphylococci in medical students. J Infect Chemother 13:63–66
Ilic SB, Konstantinovic SS, Todorovic ZB (2005) UV/VIS Analysis and antimicrobial activity of Streptomyces isolates. FACTA Universitatis: Medicine and Biology 12:44–46
Katsuda K, Hata E, Kobayashi H, Kohmoto M, Kawashima K, Tsunemitsu K, Eguchi M (2005) Molecular typing of Staphylococcus aureus isolated from bovine mastitic milk on the basis of toxin genes and coagulase gene polymorphisms. Vet Microbiol 105:301–305
Koksal F, Yasar H, Samasti M (2007) Antibiotic resistance patterns of coagulase negative staphylococcus strains isolated from blood cultures of septicemic patients in Turkey. Microbiol Res. doi:10.1016/j.micres.2007.03.004
Lam KS (2006) Discovery of novel metabolites from marine Actinomycetes. Curr Opin Pharmacol 9:245–251
Lis DO, Pacha JZ, Idzik D (2009) Methicillin resistance of airborne coagulase-negative staphylococci in homes of persons having contact with a hospital environment. Am J Infect Control 37:177–182
Luthje P, Schwarz S (2007) Molecular basis of resistance to macrolides and lincosamides among staphylococci and streptococci from various animal sources collected in the resistance monitoring program BfT-GermVet. Int J Antimicrob Agents 29:528–535
Malik S, Christensen H, Peng H, Barton MD (2007) Presence and diversity of the b-lactamase gene in cat and dog staphylococci. Vet Microbiol 123:162–168
Mellouli L, Ameur-Mehdi RB, Sioud S, Salem M, Bejar S (2003) Isolation, purification and partial characterization of antibacterial activities produced by a newly isolated Streptomyces sp. US24 strain. Res Microbiol 154:345–352
Moodley A, Guardabassi L (2009) Clonal spread of methicillin-resistant coagulase-negative staphylococci among horses, personnel and environmental sites at equine facilities. Vet Microbiol 137:397–401
National Committee for Clinical Laboratory Standards (2003) Performance Standards for Antimicrobial Disk Susceptibility Tests; Approved StandardM2-A8.NCCLS,Wayne, Pa
Nithya C, Pandian SK (2009) Isolation of heterotrophic bacteria from Palk Bay sediments showing heavy metal tolerance and antibiotic production. Microbiol Res. doi:10.1016/j.micres.2009.10.004
Oskay M, Tamer AU, Azeri C (2004) Antibacterial activity of some actinomycetes isolated from farming soils of Turkey. Afr J Biotechnol 3:141–146
Quinn PJ, Carter ME, Markey BK, Carter GR (1994) Clinical veterinary microbiology. Wolfe Pub, London, pp 125–126
Resch M, Nagel V, Hertel C (2008) Antibiotic resistance of coagulase-negative staphylococci associated with food and used in starter cultures. Int J Food Microbiol 127:99–104
Ruscher C, Becker A, Wleklinski C, Soba A, Wieler LH, Walther B (2009) Prevalence of Methicillin-resistant Staphylococcus pseudintermedius isolated from clinical samples of companion animals and equidaes. Vet Microbiol 136:197–201
Schultz PJ, Torres AH, DeGraves FJ, Gebreyes WA, Patchanee P (2009) Antimicrobial resistance and genotypic characterization of coagulase-negative staphylococci over the dry period. Vet Microbiol 134:55–64
Seyler TS, Jaeger B, Bockelmann W, Noordman WH, Geis A, Heller KJ (2004) Molecular identification and differentiation of Staphylococcus species and strains of cheese origin. Systems Applied Microbiology 27:211–218
Stackebrandt E, Witt D, Kemmerling C, Kroppenstedt R, Liesack W (1991) Designation of streptomycete 16S and 23S rRNA-based target regions for oligonucleotide probes. Appl Environ Microbiol 57:1468–1477
Sudagidan M, Yenidunya AF, Gunes H (2005) Identification of staphylococci by 16S internal transcribed spacer rRNA gene restriction fragment length polymorphism. J Med Microbiol 54:823–826
Taechowisan T, Lu C, Shen Y, Lumyong S (2005) Secondary metabolites from endophytic Streptomyces aureofaciens CMUAc130 and their antifungal activity. Microbiology 151:1691–1695
Thakur D, Yadav A, Gogoi BK, Bora TC (2007) Isolation and screening of Streptomyces in soil of protected forest areas from the states of Assam and Tripura, India, for antimicrobial metabolites. J Mycolog Med 17:242–249
Acknowledgements
The authors are indebted to Prof. Dr. Helga Gerlach and Dr. Helene Pendl for critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sadigh-Eteghad, S., Dehnad, A., Shanebandi, D. et al. Identification and characterization of a Streptomyces sp. isolate exhibiting activity against multidrug-resistant coagulase-negative Staphylococci. Vet Res Commun 35, 477–486 (2011). https://doi.org/10.1007/s11259-011-9491-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11259-011-9491-9